Abstract
CarD, a global transcriptional regulator in Myxococcus xanthus, interacts with CarG via CarDNter, its N-terminal domain, and with DNA via a eukaryotic HMGA-type C-terminal domain. Genomic analysis reveals a large number of standalone proteins resembling CarDNter. These constitute, together with the RNA polymerase (RNAP) interacting domain, RID, of transcription–repair coupling factors, the CarD_TRCF protein family. We show that one such CarDNter-like protein, M. xanthus CdnL, cannot functionally substitute CarDNter (or vice versa) nor interact with CarG. Unlike CarD, CdnL is vital for growth, and lethality due to its absence is not rescued by homologs from various other bacteria. In mycobacteria, with no endogenous DksA, the function of the CdnL homolog mirrors that of Escherichia coli DksA. Our finding that CdnL, like DksA, is indispensable in M. xanthus implies that they are not functionally redundant. Cells are normal on CdnL overexpression, but divide aberrantly on CdnL depletion. CdnL localizes to the nucleoid, suggesting piggyback recruitment by factors such as RNAP, which we show interacts with CdnL, CarDNter and RID. Our study highlights a complex network of interactions involving these factors and RNAP, and points to a vital role for M. xanthus CdnL in an essential DNA transaction that affects cell division.
INTRODUCTION
The programmed cell differentiation and multicellular developmental process that occurs on starvation to form fruiting bodies is a striking feature of myxobacteria, and Myxococcus xanthus serves as a prokaryotic model for this complex process (1–3). Temporal and spatial control of gene expression during M. xanthus development is achieved, among others, via a number of eukaryotic-like signal transduction proteins and transcriptional factors (4–8). One such factor, CarD, is a global regulator in M. xanthus that also acts in light-induced carotenogenesis and other processes (6,9,10).
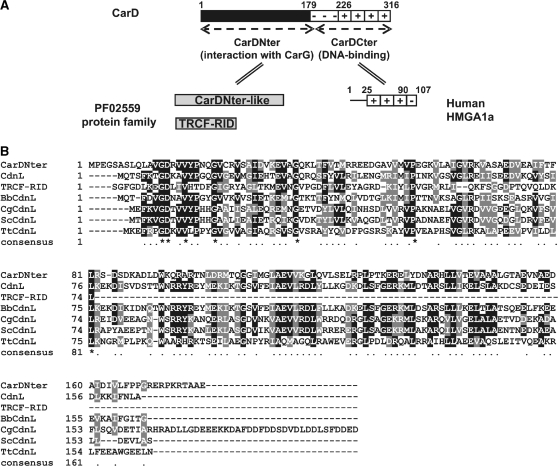
CarD has a rather unique two-domain architecture comprising a C-terminal DNA-binding domain resembling eukaryotic high-mobility group A (HMGA) proteins and an N-terminal domain whose sequence homologues are found exclusively in bacteria (Figure 1A; 7,11,12). HMGA are small, relatively abundant, non-histone chromatin components in eukaryotes that are essential in many DNA transactions due to their roles in chromatin remodeling and in the assembly of specific nucleoprotein complexes (13,14). The hallmarks of HMGA are multiple repeats of a basic region with ‘AT-hooks’ (the DNA-binding motif) and a flanking highly acidic region. Four AT-hooks and a contiguous highly acidic region form the CarD C-terminal domain whose physical, structural and DNA-binding properties resemble human HMGA1a (11,15). It is noteworthy that CarD and its Stigmatella aurantiaca ortholog (CarDSa) are the only two known proteins in prokaryotes with HMGA-like domains (12). A third myxobacterium, Anaeromyxobacter dehalogenans, also has a CarD analog, CarDAd, whose C-terminal domain resembles histone H1 rather than HMGA. Nonetheless, the two domains could be interchanged in CarD with no loss of function (15).
Figure 1.
M. xanthus CarD and the PF02559 protein family. (A) Schematic showing CarD domain architecture. Residue numbers (on top) demarcate the distinct regions. The CarD N-terminal domain, CarDNter, defines the PF02559 family that includes a number of CarDNter-like proteins and TRCF-RID. CarDCter denotes the HMGA-like CarD C-terminal domain, with human HMGA1a shown for comparison (‘+’ indicate the basic AT-hooks and ‘−’ the acidic region). (B) Sequence alignment of representative PF02559 family members. NCBI accession codes and the corresponding bacterium are indicated in brackets: CarDNter (CAA91224; M. xanthus); CdnL (YP_630846; M. xanthus); TRCF-RID, the segment corresponding to residues 514–587 of TRCF (YP_629274; M. xanthus); BbCdnL (NP_969149; B. bacteriovorus); CgCdnL (YP_001139479; C. glutamicum); ScCdnL (NP_628406; S. coelicolor); TtCdnL (YP_005787; T. thermophilus). Residues are shaded black (with an asterisk in the consensus line below) when identical in the majority of the aligned sequences, or grey when similar.
CarD, CarDSa and CarDAd share an N-terminal domain that is absent in eukaryotic HMGA (Figure 1A). This 179-residue domain in CarD, CarDNter, has no DNA-binding activity and is absolutely required for CarD to interact specifically with CarG, a zinc-bound protein that participates in every known CarD-regulated process without binding DNA directly (8,15). Given that CarD and CarG work as a single regulatory unit, one always coexists with the other, their occurrence being confined exclusively to myxobacteria of the suborder Cystobacterineae (8,12,15). By contrast, standalone proteins resembling CarDNter in size and sequence, with no identifiable DNA-binding domain (as opposed to CarD), are widely distributed in bacteria (12). These have been grouped as the CarD_TRCF (PF02559) protein family that, based on sequence similarity, also includes the RNA polymerase (RNAP) interacting domain, RID, of transcription–repair coupling factors (TRCFs) (Figure 1; 16,17). Despite the ever-increasing number of CarDNter-like proteins identified due to large-scale sequencing of bacterial genomes (>400, so far; Table 1) their functions remain largely uncharacterized.
Table 1.
Distribution of CarDNter-like proteins among bacteria and comparison with CdnL
| Taxonomical group | Length (number of residues) | Identity to CdnL (%) | Similarity to CdnL (%) |
|---|---|---|---|
| δ-Proteobacteria | 125–188 | 33–100 | 55–100 |
| α-Proteobacteria | 143–456 | 27–39 | 49–59 |
| Actinobacteria | 137–301 | 24–39 | 47–68 |
| Firmicutes | 148–176 | 24–47 | 47–70 |
| Deinococcus-Thermus | 145–178 | 24–30 | 44–54 |
| Spirochaetales | 161–208 | 26–37 | 50–64 |
| Bacteroidetes | 158 | 36 | 61 |
A total of 449 members were found in a BLAST search analysis of the 3179 bacterial genomes available at http://www.ncbi.nlm.nih.gov (complete—912, assembly—1127, unfinished—1140) using CdnL as query. Genomes of β-, ε- and γ-Proteobacteria, Chlamydiae and Cyanobacteria lack CarDNter-like proteins.
Our analysis of the M. xanthus CarDNter-like protein is reported here. We have named this protein CdnL (for CarD N-terminal like) to distinguish it from CarD, the term we originally coined for the latter based on its role in light-induced carotenogenesis (6,7). A fundamental difference between CarD and CdnL (or its homologs) is that the latter lacks an identifiable DNA-binding domain. Very recently, a protein in Mycobacterium tuberculosis that was called CarD, but is really like CdnL (12), was reported to be essential for bacterial growth and persistence and shown to be involved in downregulating rRNA transcription in response to starvation in a manner somewhat similar to that of DksA in Escherichia coli (18). It was also concluded that the mycobacterial CdnL could functionally replace DksA in E. coli, in spite of no apparent sequence similarity between the two proteins, suggesting that bacteria lacking DksA could employ CdnL, when present, for related functions. Genome data, however, reveal the existence of bacteria in which both DksA and CdnL homologs exist, as well as examples with neither. Moreover, M. xanthus (as also S. aurantiaca and A. dehalogenans) contains not only CdnL, whose function remains to be characterized (12), but also CarD and DksA (19). This simultaneous existence of all three proteins in one bacterium therefore affords a unique case scenario to investigate the interplay among them. Despite its sequence similarity with CarDNter and classification in the same protein family, we show in this study that CdnL is functionally distinct and, unlike CarD, is essential for M. xanthus growth and viability. This, together with our earlier finding that DksA is also indispensable in M. xanthus (19), provides strong evidence that CdnL and DksA are not functionally redundant. Lack of CdnL could not be complemented by homologues from several other bacteria or by CarDNter, indicating that the requirement for CdnL is highly specific. Although it lacks a DNA-binding domain, CdnL localized to the nucleoid. A likely agent mediating the delivery to the nucleoid is RNAP, as we found that it interacts with CdnL. Despite being functionally distinct, CarDNter interacted with the same region of RNAP, thereby highlighting a complex network of overlapping interactions that links RNAP to CarD and CdnL, with those involving the latter being essential for viability.
MATERIALS AND METHODS
Strains, growth conditions and isolation of genomic DNA
Strains used in this study are listed in Supplementary Table S1. M. xanthus vegetative growth was carried out at 33°C in rich CTT medium supplemented, as required, with antibiotic (kanamycin, Km, at 40 μg ml−1; tetracycline, Tc, at 10 μg ml−1) and 0.75 µM vitamin B12 (19). Fruiting body development was induced on TPM agar as previously described (8). Escherichia coli DH5α (for plasmid constructions) and BL21-(DE3) (for protein overexpression) were grown in Luria broth at 37 or 25°C. Thermus thermophilus, Bdellovibrio bacteriovorus (DSMZ culture collection, Braunschweig, Germany) and Corynebacterium glutamicum (Spanish culture collection, CECT-Valencia, Spain) were grown as recommended and genomic DNA was isolated using Wizard genomic DNA purification kit (Promega). Streptomyces coelicolor genomic DNA was generously provided by Dr. Ramón Santamaría Sánchez (University of Salamanca, Spain).
Plasmid and strain construction
Plasmids used in this study are listed in Supplementary Table S2. Standard protocols and kits were used in preparing and manipulating plasmid DNA. Genomic DNA was isolated using WizardTM Genomic DNA Purification Kit (Promega) or Instagene (BioRad). The PCR overlap extension method was employed for site-directed mutagenesis (20). All plasmid constructs were verified by DNA sequencing. Plasmid pMR2768 contains the carDΔN allele, in which the region coding for residues 2–178 of CarD has been deleted and replaced by an EcoRI site, flanked by about 1.2-kb genomic DNA on either side (12). cdnL (not including its stop codon) was PCR-amplified with 5′ and 3′ EcoRI sites and inserted into pMR2768 in-frame with the segment encoding CarD residues 179–316 to yield pMR2859, which has the chimeric gene under the control of the carD promoter. pMAR240 (KmR) allows transcriptional fusions to a reporter lacZ gene. It contains 1.38-kb M. xanthus DNA that permits integration at a chromosomal site with no promoter activity (21). A 180-bp PCR-amplified DNA fragment corresponding to the cdnL promoter region (PcdnL) was introduced into the EcoRI site of pMAR240 (KmR) to generate pMR2953, with the PcdnL::lacZ transcriptional fusion for promoter analysis. To construct pMR2873, used in generating a complete in-frame deletion of cdnL, regions flanking cdnL in the genome were first PCR-amplified from genomic DNA: (i) a ∼750-bp DNA fragment upstream of cdnL with a 5′ HindIII site and a 3′ XbaI site; (ii) a ∼770-bp DNA fragment downstream of cdnL with a 5′ XbaI site and a 3′ BamHI site. The two fragments were ligated at the XbaI site and cloned into the HindIII-BamHI sites of plasmid pBJ114 (22), supplied with GalS and KmR markers for negative and positive selection, respectively. In pMR2873, cdnL is replaced by an XbaI site but retains the flanking genomic DNA regions. The coding region for cdnL variants or a given PF02559 member (PCR-amplified from genomic DNA) can be inserted into the XbaI site to yield constructs for complementation analysis. pMR2928 was constructed by cloning cdnL plus its promoter region (PcdnL) into the PstI/KpnI sites of pMR2915, which contains a 1.38 kb region for integration at an unlinked locus in M. xanthus and TcR for selection (19). pMR2991, with the 1.38-kb M. xanthus DNA for chromosomal integration and carH expressed from a constitutive promoter, is designed for vitamin B12-dependent conditional expression of candidate genes (19). Cloning PCR-amplified cdnL (with ribosomal binding site) into the XbaI site of pMR2991 yielded pMR3015, in which cdnL, expressed from promoter PB, is repressed by CarH if B12 is present. To generate pMR3000, a DNA segment corresponding to two tandem rRNA promoters was isolated from pMR2831 (23) digested with PstI/KpnI and inserted together with cdnL (PCR-amplified with 5′ KpnI and 3′ EcoRI sites) into the PstI/EcoRI sites of pMR2915. pMR3164, containing the PcdnL::cdnL-eGFP fusion, was constructed by inserting eGFP with 5′ NheI and 3′ KpnI sites (PCR-amplified from pEGFP-C2, Clontech) and PcdnL::cdnL with 5′ PstI and 3′ NheI sites into the PstI/KpnI sites of pMR2915.
Plasmids were introduced into M. xanthus by electroporation and transformants with plasmids integrated into the chromosome by homologous recombination were selected on CTT plates with the appropriate antibiotic. A galK gene provided in some plasmids allows negative selection by conferring sensitivity to galactose (GalS). To evict integrated plasmids with a galK gene, cells were grown for several generations with no antibiotic and then plated on CTT supplemented with 10 mg/ml galactose to select against GalS plasmid-bearing cells. Plasmid integration or eviction was verified in each case by PCR analysis of genomic DNA.
Plasmid pMR3400, for overexpressing CdnL fused to a His6 tag at its C-terminus (CdnL-His6), was constructed as follows. First, cdnL was PCR-amplified using as template DNA pET11-cdnL (with cdnL cloned at the NdeI and BamHI sites of the pET11b expression vector) and as oligonucleotides: (i) one that hybridizes at the T7 promoter region, in the pET11b backbone upstream of cdnL; (ii) another that hybrizes at the C-terminus of cdnL, excluding the stop codon, and generates a 3′ HindIII site. The PCR product was then cut with XbaI and HindIII and cloned into XbaI-HindIII digested pET28b (Novagen) to generate pMR3400.
β-Galactosidase activity assays
β-Galactosidase activity was qualitatively assessed from blue colour development on CTT or LB plates with X-gal (40 µg/ml), and measured (in nanomoles of o-nitrophenyl β-d-galactoside hydrolysed/min/mg protein) for liquid cultures of M. xanthus or E. coli and for M. xanthus developing cells as described before (8).
5′ RACE analysis
5′ RACE (rapid amplification of cDNA ends) was performed as follows. Total RNA was isolated from cells grown to OD550nm ≈ 1.0 using High Pure RNA isolation kit and treated with recombinant DNase I (Roche) to eliminate genomic DNA. RNA recovered in RNase-free water (pre-treated with 0.2% diethylpyrocarbonate, DEPC) was again DNase I-treated and reverse-transcribed in the presence of RNase inhibitor Protector using the 5′/3′RACE 2nd Generation kit (Roche) with an oligonucleotide that hybridizes to the 3′ end of cdnL as primer. The cDNA product was purified, tagged with a 5′ poly-A adaptor and used as PCR template with dT-Anchor and a cDNA-specific primer that hybridizes 165 bp downstream of the cdnL initiation codon, followed by a second PCR reaction with PCR Anchor replacing dT-Anchor. The product was purified (Illustra GFX kit, GE Health Sciences) and sequenced.
Two-hybrid analysis
The bacterial two-hybrid system used is based on the functional complementation between the T25 and T18 fragments of the catalytic domain of Bordetella pertussis adenylate cyclase (CyaA) when two test proteins interact (24). Constructs encoding fusions of CarD and CarD(1–179) to the C-terminus of the CyaA T25 fragment in pKT25, and CarG fused to the N-terminus of the CyaA T18 fragment in pUT18 have been described before (8). Other constructs used in two-hybrid analysis were generated by PCR-amplifying the coding region of interest and cloning it into the XbaI and BamHI sites of pKT25, pUT18 or pUT18C. Given pairs of pUT18 (or pUT18C) and pKT25 constructs were introduced into E. coli strain BTH101 (cya−) by electroporation, pairs with only one construct expressing the fusion protein serving as negative controls. Interaction was assessed from reporter lacZ activity as described above.
Protein purification
We described elsewhere the purification of M. xanthus RNAP (25) and His6-DksA (19). For CdnL-His6 purification, E. coli BL21-(DE3) freshly transformed with pMR3400 was grown in 10 ml of LB-kanamycin medium to A600 = 0.6 at 37°C and then 5 mL were innoculated into 500 ml of fresh medium. After growth to A600 = 0.6 at 37°C, 0.5 mM IPTG was added to induce cdnL expression overnight at 25°C. Analysis of cell extracts from 1 ml of culture showed that CdnL-His6 was overexpressed as a soluble protein. The protein was purified using TALON metal affinity resin (Clontech) and the accompanying native protein purification protocol. The imidazole was eliminated by extensive dialysis in buffer A (50 mM phosphate buffer, pH 7.5, 2 mM β-mercaptoethanol) plus 150 mM NaCl and the sample was further purified by passing through a phosphocellulose column (to which some contaminants, but not CdnL, bind) equilibrated with the same buffer.
Western, pull-down and size exclusion analysis
Rabbit polyclonal anti-CdnL antibodies were obtained using the same procedures as for anti-CarD antibodies (11). Immunoblots of whole cell extracts were carried out as before (12). To compare relative CdnL levels, total protein amounts in a given extract were estimated (26) and fixed amounts were analysed in immunoblots using anti-CdnL antibodies.
In the pull-down assays to check interaction between M. xanthus RNAP and CdnL-His6 (or His6-DksA), 200 µl of TALON resin in two 2 ml SigmaPrep (Sigma-Aldrich) columns were washed three times with 600 µl of low salt buffer (buffer A + 50 mM NaCl). 500 µl of His6-tagged protein (∼25 nmol) was applied to one column but not to the other, which served as control. After incubation for 1 h at 4°C with gentle rotary agitation, the resin in each column was washed with 20 volumes of low salt buffer. To each column 20 pmol of RNAP (200 µl) was added and incubated for 1–2 h at 4°C with gentle rotary agitation. After collecting the flow-through from each column separately, the resin was extensively washed with low salt buffer and subsequently eluted in five 200 µl fractions with high salt buffer (buffer A + 500 mM NaCl). These and the low salt flow-through (after RNAP incubation) were subjected to SDS–PAGE on a 10% NuPAGE Novex Bis-Tris (Invitrogen) precast gel and analysed by silver staining (ProteoSilver Silver Stain kit, Sigma).
Size-exclusion analysis was carried out using a Superdex200 analytical HPLC column equilibrated with 150 mM NaCl, 50 mM phosphate buffer pH 7.5, 2 mM β-mercaptoethanol and calibrated as described elsewhere (11).
Microscopy
Ten-microliter aliquots (∼106 cells) of cells grown in CTT in the light at 33°C/300 rpm to OD550 ≈1.0 were added to 10 ml of fresh CTT and grown at 33°C/300 rpm in separate culture flasks—one in the light (permissive conditions) and the other in the dark with 0.75 µM B12 present (restrictive conditions). One hundred micoliters of each culture were taken at different times during growth (monitored at OD550), mixed with the fluorescent dye 4′-6-diamino-2-phenylindole (DAPI; 350 nm excitation maximum, 461 nm emission maximum) to a final concentration of 1 ng/µl and a 4 µl drop applied to a glass plate coated with 3 aminopropyl-triethoxysilane. This was examined with a Nikon Eclipse 80i microscope unit equipped with a Plan Apo VC 100 × /1.40 oil immersion objective, a Hamamatsu ORCA-AG CCD camera and Metamorph 4.5 image-processing software (Universal Imaging Group). For CdnL-eGFP in vivo localization studies, 100 µl of a cell culture grown in CTT to OD550 ≈1.0 were washed in TPM, treated with DAPI, and a 4 µl drop applied to a glass plate coated with 3-aminopropyl-triethoxysilane was examined using the equipment described above.
RESULTS
CdnL neither replaces CarDNter in CarD function nor interacts with CarG
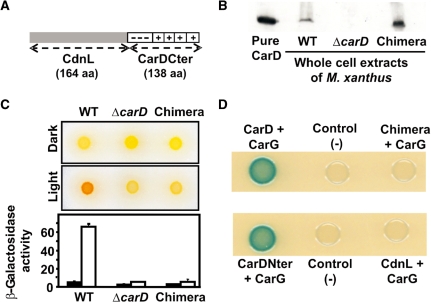
The protein family database (http://pfam.sanger.ac.uk; 27) classifies CarDNter and CdnL as members of PF02559 based on sequence similarity. Whether this correlates or not with similar roles requires to be examined. Unlike CdnL, whose function is unknown, various CarD-dependent cellular processes have been identified, and each one of these requires CarDNter to interact with CarG (8,15). We therefore compared CarDNter and CdnL at a functional level by checking if a CarD chimera with CdnL replacing CarDNter (Figure 2A) could act in light-induced carotenogenesis, one well-studied CarD-dependent process in M. xanthus. For this, a plasmid expressing the chimera (pMR2859) was introduced into the ΔcarD M. xanthus strain (MR1900) and merodiploids resulting from plasmid integration into the chromosome by homologous recombination were selected. These merodiploids expressed stable chimeric protein as verified in western blots of cell extracts using monoclonal antibodies specific for the CarD C-terminal region, CarDCter (Figure 2B; 11). However, in contrast to the wild-type strain that turns red in the light due to carotenoid synthesis, the merodiploids exhibited the yellow Car− colour phenotype of the ΔcarD strain, suggesting that the chimeric protein is not functional. In agreement with this, a haploid derivative bearing the chimeric allele (MR1968) was also phenotypically Car− (Figure 2C). In light-induced carotenogenesis, CarD is required to activate the photo-inducible PQRS promoter of the regulatory carQRS operon (7,8,10). Figure 2C (bottom) shows that expression of a PQRS-lacZ transcriptional fusion in the light was as impaired for the strain bearing the chimeric allele as for that with the ΔcarD allele. Since CarD activity in vivo requires CarDNter to interact with CarG (8), we next checked whether or not this interaction is maintained for CdnL. Neither CdnL nor the CdnL-CarDCter chimera interacted with CarG in the CyaA-based bacterial two-hybrid system (Figure 2D; see ‘Materials and Methods’ section) or in size-exclusion experiments with pure proteins (data not shown). The lack of this crucial interaction could therefore explain why CdnL cannot replace CarDNter in CarD function.
Figure 2.
CdnL cannot replace CarDNter in CarD function. (A) Schematic of the CarD chimera with CarDNter (residues 1–178) replaced by CdnL. (B) Western blot of pure CarD (lane 1) and cell extracts of strains expressing CarD (‘WT’; lane 2), the CdnL-CarDCter chimera (‘Chimera’, lane 4), or no CarD (‘ΔcarD’; lane 3) probed with monoclonal anti-CarD antibodies. (C) Top: colour phenotypes of cell spots after growth for 1 day on CTT plates in the dark or in the light. Bottom: expression levels of the reporter PQRS::lacZ probe in strains bearing the indicated carD allele. Cell cultures were grown to early exponential phase in the dark, divided into two, grown for a further 8 h, one in the dark (filled bars) and the other in the light (empty bars). The specific β-galactosidase activities (in nanomoles of o-nitrophenyl β-d-galactoside hydrolyzed/min/mg protein) are from two or more independent measurements. (D) Bacterial two-hybrid analysis of cell spots expressing the protein pair indicated on plates containing X-gal. Cells expressing only one fusion protein served as the negative control.
CdnL is essential for M. xanthus viability
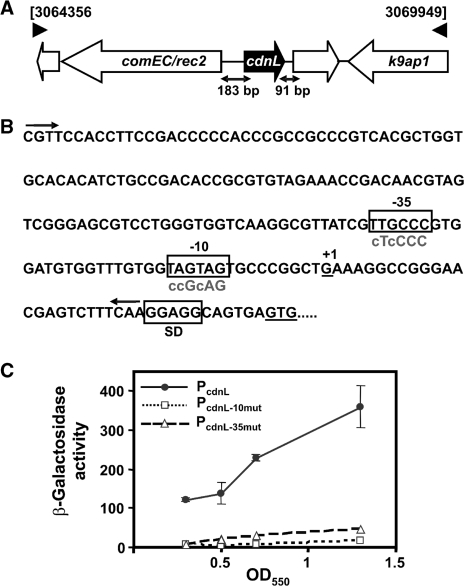
Gene cdnL (NCBI accession code: YP_630846; genome locus-tag MXAN_2627) is flanked upstream by a comE/rec2-like gene encoding a putative channel protein for DNA uptake/competence transcribed in the opposite orientation and downstream by a gene (MXAN_2628) encoding a hypothetical protein transcribed in the same direction (Figure 3A). Among myxobacteria whose genome has been sequenced, a similar comE/rec2-like gene is found upstream of cdnL in S. aurantiaca and A. dehalogenans (which belong, like M. xanthus, to the Cystobacterineae suborder) but not in Sorangium cellulosum (Sorangineae suborder) or Plesiocystis pacifica (Nannocystineae suborder). A MXAN_2628-like gene downstream of cdnL is only found in S. aurantiaca (Supplementary Figure S1).
Figure 3.
Genome context, promoter elements and expression of cdnL. (A) cdnL genome context. Genes are represented by the thick arrows. Those unlabeled encode hypothetical proteins of unknown functions. comEC/rec2 encodes a predicted DNA-internalization competence protein and k9ap1 is Pk9 associate protein 1, a predicted FHA (forkhead associated) domain signaling protein. (B) DNA sequence of the cdnL promoter region. The –10 and –35 promoter elements and the Shine-Dalgarno ribosome binding site are boxed; ‘+1’ is the transcription start site determined by 5′-RACE. The two arrows bracket the DNA segment fused to a reporter lacZ gene used for analyzing promoter activity. Lowercase letters in the sequences below the boxed –10 and –35 regions indicate bases that were mutated in the corresponding hexamers. (C) Specific β-galactosidase activities in nmol of o-nitrophenyl β-d-galactoside hydrolyzed/min/mg protein (mean of three independent measurements) of reporter lacZ probe with native (filled circle) or mutated PcdnL in the –35 (open triangle) or –10 (open square) promoter regions for different cell densities during vegetative growth.
In order to identify the function of cdnL in M. xanthus, we first attempted to delete the gene. A construct (pMR2873) bearing a complete in-frame deletion of cdnL (ΔcdnL allele) flanked by ∼0.75 kb genomic DNA was electroporated into the wild-type M. xanthus strain and stable merodiploids produced by plasmid integration into the chromosome by homologous recombination were selected. A GalS KmR transformant (MR1495) was grown for several generations in the absence of kanamycin and cells that had lost the plasmid were isolated on plates containing galactose. Out of 75 randomly picked GalR KmS colonies analysed by PCR none had the ΔcdnL allele, suggesting that cdnL may be indispensable in M. xanthus.
Hence, we next checked if the endogenous cdnL gene could be deleted if a second, functional copy of cdnL, expressed from its natural promoter (PcdnL), was supplied at a heterologous site. Given that comE/rec2 is transcribed divergently, PcdnL would be expected to lie in the 183-bp intergenic DNA stretch upstream of cdnL. Using 5′ RACE, the cdnL transcription start site was identified 36-bp upstream of the translational start codon, so that the sequences TTGCCC and TAGTAG could correspond to the –35 and the –10 regions, respectively, of PcdnL (Figure 3B). That these hexamers, which resemble those found at various M. xanthus promoters dependent on the major σA factor, are indeed critical elements of PcdnL was confirmed by site-directed mutagenesis. First, the intergenic DNA stretch between comE/rec2 and cdnL was fused to a reporter lacZ gene (PcdnL::lacZ) and its expression was examined in vivo. Reporter lacZ expression confirmed promoter activity of the DNA segment used during vegetative growth, with β-galactosidase activity levels increasing gradually from early exponential phase (OD550 = 0.3) to stationary phase (OD550 = 1.3–1.4) (Figure 3C). Mutating the putative –35 or –10 region abolished lacZ expression (Figure 3B and C), thereby demonstrating these to be crucial promoter elements. We thus used the DNA segment containing PcdnL to drive expression of cdnL at a heterologous position in the genome and check whether deletion of cdnL at its natural site could now be obtained. For this, a plasmid with the PcdnL-cdnL segment (pMR2928) was introduced into the merodiploid with the wild-type and ΔcdnL alleles and haploid segregants were obtained as above. PCR-analysis of genomic DNA from 25 of these colonies chosen at random revealed 15 to have the ΔcdnL allele. This demonstrates that the PcdnL segment chosen contains the necessary elements for cdnL expression at functional levels and reinforces the idea that cdnL is an essential gene.
Depletion of CdnL results in aberrant cell division but overexpression has no apparent effect on cell growth or morphology
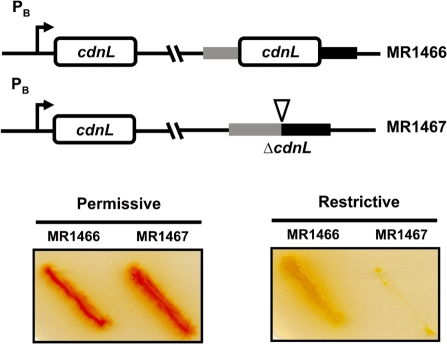
As a further check that cdnL is vital in M. xanthus and to gain insights into its possible cellular function we expressed it conditionally. For this, we used a system that we developed recently in which the gene of interest is placed under the control of the light-inducible PB promoter (19). In this system, PB is expressed in the light (permissive conditions) and repressed in the dark by CarH and vitamin B12 (restrictive conditions). Strains conditionally expressing cdnL at a heterologous site and with either wild-type cdnL (MR1466) or the ΔcdnL allele (MR1467) at the native chromosomal locus were constructed as depicted in Supplementary Figure S2. On plates, MR1466 grew well under permissive or restrictive conditions, with colony colour being red under permissive conditions due to light-induced carotenogenesis (Figure 4). By contrast, MR1467 grew well (and was also red) under permissive conditions but its growth was almost completely arrested under restrictive conditions (Figure 4). Thus, conditional expression of cdnL confirms the gene to be essential in M. xanthus.
Figure 4.
Conditional expression of cdnL shows it is essential. (Top) Schematic for M. xanthus strains conditionally expressing cdnL. (Bottom) Cells grown on CTT plates in the light were streaked on CTT plates ± B12, then incubated at 33°C for 2 days under permissive (plates without B12 incubated in the light) or restrictive (plates with B12 incubated in the dark) conditions. Note the lack of growth of MR1467 under restrictive conditions. The red colour in the light is due to carotenogenesis.
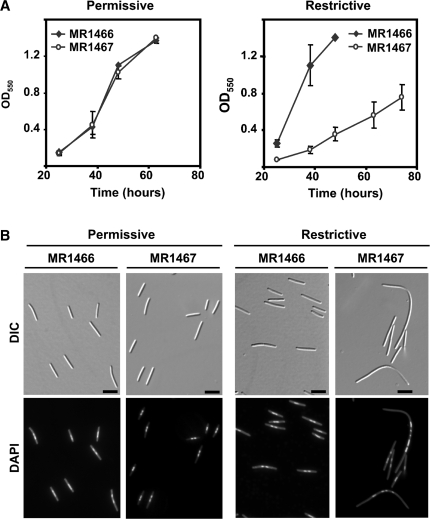
Cellular morphology under permissive conditions and on shifting to restrictive conditions was examined using differential interference contrast (DIC) and fluorescence microscopy of DAPI-stained cells to visualize the nucleoid. In agreement with the behavior observed on plates, MR1467 grew in liquid culture at about the same rate as MR1466 under permissive conditions (Figure 5A). Moreover, it exhibited normal cellular lengths (∼4–8 µm), just like MR1466, with cells containing one or two nucleoids at the mid-cell section (Figure 5B). However, on shifting to restrictive conditions, the growth rate of MR1467 was severely diminished relative to MR1466 (Figure 5A). Also, while the normal cellular morphology persisted for MR1466, ∼65% of MR1467 cells examined were considerably more elongated (∼30% of these were >15 µm in length and some reached up to 95 µm). Moreover, DAPI fluorescence of these abnormally long, filamentous cells revealed several rather well-defined foci preferentially distributed in the central zone, with the pole regions being essentially devoid of DAPI-stained foci (Figure 5B). Such filamentous cell morphology is typically associated with aberrant cell division. Thus, consistent with it being essential in M. xanthus, CdnL depletion is deleterious to growth and this directly or indirectly affects cell division.
Figure 5.
Growth rate and cellular morphology of cells conditionally expressing cdnL. (A) Growth curves for MR1466 and MR1467 under permissive (left) or restrictive (right) conditions. Cells were grown in the light at 33°C, 300 rpm, and aliquots of ∼106 cells were innoculated into 10 ml CTT in separate flasks. One was grown in the light (permissive conditions) and one in the dark with 0.75 µM B12 (restrictive conditions) at 33°C, 300 rpm; the abscissa indicates time (in hours) after the shift in growth conditions. (B) Cellular morphology of cell samples from cultures grown as in A, examined at an OD550 ∼0.8 after DAPI-staining by DIC and fluorescence microscopy. Note the unusually elongated cells indicative of aberrant cell division on shifting to restrictive conditions for the strain conditionally expressing cdnL and the extended span of the pole regions lacking DAPI-stained foci (scale marker: 5 μm).
What then, if any, are the consequences of overexpressing CdnL? We have previously overexpressed genes of interest in M. xanthus by placing them under the control of two tandem, typically strong, rRNA promoters (23,28). A construct (pMR3000) was thus generated for driving cdnL expression from the same two rRNA promoters. Introduction of pMR3000 into the wild-type strain by electroporation yielded colonies containing two copies of cdnL, one under the control of its natural promoter and the other under the control of the rRNA promoters (Supplementary Figure S3A). That these recombinants express significantly higher levels of CdnL than the wild-type was confirmed in western blots of cell extracts using polyclonal anti-CdnL antibodies (Supplementary Figure S3B). However, unlike depletion of CdnL, its overexpression did not cause any apparent effect on growth rate or cellular morphology (Supplementary Figure S3C and D).
CdnL localizes to the nucleoid in vivo suggesting a role in an essential DNA transaction
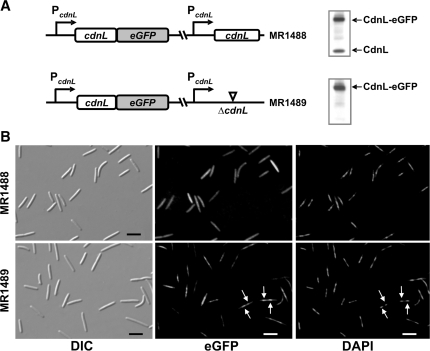
The results described so far indicate that CdnL, expressed all along vegetative growth, differs functionally from CarDNter and plays a vital role in M. xanthus, its depletion causing aberrant cell division. Sequence analysis indicates that CdnL, besides lacking an obvious DNA-binding domain, is acidic (theoretical pI = 5.4). Consistent with this, we have been unable to detect non-specific binding of CdnL to polyanions such as phosphocellulose. Thus, a priori CdnL would not be expected to localize to the nucleoid in vivo. To check this, we fused cdnL to the coding sequence for the enhanced green fluorescent protein (eGFP; 29). A plasmid bearing the PcdnL::cdnL-eGFP gene fusion was introduced into the merodiploid strain with the wild-type and ΔcdnL alleles (where it integrates at a heterologous site). Haploid segregants bearing the PcdnL::cdnL-eGFP fusion and either the wild-type cdnL allele (MR1488) or the ΔcdnL one (MR1489) were obtained at about equal frequency (Figure 6A). Immunoblots of whole cell extracts using anti-CdnL antibodies confirmed that MR1488 expressed both CdnL and CdnL-eGFP, and MR1489 only CdnL-eGFP (Figure 6A). Since MR1489 exhibited normal growth rate and cell morphology, the CdnL-eGFP fusion must be functional in vivo.
Figure 6.
CdnL subcellular localization. (A) Scheme of cdnL alleles in MR1488 and MR1489 (left). Western blots of the corresponding cell extracts probed with polyclonal anti-CdnL antibodies are shown on the right. (B) Subcellular localization of CdnL-eGFP in MR1488 and MR1489. Cells were grown to mid-log phase and examined by DIC (left panels) and fluorescence microscopy for CdnL-eGFP (middle panels) or DAPI-stained nucleoid (right panels). Arrows point to the more intense CdnL-eGFP foci in MR1489, which coincide with the nucleoid (scale marker: 5 μm).
Even though CdnL is not expected to bind DNA per se, as noted earlier, the green CdnL-eGFP fluorescence in MR1489 was found to coincide with the blue DAPI stain associated to the nucleoid (Figure 6B). On the other hand, this preferential localization to the nucleoid was less apparent in MR1488 cells, which express CdnL-eGFP as well as CdnL, possibly because of an overall excess of CdnL (GFP-free plus GFP-linked) and because the two forms would compete for associating with the nucleoid (Figure 6B). Overall, our data suggest that CdnL piggybacks to the nucleoid through interaction with a DNA-binding protein and that its essential role may be in a vital DNA transaction.
CdnL and CarDNter interact with the β subunit of M. xanthus RNAP
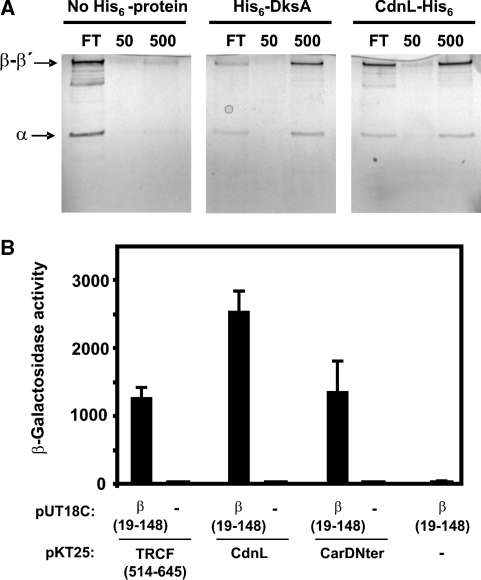
As mentioned earlier, PF02559 includes not only the family of CarDNter-like proteins but also TRCF-RID (Figure 1). A direct physical interaction between RID and a specific region of the RNAP β subunit has been demonstrated in E. coli (17). This interaction mediates removal of transcription elongation complexes stalled at DNA lesions and recruitment of the DNA repair machinery (16,17). We therefore checked whether CdnL could also interact with RNAP by determining the ability of purified CdnL-His6, immobilized on a metal affinity resin, to retain RNAP. Using this pull-down assay we had previously shown that M. xanthus DksA, like E. coli DksA, directly binds RNAP in vitro (19). Hence, resin bound to His6-DksA was used as a positive control in the pull-down assay, while protein-free resin served as the negative control. RNAP loaded onto a column containing resin without any immobilized protein, appeared almost entirely in the low salt flow through, as expected. By contrast, a significant fraction of RNAP was retained at low salt by CdnL-His6-bound resin and eluted only at high salt, just as was observed with the His6-DksA-bound resin (Figure 7A). This therefore shows that CdnL, like DksA, can interact directly with RNAP.
Figure 7.
CdnL and CarDNter physically interact with M. xanthus RNAP. (A) Analysis of RNAP retention by CdnL-His6 immobilized on TALON metal affinity resin. Shown are silver-stained 10% SDS–PAGE gels of the flowthrough (FT), low salt (50 mM) and high salt (500 mM) eluates on passing M. xanthus core RNAP through a column of TALON resin alone (‘No His6-protein’), TALON bound to His6-DksA as the positive control, or TALON bound to CdnL-His6. The α, β and β′ RNAP subunits are indicated. (B) Bacterial two-hybrid analysis of the interaction of M. xanthus RNAP β19–148 region (in plasmid pUT18C) with the TRCF segment (514–645), CdnL, or CarDNter (in pKT25) as indicated. Cells expressing only one fusion protein are negative controls.
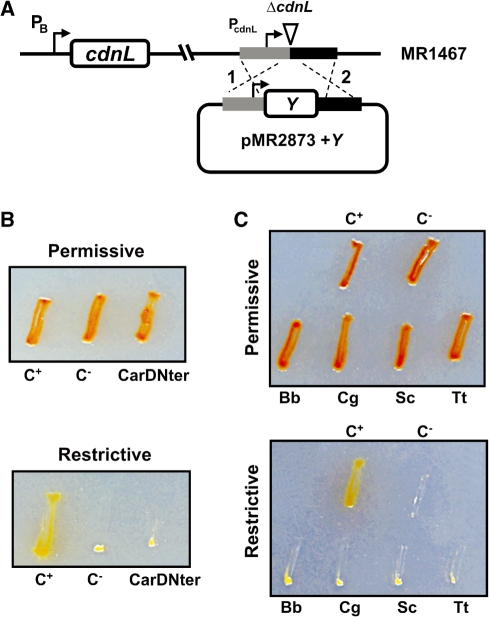
Interaction of RNAP with TRCF-RID in E. coli has been mapped to a domain at the N-terminal part of RNAP β subunit (17). The equivalent regions of M. xanthus TRCF and RNAP β subunit (TRCF514–645 and β19–148; Supplementary Figure S4) also interacted with each other, as was confirmed using a bacterial two-hybrid system: the reporter lacZ probe was expressed in cells producing both fusion proteins but not in those producing only one (Figure 7B). This two-hybrid analysis further revealed that β19–148 exhibited a direct physical interaction not only with CdnL but also with CarDNter (Figure 7B). Thus, the ability of TRCF-RID to interact with β19–148 is shared by both CdnL and CarDNter. Can CarDNter then functionally replace CdnL even though the latter cannot replace the former in CarD function? To check this we cloned the CarDNter coding sequence in the plasmid bearing the ΔcdnL allele (pMR2873) and then electroporated the construct into the strain conditionally expressing cdnL (Figure 8A). A similar construct but with cdnL introduced into pMR2873 served as the positive control. Transformants resulting from chromosomal integration of the plasmid by homologous recombination were obtained under permissive conditions and then checked for the ability to grow under restrictive conditions. None of the transformants with the CarDNter coding sequence grew under restrictive conditions in contrast to those with cdnL reintroduced (Figure 8B) indicating that CarDNter cannot substitute for CdnL in M. xanthus, thereby reaffirming that the two are functionally distinct.
Figure 8.
M. xanthus CdnL cannot be replaced by CarDNter or by CdnL homologs from other bacteria. (A) Scheme showing the strategy used for complementation analysis. A plasmid derived from pMR2873, with ‘Y’ (CarDNter, CdnL, or a given CdnL homolog) under PcdnL control and DNA segments flanking cdnL upstream (grey) and downstream (black) in the genome, was introduced into the MR1467 strain conditionally expressing cdnL. Merodiploids resulting from plasmid integration by recombination at either ‘1’ or ‘2’ would exhibit constitutive expression of the inserted variant and conditional expression of the cdnL allele at the heterologous site. (B) Complementation analysis with CarDNter. The test strain ‘CarDNter’ resulted from using pMR2873 with ‘Y’ = CarDNter coding sequence; C+, is the positive control derived from using pMR2873 with ‘Y’ = cdnL, and the negative control C− is the recipient strain MR1467. (C) Complementation analysis with BbCdnL, CgCdnl, ScCdnL, and TtCdnL. Test strains were generated using the pMR2873 with ‘Y’ = cdnLBb (‘Bb’), cdnLCg (‘Cg’), cdnLSc (‘Sc’) or cdnLTt (‘Tt’). C+ and C− are as in B. In B and C, cells grown on CTT plates in the light were streaked on CTT plates ± B12, then incubated at 33°C for 2 days under permissive (plates without B12 incubated in the light) or restrictive conditions (plates with B12 incubated in the dark). The red colour in the light is due to carotenogenesis.
CdnL cannot be substituted by homologs from other bacteria
Heterologous complementation can provide insights into gene function and specificity. Hence, we examined if CdnL in M. xanthus could be functionally replaced by homologs from other bacteria choosing as test cases those shown in Figure 1B. The genomic contexts of these homologs and of cdnL in myxobacteria (Supplementary Figure S1) indicate that cysS, encoding cysteinyl tRNA-synthetase and therefore essential for protein synthesis, is frequently encountered in the neighborhood of cdnL. Two other genes, ispD and ispF, which encode 2-C-methyl-d-erythritol 4-phosphate and 2,4-cyclodiphosphate cytidylyl transferases, respectively, of the mevalonate-independent isoprenoid biosynthetic pathway, are also often located in the genomic vicinity of cdnL. These exist in most eubacteria but not in M. xanthus, and have been shown to be essential in E. coli, Bacillus subtilis and M. tuberculosis (30–32).
The homologs chosen for our complementation studies vary in: (i) the percentage sequence identity/similarity to CdnL (60/76% for the 164-residue BbCdnL from B. bacteriovorus, a δ-proteobacterium like M. xanthus; 32/64% for the 160-residue ScCdnL from S. coelicolor and 33/63% for the 198-residue CgCdnL from C. glutamicum, both of which are Actinobacteria; 28/52% for the 164-residue TtCdnL from T. thermophilus, of the Deinococcus-Thermus group); (ii) the GC-content (72% in S. coelicolor and 70% in T. thermophilus, comparable to 69% in M. xanthus; 54% in C. glutamicum and 51% in B. bacteriovorus); (iii) the presence of dksA (B. bacteriovorus, S. coelicolor) or its absence (C. glutamicum, T. thermophilus), according to genome annotation. Functional complementation by these homologs in M. xanthus was checked using the strategy employed in the previous section. A plasmid construct with the coding sequence of each homolog in pMR2873 (Figure 8A) was introduced into the strain conditionally expressing cdnL and transformants were selected under permissive conditions. When examined under restrictive conditions, no growth was observed for any of the transformants with the CdnL homologs (Figure 8C). Since we could detect BbCdnL (the homolog most similar to CdnL, yet also the one most divergent in the GC-content and so codon usage) in M. xanthus cell extracts using anti-CdnL antibodies (results not shown), we surmise that the CdnL homologs are expressed in M. xanthus. Hence, none of the homologs examined appears to be capable of functionally replacing CdnL in M. xanthus suggesting that the requirement for CdnL is specific.
DISCUSSION
Genomic analysis has led to the identification of a large family of bacterial proteins of unknown function with sequence similarity to CarDNter, the N-terminal domain of the global M. xanthus transcriptional regulator CarD, which is implicated in light-induced carotenogenesis (hence the name), starvation-induced multicellular development and other processes (6,9,12,15). In CarD and its orthologs, identified thus far only in myxobacteria, CarDNter is linked to a C-terminal domain resembling eukaryotic HMGA or histone H1 that confers DNA binding essential for activity (7,11,15). Thus, an important difference between CarD and the CarDNter-like proteins is the absence in the latter of a DNA-binding domain. This, together with our demonstration here that CarD and the one CarDNter-like protein in M. xanthus are functionally dissimilar, highlights the necessity of differentiating CarD from the CarDNter-like proteins in the PF02559 protein family. Therefore, referring to these latter proteins as CdnL (as in this study), rather than CarD, would be more apt. Hence, to avoid confusion, we propose that the protein recently reported as CarD in mycobacteria (18) be renamed CdnL, and that this be the term used for equivalent proteins in other bacteria.
Interactions with CarG, besides DNA, are crucial for CarD activity (8,15). The inability of CdnL to interact with CarG could then account for why CdnL could not functionally replace CarDNter, the domain that mediates interaction with CarG. Neither could CarDNter substitute for CdnL function. Also, whereas carD (or carDNter) can be deleted with no loss of viability despite its role as a global regulator (6,8,12), CdnL is indispensable for M. xanthus growth and survival. A function for CdnL in an essential DNA transaction is suggested by our finding that in vivo it localizes to the nucleoid. That this occurs despite an intrinsic inability to bind DNA implies the existence of factors that interact with CdnL and piggyback it to the nucleoid. One likely vehicle for CdnL recruitment to the nucleoid is RNAP since we find that it interacts physically with CdnL. The sequence similarity of CdnL to RID, the TRCF module that interacts with RNAP, allowed us to map CdnL interaction to the same N-terminal segment of the M. xanthus RNAP β subunit as RID. Similarity with RID extends to other PF02559 family members. This led us to confirm that, like RID or CdnL, CarDNter also interacted with the same region of β, in line with the established role of CarD in transcriptional regulation. An implication of our findings is that CdnL, CarD and TRCF would compete with each other for binding to the same region of RNAP in vivo. Clearly, this competition would be subject to additional levels of control, such as their relative concentrations, affinities for RNAP, and interactions with distinct factors that would confer specificity to each protein for its function. This is emphasized by the interaction of CarDNter, but not CdnL, with CarG, and by our finding that not just CarDNter but also several sequence homologs from diverse bacteria failed to complement the lack of CdnL.
The interaction with RNAP implicates CdnL in a possibly vital role in M. xanthus transcription. The CdnL homolog in mycobacteria, also essential, was recently implicated in the control of rRNA transcription at steady state and during diverse cellular stresses such as starvation, in the early stages of which there is as much as a 20-fold enhancement in cdnL expression (18,33). This CdnL was also shown to bind RNAP, but its mapping to the β subunit was deduced from observations with TtCdnL, since mycobacterial CdnL itself failed to exhibit such an interaction in the two-hybrid analysis employed. Control of rRNA transcription in response to starvation has been extensively studied in E. coli (without any CdnL homolog) where it is mediated by DksA, which interacts with the RNAP secondary channel and enhances the inhibitory effects of (p)ppGpp on rRNA promoters during the stringent response (34–38). Escherichia coli DksA differs markedly from CdnL in amino acid sequence, in not being essential, at least for growth in rich media (39), and in its RNAP-binding mode. Yet, surprisingly, mycobacterial CdnL could partially complement the lack of DksA in E. coli and interact with E. coli RNAP, even as DksA neither rescued lethality of CdnL-depleted mycobacteria (which lack endogenous DksA) nor interacted with mycobacterial RNAP (18). Thus, the molecular mechanisms of action of DksA and CdnL are likely to be different. Also, additional mechanisms must be available given that normal growth is not impaired on knocking out the cdnL homolog in B. subtilis (40), which has no DksA (41); and bacteria with neither CdnL nor DksA exist. Moreover, several bacterial species have both CdnL and DksA suggesting that the two may not be functionally redundant. Indeed, we show this is the case in M. xanthus, which not only has CdnL and DksA, but also CarD. Besides being functionally distinct from CarDNter, our finding that CdnL is vital in M. xanthus, as is DksA (19), is convincing evidence that CdnL and DksA are not functionally redundant.
In M. xanthus, (p)ppGpp accumulation in response to amino acid starvation initiates the early gene expression pattern that leads to fruiting body development, suggesting a connection between this process and a stringent response (42–46). Thus, disrupting the relA homolog in M. xanthus not only blocked (p)ppGpp accumulation but also fruiting body formation. Given that CdnL and DksA are essential, and no system is as yet available for conditional expression during starvation-induced development in M. xanthus (19), examining whether they are involved in the stringent control and, if so, the interplay between them, must await future studies. It should be noted, however, that whereas carD expression is transiently activated early during development, consistent with its known role in this process (6), our analysis of PcdnL expression did not reveal any marked upregulation early (Supplementary Figure S5) or later in development. This also contrasts with the sharp upregulation early on starvation reported for the mycobacterial cdnL (18).
Why are CdnL and DksA simultaneously required for normal vegetative growth in M. xanthus? Stringent response per se does not appear to be essential for growth, since M. xanthus relA mutants are viable (45). Thus, while CdnL and DksA may be involved in stringent control in M. xanthus, each could have additional and distinct functions that are vital. Interestingly, depletion of CdnL in M. xanthus provokes an abnormal elongated cell morphology typically associated with aberrant cell division, like the filamentous phenotype caused by the depletion of DksA that we reported previously (19). In E. coli, this aberrant cell morphology and compromised cell viability was not observed on eliminating DksA alone, but was when done in combination with other RNAP modulators (TRCF or GreA) and the RuvABC resolvase (47,48). It was proposed that eliminating factors that modulate RNAP or dislodge it from the DNA, when DNA repair is also compromised, enhances accumulation of stalled RNAP. At strong promoters, like those of rRNA operons, piled-up RNAP arrays would pose a formidable block to the replication machinery leading to severely reduced viability. Since we observe impaired cell growth in M. xanthus on limiting CdnL or DksA alone (in a repair-proficient background), the cell division phenotype may be unrelated to their possible function in downregulating rRNA transcription. Alternatively, their essential role might result from controlling expression of a gene(s) essential for M. xanthus viability. Also, a direct involvement of CdnL and/or DksA in cell division cannot be ruled out. Therefore, the next challenge is to understand if and how CdnL and DksA control rRNA transcription in M. xanthus, and to identify what makes each of them essential in this bacterium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación-Spain (BFU2006-14524 and BFU2009-12445-C02-01 to M.E.-A., BFU2008-00911 and BFU2009-12445-C02-02 to S.P.); and Fundación Séneca-Murcia (08748/PI/08 to F.J.M.). Funding for open access charge: Ministerio de Ciencia e Innovación-Spain.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J. A. Madrid for technical assistance and C. Flores for DNA-sequencing.
REFERENCES
- 1.Dawid W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 2000;24:403–427. doi: 10.1111/j.1574-6976.2000.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser D. Signaling in myxobacteria. Annu. Rev. Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 3.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007;5:862–72. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 4.Kroos L. Eukaryotic-like signaling and gene regulation in a prokaryote that undergoes multicellular development. Proc. Natl Acad. Sci. USA. 2005;102:2681–2682. doi: 10.1073/pnas.0500157102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl Acad. Sci. USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolás FJ, Ruiz-Vázquez RM, Murillo FJ. A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 1994;8:2375–2387. doi: 10.1101/gad.8.19.2375. [DOI] [PubMed] [Google Scholar]
- 7.Nicolás FJ, Cayuela ML, Martínez-Argudo IM, Ruiz-Vázquez RM, Murillo FJ. High mobility group I(Y)-like DNA-binding domains on a bacterial transcription factor. Proc. Natl Acad. Sci. USA. 1996;93:6881–6885. doi: 10.1073/pnas.93.14.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peñalver-Mellado M, García-Heras F, Padmanabhan S, García-Moreno D, Murillo FJ, Elías-Arnanz M. Recruitment of a novel zinc-bound transcriptional factor by a bacterial HMGA-type protein is required for regulating multiple processes in Myxococcus xanthus. Mol. Microbiol. 2006;61:910–926. doi: 10.1111/j.1365-2958.2006.05289.x. [DOI] [PubMed] [Google Scholar]
- 9.Galbis-Martínez M, Fontes M, Murillo FJ. The high mobility group A-type protein CarD of the bacterium Myxococcus xanthus as a transcription factor for several distinct vegetative expressed genes. Genetics. 2004;167:1585–1595. doi: 10.1534/genetics.104.029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elías-Arnanz M, Fontes M, Padmanabhan S. Carotenogenesis in Myxococcus xanthus: a complex regulatory network. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. Washington, DC: ASM Press; 2008. pp. 211–225. [Google Scholar]
- 11.Padmanabhan S, Elías-Arnanz M, Carpio E, Aparicio P, Murillo FJ. Domain architecture of a high mobility group A-type bacterial transcriptional factor. J. Biol. Chem. 2001;276:41566–41575. doi: 10.1074/jbc.M106352200. [DOI] [PubMed] [Google Scholar]
- 12.Cayuela ML, Elías-Arnanz M, Peñalver-Mellado M, Padmanabhan S, Murillo FJ. The Stigmatella aurantiaca homolog of Myxococcus xanthus HMGA-type transcription factor CarD: insights into the functional modules of CarD and their distribution in bacteria. J. Bacteriol. 2003;185:3527–3537. doi: 10.1128/JB.185.12.3527-3537.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Heras F, Padmanabhan S, Murillo FJ, Elías-Arnanz M. Functional equivalence of HMGA- and histone H1-like domains in a bacterial transcriptional factor. Proc. Natl Acad. Sci. USA. 2009;106:13546–13551. doi: 10.1073/pnas.0902233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 17.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 18.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Moreno D, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. A vitamin B12-based system for conditional expression reveals dksA to be an essential gene in Myxococcus xanthus. J. Bacteriol. 2009;191:3108–3119. doi: 10.1128/JB.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Argudo I, Ruiz-Vázquez RM, Murillo FJ. The structure of an ECF-sigma-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol. Microbiol. 1998;30:883–893. doi: 10.1046/j.1365-2958.1998.01129.x. [DOI] [PubMed] [Google Scholar]
- 22.Julien B, Kaiser D, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl Acad. Sci. USA. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Marín MC, Padmanabhan S, Polanco MC, Murillo FJ, Elías-Arnanz M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol. Microbiol. 2008;67:804–819. doi: 10.1111/j.1365-2958.2007.06086.x. [DOI] [PubMed] [Google Scholar]
- 24.Karimova G, Ullmann A, Ladant D. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 2000;328:59–73. doi: 10.1016/s0076-6879(00)28390-0. [DOI] [PubMed] [Google Scholar]
- 25.López-Rubio JJ, Padmanabhan S, Lázaro JM, Salas M, Murillo FJ, Elías-Arnanz M. Operator design and mechanism for CarA repressor-mediated down-regulation of the photoinducible carB operon in Myxococcus xanthus. J. Biol. Chem. 2004;279:28945–28953. doi: 10.1074/jbc.M403459200. [DOI] [PubMed] [Google Scholar]
- 26.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 27.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Marín MC, López-Rubio JJ, Murillo FJ, Elías-Arnanz M, Padmanabhan S. The N-terminus of Myxococcus xanthus CarA repressor is an autonomously folding domain that mediates physical and functional interactions with both operator DNA and antirepressor protein. J. Biol. Chem. 2004;279:33093–33103. doi: 10.1074/jbc.M405225200. [DOI] [PubMed] [Google Scholar]
- 29.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 30.Campbell TL, Brown ED. Characterization of the depletion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase in Escherichia coli and Bacillus subtilis. J. Bacteriol. 2002;184:5609–5618. doi: 10.1128/JB.184.20.5609-5618.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buetow L, Brown AC, Parish T, Hunter WN. The structure of Mycobacteria 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase, an essential enzyme, provides a platform for drug discovery. BMC Struct. Biol. 2007;7:68. doi: 10.1186/1472-6807-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eoh H, Brown AC, Buetow L, Hunter WN, Parish T, Kaur D, Brennan PJ, Crick DC. Characterization of the Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase: potential for drug development. J. Bacteriol. 2007;189:8922–8927. doi: 10.1128/JB.00925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connolly LE, Cox JS. CarD tricks and magic spots: mechanisms of stringent control in mycobacteria. Cell Host Microbe. 2009;6:1–2. doi: 10.1016/j.chom.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel-structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 36.Potrykus K, Vinella D, Murphy H, Szalewska-Palasz A, D'Ari R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 2006;281:15238–48. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- 37.Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang PJ, Craig E. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krásný L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J. Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manoil C, Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J. Bacteriol. 1980;141:305–315. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- 45.Harris BZ, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in. Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford EW, Jr, Shimkets LJ. The stringent response in Myxococcus xanthus is regulated by SocE and the CsgA C-signaling protein. Genes Dev. 2000;14:483–492. [PMC free article] [PubMed] [Google Scholar]
- 47.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Rep.(Amst.) 2007;6:981–993. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.