Abstract
Here we describe a northern blot procedure that allows the detection of endogenous RNAs as small as ∼14 nt in total RNA extracts from bacteria. RNAs that small and as part of total bacterial RNA extracts usually escape detection by northern blotting. The approach combines LNA probes 5′-digoxigenin-endlabeled for non-radioactive probe detection with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide-mediated chemical crosslinking of RNAs to nylon membranes, and necessitates the use of native PAGE either with the TBE or MOPS buffer system.
INTRODUCTION
The discovery of small regulatory RNAs in all domains of life and their multi-faceted key roles in cell biology has propelled advances in technologies for their analysis. This includes the use of LNA probes in hybridization approaches to increase sensitivity (1,2), development of the looped-primer RT–PCR technique to quantify mature miRNAs (3), as well as high-throughput methods such as RNomics to generate cellular RNA libraries for deep sequencing (4) or miRNA profiling arrays (5). Despite these advances, the classic northern blot technique has remained irreplaceable, since the approach allows the researcher to visualize and roughly quantify cellular levels of RNAs and their processing intermediates relative to endogenous RNA standards (e.g. 5S rRNA). The northern blot has even gained importance owing to the growing number of small RNA candidates identified by high-throughput and bioinformatic methods, which need to be verified experimentally. Recently, Hamilton and co-workers (6,7) reported the use of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) to covalently couple small RNAs via their 5′-phosphates to amino groups at the surface of nylon membranes. Relative to the standard UV crosslinking to nylon membranes, this reportedly 5′-end-selective attachment enhanced the detection of RNAs <40 nt 25- to 50-fold. The likely reason is that UV crosslinking to the free amines of nylon membranes occurs via the bases (primarily uracil) which, in contrast to immobilization via the phosphate, sterically blocks accessibility of the immobilized nucleic acid for hybridization, particularly in the case of small RNAs owing to their limited interaction surface. Furthermore, the number and positions of UV-crosslinked bases vary depending on the particular RNA sequence and the applied UV crosslinking conditions (length of irradiation, UV light intensity), making UV crosslinking a process that is hard to control (6,7).
We were faced with the limitations of standard northern blot protocols when trying to detect small so-called product RNAs (pRNAs, ∼14 nt in length) in total RNA extracts from Bacillus subtilis; pRNAs are short transcripts synthesized by bacterial RNA polymerases in an RNA-dependent RNA polymerization reaction using 6S RNA as template. 6S RNA, ∼200 nt in size, has been identified as a growth-phase dependent riboregulator of bacterial transcription (8). In Escherichia coli, this RNA acts by complex formation with the σ70 housekeeping RNA polymerase (RNAP) holoenzyme (Eσ70) and accumulates during stationary phase. 6S RNA is primarily helical, with a large single-stranded loop in the center. This highly conserved secondary structure is essential for the ability of 6S RNA to form stable complexes with Eσ70 and to serve as a template for the synthesis of pRNAs (9,10). Synthesis of these short transcripts leads to dissociation of 6S RNA–RNAP complexes. In contrast to E. coli with only one 6S RNA gene, the B. subtilis genome harbors two 6S RNA homologs, termed 6S-1 and 6S-2 (11,12). Both RNAs were co-immunoprecipitated with the σA housekeeping RNAP holoenzyme using antibodies against σA or the α subunit (13). Our identification of 8- to 15-mer pRNAs derived from transcription on 6S-1 RNA as template in B. subtilis by a deep sequencing approach (unpublished results), but the failure to detect those by northern blotting, prompted us to explore improved northern blot protocols tailored to the detection of cellular RNAs as small as 14 nt.
MATERIALS AND METHODS
Strains and growth conditions
Bacillus subtilis 168 and PY79, E. coli MG1655, and 6S RNA knockout strains B. subtilis PY79 ΔbsrA and E. coli MG1655 ΔssrS were grown in LB medium at 37°C. In preparation for outgrowth experiments, 50 ml of fresh medium were inoculated with an overnight culture to an OD600 of 0.05; cells were then grown to stationary phase for ∼24 h. Then 20 ml of stationary culture were diluted with 80 ml fresh pre-warmed LB medium to induce outgrowth at 37°C under shaking; total RNA was extracted 3 min after outgrowth had been started. Construction and phenotype of the 6S-1 RNA knockout strain B. subtilis PY79 ΔbsrA and the 6S RNA knockout strain E. coli MG1655 ΔssrS will be reported elsewhere (manuscript in preparation).
RNA extraction
Cellular total RNA was prepared using the hot phenol method (14). Briefly, B. subtilis or E. coli cell pellets were resuspended in extraction buffer (10 mM sodium acetate pH 4.8, 150 mM sucrose) and incubated with 0.1 volumes of lysozyme (20 mg/ml, Roth, Karlsruhe, Germany) for 10 min at room temperature. 10% SDS was added to a final concentration of 1% prior to vigorous vortexing. After addition of 1 volume of phenol (preheated to 65°C) and vortexing, the mixture was incubated for 5 min at 65°C and 5 min on ice, followed by centrifugation at 4°C for 30 min (8220g). Phenol extraction was repeated, followed by chloroform (1+1) extraction and ethanol precipitation. RNA concentration was determined by UV spectroscopy.
Northern blotting
For northern blotting, 6 or 10 µg of total cellular RNA were loaded per gel lane. Mature B. subtilis 6S-1 RNA (190 nt) and E. coli 6S RNA (186 nt) included as controls (Figure 1) were synthesized by in vitro runoff transcription using T7 RNA polymerase. Chemically synthesized pRNA oligonucleotides 5′-GUU CGG UCA AAA CU-3′ (B. sub 6S-1p wt), 5′-GUU CGG UCA CGA CU-3′ (B. sub 6S-1p mut) and 5′-AUC GGC UCA GGG GA-3′ (E. coli 6Sp wt) were obtained from Noxxon (Berlin, Germany). 14- or 16-mer probes complementary to pRNAs included 5′-digoxigenin-aGt tTt gAc cGa Ac-3′ (probe for B. subtilis wt 6S-1 pRNA), 5′-digoxigenin-aGt cgT gAC cga Ac-3′ (probe for B. subtilis mutant 6S-1 pRNA), 5′-digoxigenin-tcC cCt gAg cCg At-3′ (E. coli 6S pRNA probe 1), 5′-digoxigenin-tcc ccT gag ccg At-3′ (E. coli 6S pRNA probe 2) and 5′-digoxigenin-agT ccc ctg agc cgA t-3′ (E. coli 6S pRNA probe 3), with uppercase letters indicating LNA and lowercase letters DNA residues (Figures 1 and 7); 5′-digoxigenin-labeled, LNA-containing probes were obtained from Exiqon (Vedbaek, Denmark). For specific detection of B. subtilis and E. coli 5S rRNA loading controls and for E. coli 6S RNA, antisense transcripts covering the respective full-length RNA and internally labeled with digoxigenin-UTP were synthesized from PCR templates by T7 RNA polymerase (according to the DIG RNA Labeling Mix protocol provided by Roche Diagnostics, Mannheim, Germany). For native PAGE, samples were adjusted to 1× native loading buffer by mixing with 6× native loading buffer [0.25% (w/v) bromophenol blue, 0.25% (w/v) xylene cyanol blue, 30% (v/v) glycerol]. For denaturing PAGE, samples were adjusted to 1× denaturing loading buffer [0.01% (w/v) bromophenol blue, 0.01% (w/v) xylene cyanol blue, 1.3 M urea, 33% (v/v) formamide, 1× TBE]. Before gel loading, samples were heated to 95°C for 3 min, followed by immediate cooling on ice. Generally, 10 or 20% polyacrylamide gels were used for RNA separation under native or denaturing conditions. For native PAGE, gel solutions as well as electrophoresis buffers either contained 89 mM Tris, 89 mM borate and 2 mM EDTA (TBE buffer system) or 20 mM MOPS–NaOH, pH 7, (MOPS–NaOH buffer system). For denaturing PAGE, gel solutions as well as electrophoresis buffers either contained 89 mM Tris, 89 mM borate, 2 mM EDTA and 8 M urea (TBE buffer system) or 20 mM MOPS–NaOH, pH 7 and 2% formaldehyde (MOPS–NaOH buffer system). RNA was blotted on positively charged nylon membranes (Roche Diagnostics) overnight using a semi-dry blotter with 3.75 mA/cm2 and 0.5× TBE as transfer buffer. To add a 5′-phosphate to the chemically synthesized pRNAs, the oligonucleotide was incubated with ATP and T4 polynucleotide kinase (Fermentas, St. Leon-Rot, Germany) according to the manufacturer’s instructions. Efficiency of phosphorylation was analyzed on a 20% denaturing polyacrylamide gel (TBE buffer system), with the 5′-phosphorylated oligonucleotide migrating faster than the 5′-OH variant owing to its extra negative charge; pRNAs with 5′-OH and 5′-phosphate were subjected to preparative co-electrophoresis, followed by gel excision and gel elution in 1 M Na(OAc) pH 4.9. Finally, pRNAs were concentrated by ethanol precipitation.
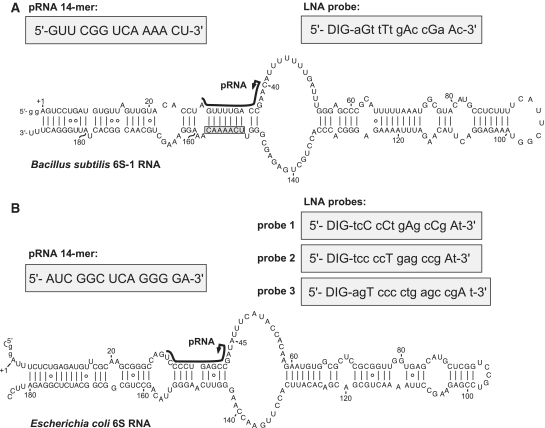
Figure 1.
Secondary structure presentation of (A) mature B. subtilis 6S-1 RNA (190 nt), adapted from (11), and (B) of E. coli 6S RNA according to (9); both RNAs were transcribed with two artificially added G residues (lowercase letters) for reasons of efficient synthesis by T7 RNA polymerase. In both panels, the black lines along the sequence indicate the region of B. subtilis 6S-1 RNA and E. coli 6S RNA that serve as a template for the synthesis of small transcripts (product RNAs = pRNAs) by RNA polymerase (this study, 9); arrows mark the starting point of pRNA transcription; the chemically synthesized pRNA mimics are depicted in the grey boxes on the left above the 6S RNA structures. In panel A, 6S-1 RNA nt 151–157 complementary to nt 2–8 of the LNA probe are boxed. LNA probes for pRNA detection are shown in grey boxes on the right above the secondary structures; DIG, digoxigenin attached to the 5′-end via a C6 linker; small letters depict DNA residues, capital letters LNA residues.
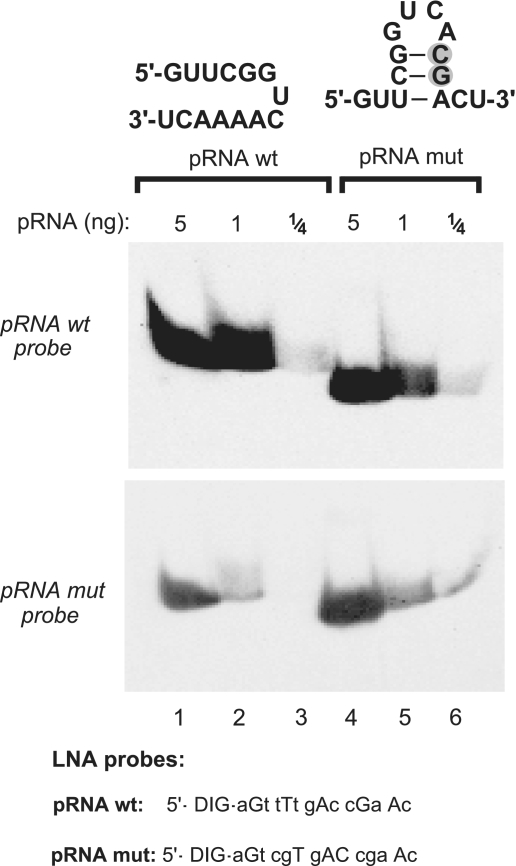
Figure 7.
Evaluation of the influence of potential intramolecular RNA secondary structures on detection efficiency. A mutated version of the 6S-1 pRNA (pRNA mut with two consecutive point mutations, grey-shaded; lanes 4–6), designed to have the potential to form a hairpin with a 3-bp stem, migrated somewhat faster than wt pRNA (lanes 4–6 versus 1–3) in a 20% native PAA gel (TBE buffer system), suggesting that the hairpin indeed formed at least transiently during electrophoresis. The two pRNA variants (carrying 5′-OH termini) were mutually detected with LNA probes either complementary to wt pRNA (top panel) or to pRNA mut (bottom panel), with LNA probes specified below the blots (uppercase letters indicate LNA and lowercase letters DNA residues). Films were exposed to the blot for 35 min.
Hybridization and detection
Classical RNA immobilization was performed by incubation of the wet membrane at 80°C for 30 min and storage at 4°C. Alternatively, EDC crosslinking was utilized to detect very small RNAs, exactly as described (7). Briefly, 245 µl of 12.5 M 1-methylimidazole (Sigma Aldrich, Taufkirchen, Germany) were added to 9 ml of double-distilled water and the pH was adjusted to 8.0 using HCl. Directly prior to use, 0.75 g EDC [1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, Sigma Aldrich] were added and the volume was adjusted to 24 ml with double-distilled water. The wet membrane was placed on a Whatman paper soaked with EDC solution (160 mM EDC, 130 mM 1-methylimidazole pH 8.0), wrapped in plastic foil and incubated for 2 h at 60°C. After careful washing with water, the membrane was stored at 4°C. For pre-hybridization, 7 ml of hybridization solution (termed ‘DIG Easy Hyb Granules’, Roche Diagnostics) were heated to 50°C (in the case of DIG-LNA/DNA mixmer probes) or 68°C (in the case of T7 transcript probes >100 nt) and added to the membrane in a hybridization tube (avoid sticking of the membrane to the tube wall), followed by slow rotation in an hybridization oven for 2 h at 50 or 68°C, respectively. Probes, either 10 µl T7 transcript or 300 pmol DIG-LNA/DNA mixmer, were denatured for 3 min at 95°C, followed by immediate transfer on ice. The probe was then added to 7 ml pre-heated (50 or 68°C) hybridization solution to replace the pre-hybridization solution. Hybridization was conducted for at least 6 h, usually overnight, at 50 or 68°C under slow rotation in a hybridization oven. In some cases (Figure 8A) the (pre-)hybridization temperature for LNA/DNA mixmer probes was increased (depending on the probe’s G/C content and extent of LNA modification) in order to suppress non-specific signals. Immunological detection of RNA was performed according to the instructions of the DIG Northern Starter Kit (Roche Diagnostics). X-ray films (Kodak BioMax Light Film, Sigma Aldrich) were exposed to the membrane for chemiluminescence detection (usually 20–45 min). Developed X-ray films were scanned (BioRad GS-800) for documentation.
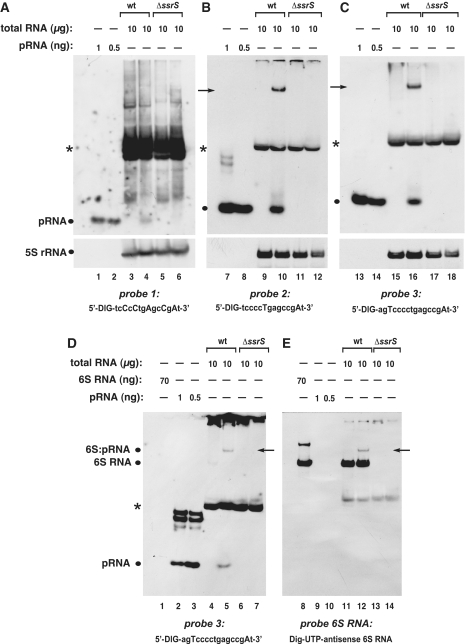
Figure 8.
Specificity of different probes for the detection of pRNAs in RNA extracts from E. coli. (A–C) Detection of pRNAs derived from E. coli 6S RNA in total RNA extracts from the wild-type (wt) strain MG1655 (lanes 3, 4, 9, 10, 15 and 16) versus RNA extracts derived from a MG1655 6S RNA knockout (ΔssrS) strain (lanes 5, 6, 11, 12, 17 and 18) using three different probes shown below each panel (uppercase letters indicate LNA and lowercase letters DNA residues). Deviating from our standard procedure (‘Materials and Methods’ section), RNA samples were heated in denaturing loading buffer for 5 min at 95°C immediately before gel loading. In analogy to B. subtilis (Figure 6), RNA harvested from cells in stationary phase (lanes 3, 5, 9, 11, 15 and 17) or during outgrowth (lanes 4, 6, 10, 12, 16 and 18) was analyzed. The chemically synthesized E. coli pRNA 14-mer (Figure 1B) was loaded as size control in lanes 1, 2, 7, 8, 13 and 14 (pRNA positions marked by dots at the left margin of each panel). NC filters were stripped after hybridization with pRNA probes and hybridized with a 5S rRNA-specific probe (as loading control) shown at the bottom of each panel. A pRNA signal was seen under outgrowth conditions for wt bacteria (lanes 4, 10 and 16), but not for ΔssrS bacteria (lanes 6, 12, 18). Probe 1 (A) was the least specific and yielded the highest background among the three different probes, which we attribute to the high number of five LNA residues, two of which are in the tetracytidylate stretch that is able to basepair with tetraguanylates. Signals indicated by arrows in lanes 10 and 16 are 6S RNA:pRNA hybrids (see below); asterisks mark a cross-hybridizing RNA of unknown identity. (D and E) The two panels are halves of the same blot, representing identical sample sets, but hybridized to different probes (probe 3 in D, antisense 6S RNA in E). The signals in lanes 5 and 12, indicated by arrows, are inferred to be 6S RNA:pRNA hybrids, as they migrate at identical position and are detected with both probes. For control, in vitro transcribed E. coli 6S RNA was loaded (lanes 1 and 8). All samples in D and E were heated for 2 min at 95°C in denaturing loading buffer immediately before gel loading. Deviating from the standard protocol (‘Materials and Methods’ section), the hybridization temperature was 72°C instead of 50°C for probe 1, and 68°C instead of 50°C for probes 2 and 3.
RESULTS
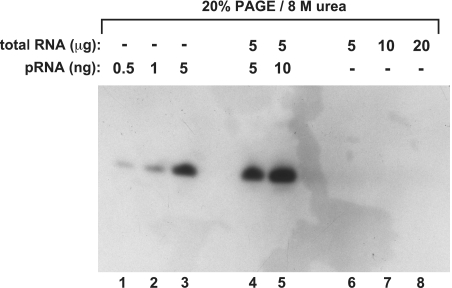
From a deep sequencing library we knew that pRNA transcripts, predominantly 8–15 nt in length, accumulate in B. subtilis cells under conditions of outgrowth from stationary phase (unpublished results, manuscript in preparation). The pRNAs are synthesized by RNA polymerase using 6S-1 RNA as the template (Figure 1A). However, we and others (Jörg Vogel, personal communication) had been unable to detect pRNA transcripts by northern blot analysis. Such an example, where we used 20% PAGE/8 M urea and classical ‘baking’ of the membrane at 80°C for RNA immobilization, is shown in Figure 2 (comparable results were obtained with UV crosslinking; data not shown). Although we employed an LNA/DNA mixmer probe (Figure 1A) for efficient hybridization, no endogenous pRNA signal could be detected in up to 20 µg of total RNA from B. subtilis cells at 3 min of outgrowth from stationary phase (Figure 2, lanes 6–8). Yet, when gels were loaded with 0.5, 1 or 5 ng of a chemically synthesized pRNA 14-mer, signals were obtained (Figure 2, lanes 1–3). Likewise, a signal of similar intensity as in lane 3 (5 ng pRNA) was observed when 5 ng of the pRNA 14-mer were added to 5 µg of total cellular RNA from outgrowth bacteria (lane 4). These findings admitted different explanations: (i) endogenous pRNA levels were below the sensitivity level of the applied northern blot setup; (ii) endogenous pRNAs might be masked in the context of total cellular RNA extracts owing to their association with 6S-1 RNA; non-specific masking could be excluded based on the results shown in Figure 2, lanes 4 and 5.
Figure 2.
Standard denaturing northern blot using the classical ‘baking’ of the membrane at 80°C for RNA immobilization. Lanes 1–3: various amounts of the chemically synthesized pRNA 14-mer (with 5′-OH terminus; Figure 1A); lanes 4 and 5: 5 or 10 ng of the chemically synthesized pRNA 14-mer mixed with 5 µg of total RNA from B. subtilis before electrophoresis and membrane transfer; lanes 6–8: samples with varying amounts of total RNA from B. subtilis for the purpose of endogenous pRNA detection. Total cellular RNA was isolated 3 min after outgrowth from stationary phase using the hot phenol method; the 5′-digoxigenin-labeled LNA–DNA mixmer shown in Figure 1A was used as probe. (for details, see ‘Materials and Methods’ section).
Native versus denaturing PAA gels combined with EDC crosslinking
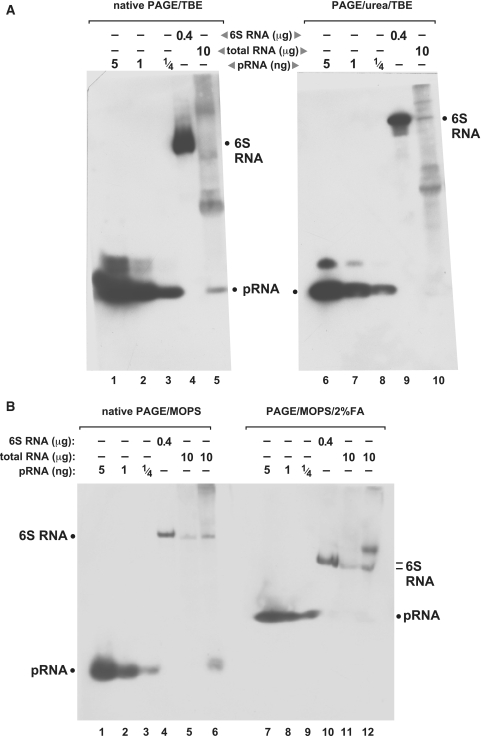
Since the oligonucleotide coupling to nylon membranes is thought to occur via primary amino groups (15), we considered the possibility that the amino groups of urea in denaturing PAA/TBE gels might compete with the membrane amines during the coupling step. Moreover, the necessity of denaturing conditions for electrophoretic separation according to size is certainly less mandatory for short RNA oligonucleotides compared with larger and more structured RNAs. To address these points, we performed northern blots with native and denaturing PAA gels, either using the 1× TBE or the MOPS buffer system, and applying EDC crosslinking for RNA coupling to the nylon membrane (Figure 3). In the case of native but not denaturing conditions, we were now able to detect endogenous pRNAs in total RNA extract from outgrowth bacteria (Figure 3A, lane 5; Figure 3B, lane 6). The endogenous pRNA signal co-migrated with the chemically synthesized pRNA 14-mer (Figure 3A and B, lanes 1–3). Assuming that the 5′-end groups (5′-OH for the chemically synthesized pRNA, 5′-triphosphate for the endogenous pRNA) had little effect on gel mobility in native 10% PAA gels, our results suggest that the bulk of endogenous pRNAs transcribed from 6S-1 RNA have sizes of ∼14 nt, in line with the prevalence of 8- to 15-mer sequence reads in our deep sequencing library (data not shown). The absence of an endogenous pRNA signal in the presence of 8 M urea (Figure 3A, lane 10 versus 5) or 2% formaldehyde (Figure 3B, lane 12 versus 6) can be explained by urea competing with amino groups at the membrane surface for reaction with EDC and formaldehyde reacting with membrane amino groups which then become unavailable to reaction with the EDC crosslinker.
Figure 3.
Comparison of northern blots performed in different buffer systems under native and denaturing conditions, using EDC crosslinking for RNA immobilization on nylon membranes. (A) Northern blots of RNA separated by native (left) and denaturing (right) PAGE using the TBE buffer system. Lanes 1–3 and 6–8: varying amounts of the chemically synthesized pRNA 14-mer (Figure 1A); lanes 4 and 9: 400 ng of in vitro transcribed B. subtilis 6S-1 RNA loaded onto the gels; the northern blot signal is attributable to the probe’s complementarity to seven consecutive nucleotides of 6S-1 RNA (nts 151–157; Figure 1A), including three LNA/RNA base pairs; lanes 5 and 10: 10 µg of total cellular RNA prepared from B. subtilis cells after 3 min of outgrowth from stationary phase (‘Materials and Methods’ section); the endogenous pRNA yielded a signal under native but not under denaturing conditions (lanes 5 versus 10). Likewise, pRNA signals in lanes 1–3 were more intense than those in lanes 6–8. (B) The same samples as in panel A were analyzed by MOPS-buffered PAGE and northern blotting in the absence (lanes 1–6) and presence (lanes 7–12) of 2% formaldehyde as denaturing agent. Additionally, total RNA prepared from stationary phase cells was loaded onto the gels (lanes 5 and 11); endogenous pRNA in the outgrowth RNA fraction was again only detectable under native conditions (compare lanes 6 and 12). Note that PAGE with MOPS and formaldehyde generally resulted in less efficient separation of RNAs according to size. Film exposition times were 20–45 min; the two part figures in panel A originate from the same blot and exposed film; exposition time in panel B slightly differed from that in panel A; criterion for exposition length was good visibility of the pRNA signal (lane 5 in panel A, lane 6 in panel B), thus often two or more films were exposed to the same blot for different periods of time to optimize blot performance.
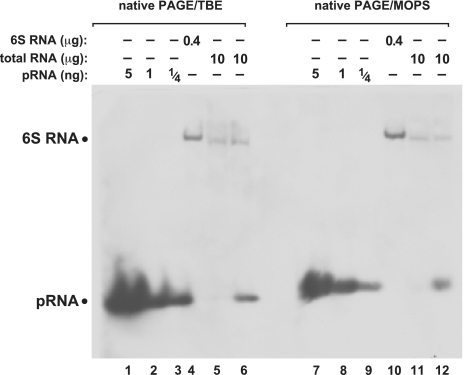
In Figure 3A, lanes 5 and 10, additional signals of RNAs larger than pRNA were observed; less additional signals were observed in the corresponding lanes 6 and 12 of Figure 3B, but here a another total RNA preparation from outgrowth bacteria was used. Thus, we attribute this difference to variations in the quality of RNA preparations. Yet, to rule out that the observed differences were related to the type of PAA buffer system, we used the same RNA preparation as in Figure 3B for native PAGE blots, either TBE or MOPS buffered (Figure 4). Here, only signals attributable to 6S-1 RNA (owing to the probe’s complementarity to seven consecutive nucleotides of 6S-1 RNA; nt 151–157 in Figure 1A) and endogenous pRNA were detected (Figure 4, lanes 6 and 12), indicating that the quality of the total RNA preparation rather than the buffer system determines the extent of non-specific signals.
Figure 4.
Comparison of native PAGE northern blots using the TBE (lanes 1–6) or MOPS (lanes 7–12) buffer systems. The two different gels were blotted simultaneously onto the same nylon membrane. For further details, see legend to Figure 3B.
Role of 5′-end groups of RNA oligonucleotides for EDC crosslinking efficiency
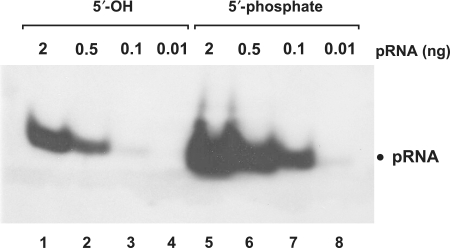
Although our chemically synthesized B. subtilis reference pRNA carried a 5′-hydroxyl group, northern blot signals were detected (e.g. Figure 4, lanes 1–3 and 7–9). This implied that EDC crosslinking to membranes had occurred despite the absence of a 5′-terminal phosphate. To further analyze this aspect, we 5′-phosphorylated this pRNA and compared northern blot signal intensity to that of the 5′-hydroxylated pRNA. Signal intensity was largely (∼10-fold) increased owing to the presence of the 5′-phosphate (Figure 5, lanes 1–3 versus 5–7). However, the presence of a 5′-phosphate is not essential, indicating that internal phosphodiesters can react as well, although with reduced efficiency.
Figure 5.
Functional groups at the 5′-terminus have a strong impact on signal strength. pRNA with a 5′-phosphate (lanes 5–8) yielded enhanced signal strength (∼10-fold) compared to the same RNA with a 5′-hydroxyl end (lanes 1–4). Gel electrophoresis: 10% native PAGE, 1× TBE; RNAs were coupled to nylon membranes by EDC crosslinking.
Verification of pRNA detection specificity
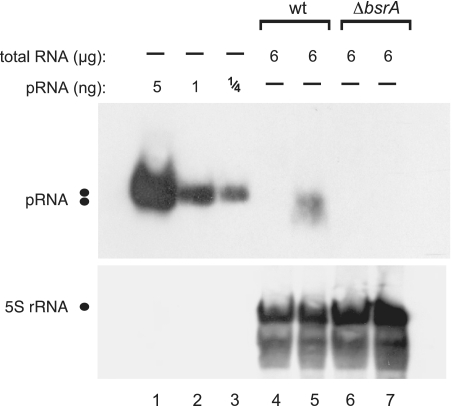
As mentioned before, we observed additional signals beyond that for pRNA in some of our initial gels (e.g. Figure 3A, lane 5). To increase confidence that we indeed detected genuine endogenous 6S-1 pRNAs, we also prepared total RNA from a B. subtilis PY79 ΔbsrA strain, in which the 6S-1 RNA gene was replaced with a spectinomycin resistance cassette (unpublished results, manuscript in preparation). In contrast to the wild-type PY79 strain, no pRNA signal was observed for total RNA derived from the PY79 ΔbsrA strain harvested under outgrowth conditions (Figure 6, lanes 5 versus 7), indicating that the signal in lane 5 indeed represents authentic 6S-1 pRNAs.
Figure 6.
Specificity of pRNA detection demonstrated by using total RNA from the B. subtilis wild-type (wt) strain PY79 (lanes 4 and 5) versus a 6S-1 RNA knockout derivative strain, PY79 ΔbsrA (lanes 6 and 7); lanes 4 and 6, total RNA from cells harvested in stationary phase; lanes 5 and 7, total RNA from cells harvested during outgrowth (for details, see ‘Materials and Methods’ section). 5S rRNA was used as an internal loading control in lanes 4–7; since we used native PAGE here, the expanded area of 5S rRNA signals may be due to different 5S rRNA conformers.
Detection of 14-mer sequence variants and influence of intramolecular RNA secondary structure
Our results raised the question if northern blot detection of a 14-mer is sensitive to sequence identity and, since we have employed native gel systems here, if potential intramolecular secondary structures might negatively affect detection sensitivity. Both questions were simultaneously addressed by designing the variant pRNA mut with two consecutive base exchanges relative to the wild-type 6S-1 pRNA; the two point mutations generated the potential of the 14-mer to form a small hairpin structure with a 3-bp stem (Figure 7, top). The two chemically synthesized pRNA variants were separated on a 20% native PAA gel (TBE buffer system), followed by northern blotting using two different 5′-digoxigenin-LNA/DNA mixmer probes, either fully complementary to wild-type pRNA or to pRNA mut. Figure 7 illustrates that, for both pRNA variants, higher detection sensitivity was obtained with the fully complementary probe, although both probes were also able to mutually recognize the corresponding mismatching pRNA variant. Detection sensitivity of the wild-type pRNA probe generally exceeded that of the pRNA mut probe, which is attributed to the different distribution of LNA modifications in the two probes (Figure 7). This illustrates the sensitivity of the method to the design of LNA probes in terms of the number and positioning of LNA residues. Of note, pRNA mut migrated somewhat faster than wild-type pRNA in the native 20% PAA gel, suggesting that its hairpin structure indeed formed during electrophoresis, at least transiently. However, the blot in Figure 7 provides no evidence for a substantial influence of hairpin formation on detection sensitivity.
Detection of small RNAs in E. coli
To test if our approach is also applicable to organisms other than B. subtilis, we isolated total RNA from the E. coli wild-type strain MG1655 and a derivative 6S RNA knockout strain (ΔssrS). Escherichia coli was chosen because the bacterium represents the major model system for studies of 6S RNA, and pRNAs of similar size (14–20 nt) have been described (9). We initially designed probe 1 (14 nt) for the detection of E. coli pRNAs, but the specific signal was weak, with a strong background of non-specific signals (Figure 8A). Since two of the five LNA modifications were in the stretch of four C residues, we were concerned that this may have led to non-specific interactions with complementary tetra-G stretches in other RNA molecules. We then designed probes 2 (14 nt) and 3 (16 nt) with only two LNA residues located outside the tetra-C stretch. This largely improved the sensitivity and specificity of the northern blot (Figure 8B and C). E. coli pRNAs were only detected under outgrowth conditions in the wild-type strain, but not in the 6S RNA knockout (ΔssrS) strain (Figure 8A–C, lanes 4, 10 and 16 versus lanes 6, 12 and 18). This is in line with the results observed for B. subtilis (Figure 6), where no pRNA signal could be detected in the 6S-1 RNA deletion strain ΔbsrA.
We further observed a hybridization signal suspected to represent pRNA complexed with E. coli 6S RNA (Figure 8B and C, arrows). To address this possibility, we ran a native 10% PAA gel, where we loaded identical sample sets onto the left and right half of the gel. After blotting to a nylone membrane, the blot was cut into the two halves with identical sample sets and hybridized either to the E. coli pRNA probe 3 or to a digoxigenin-labeled 6S RNA antisense transcript (Figure 8D and E). This experiment revealed (i) that the aforementioned band is indeed a 6S RNA:pRNA hybrid, as the signal was detected with both probes (arrows in panels D and E), (ii) that 6S RNA is present in the E. coli MG1655 wild-type, but not in ΔssrS derivative strain, also verifying that signals marked with asterisks in Figure 8A–D are not related to 6S RNA, thus representing a cross-hybridizing RNA of unknown identity. The chemically synthesized E. coli pRNA 14-mer tended to form aggregates (Figure 8D, lanes 2 and 3). We therefore added denaturing instead of native loading buffer to this RNA and heated the sample to 95°C for 5 min (Figure 8C, lanes 13 and 14). Shortening of this heating step to 2 min (Figure 8D, lanes 2 and 3) was insufficient to dissolve the RNA multimers. Noteworthy, the presence of low amounts of denaturing agents introduced into the gel via the denaturing loading buffer did not perceivably compromise EDC crosslinking.
DISCUSSION
We have developed a northern blot technique that permits to detect cellular RNAs as small as ∼14 nt in a reproducible and highly sensitive manner. This technique is novel in that it combines three elements: (i) 5′-digoxigenin-endlabeled LNA/DNA mixmer probes, (ii) EDC crosslinking of RNAs to nylon membranes, and (iii) native PAGE systems. The method was successfully applied to the detection of ‘tiny’ pRNAs from two different model systems, B. subtilis and E. coli, which leads us to conclude that the procedure will be broadly applicable. Parallel analysis of B. subtilis and E. coli 6S RNA knockout strains unequivocally confirmed specific detection of endogenous pRNAs.
Our detection of B. subtilis 6S-1 RNA with the pRNA-specific LNA/DNA mixmer probe (Figure 3A, lanes 4 and 9, Figure 3B and Figure 4), attributed to formation of seven consecutive base pairs (to 151–157 nt of 6S-1 RNA, Figure 1A), suggested that even RNAs smaller than 14 nt will be detectable by the approach. Detection of natural RNA oligonucleotides <10 nt may be interesting for the method’s application to the detection of eukaryotic small RNAs, e.g. of RNA 9-mers derived from Ago2 cleavage of siRNA passenger strands. For shorter target RNA interactions, LNA probes with an increased proportion of LNA residues (possibly combined with 2′-OCH3 instead of DNA residues), or even all-LNA variants, possibly with a single 5′-terminal non-LNA residue carrying the 5′-digoxigenin label, may preserve high detection sensitivity for target RNA oligonucleotides as short as 8-mers (16). 8- to 9-meric LNA probes have indeed been shown to form stable duplexes with their target [(16,17); for review, see (18)]. Thus, even the aforementioned RNA 9-mers derived from Ago2 cleavage of siRNA passenger strands in eukaryotic cells (19) should be detectable with our northern blot protocol, a prediction we are currently testing. Another application may be detection of short abortive transcripts (2–15 nt) synthesized by bacterial and eukaryotic RNA polymerases (20). However, successful application of such very short all-LNA probes in northern blot experiments will depend on how specific signals can be distinguished or separated from non-specific interactions. 5′-digoxigenin LNA probes may also be replaced with 5′-32P-labeled LNA probes, as efficient 5′-end labeling of LNA oligonucleotides by T4 polynucleotide kinase has been reported (21).
As expected, RNA oligonucleotides carrying a 5′-phosphate are efficient substrates for EDC crosslinking to nylon membranes. Nevertheless, RNA variants with a 5′-hydroxyl group can be crosslinked as well, although with substantially reduced efficiency (Figure 5), indicating that internal phosphodiesters are also substrates for EDC crosslinking. Thus, in the case of detection of small RNAs with 5′-OH groups derived from upstream cleavage events, a 5′-phosphorylation step before electrophoresis and membrane transfer should be included to enhance detection sensitivity.
Our results with the chemically synthesized B. subtilis pRNA and its mutant derivative (Figure 7) illustrated that intramolecular secondary structures of RNA oligonucleotides appear to have little effect on detection sensitivity. Also, one should take into account that extent and stability of intramolecular structure formation are limited for a 14-mer owing to its limited sequence space. The experiment of Figure 7 provided evidence that the number and positioning of LNA residues in LNA/DNA mixmer probes affects detection sensitivity. The detection of pRNAs in E. coli then substantiated this observation, demonstrating that the number and positioning of LNA residues profoundly affects sensitivity as well as specificity (Figure 8). Researchers are advised to restrict the number of LNA residues, for example by incorporating only 2 LNA residues at separate positions into a 14-nt long LNA/DNA mixmer. Formation of oligonucleotide dimers is another issue, but here the routinely conducted heating step before gel loading may mitigate this problem in many cases. Nonetheless, as shown for the chemically synthesized E. coli pRNA 14-mer, RNA multimerization can be a problem (Figure 8D, lanes 2 and 3). We solved this problem by adding denaturing instead of native loading buffer to the RNA gel samples and by extending the 95°C heating step to 5 min (Figure 8C, lanes 13 and 14).
Our protocol utilizes non-denaturing electrophoresis conditions. Apparently, denaturing reagents, such as formaldehyde or chaotropic compounds like urea and guanidine hydrochloride, interfere with EDC chemistry involving derivation of amino groups. Since short target RNA oligonucleotides, such as 6S RNA-derived pRNAs, can be released from complexes with complementary RNAs by a heating step, and after release do not form complex intramolecular structures that may lead to aberrant gel migration, the non-denaturing conditions used in our protocol do not entail any significant disadvantage relative to denaturing gel systems. Though we have not observed B. subtilis 6S-1 RNA:pRNA hybrids with our native PAGE-based procedure, this was clearly different for the E. coli system. Here, a fraction of endogenous 6S RNA:pRNA hybrids survived the hot phenol RNA extraction as well as the heating step (5 min at 95°C) in denaturing loading buffer (Figure 8B and C). This extraordinary stability of E. coli 6S RNA:pRNA hybrids, even under denaturing conditions, has also been observed by others (22). Detection of 6S RNA:pRNA complexes with native PAGE also illustrates a potential advantage of native versus denaturing gel systems, i.e. the increased likelihood to identify biologically relevant RNA–RNA interactions under non-denaturing conditions.
FUNDING
Deutsche Forschungsgemeinschaft (HA 1672/16-1, GK 1384). Funding for open access charge: Deutsche Forschungsgemeinschaft (GK 1384).
Conflict of interest statement. None declared.
REFERENCES
- 1.Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Várallyay E, Burgyán J, Havelda Z. Detection of microRNAs by northern blot analyses using LNA probes. Methods. 2007;43:140–145. doi: 10.1016/j.ymeth.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RHA. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 5.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pall GS, Hamilton AJ. Improved northern blot methods for enhanced detection of small RNA. Nat. Protocols. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 8.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 9.Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–1603. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- 10.Gildehaus N, Neußer T, Wurm R, Wagner R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007;35:1885–1896. doi: 10.1093/nar/gkm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willkomm DK, Hartmann RK. 6S RNA – an ancient regulator of bacterial RNA polymerase rediscovered. Biol. Chem. 2005;386:1273–1277. doi: 10.1515/BC.2005.144. [DOI] [PubMed] [Google Scholar]
- 13.Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 14.Mattatal NR, Sanderson KE. Salmonella typhimurium LT2 possesses three distinct 23S rRNA intervening sequences. J. Bacteriol. 1996;178:2272–2278. doi: 10.1128/jb.178.8.2272-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh SS, Musso GF. Covalent attachment of oligonucleotides to solid supports. Nucleic Acids Res. 1987;15:5353–5372. doi: 10.1093/nar/15.13.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elayadi AN, Braasch DA, Corey DR. Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry. 2002;41:9973–9981. doi: 10.1021/bi025907j. [DOI] [PubMed] [Google Scholar]
- 17.Mouritzen P, Noerholm M, Nielsen PS, Jacobsen N, Lomholt C, Pfundheller HM, Tolstrup N. ProbeLibrary: a new method for faster design and execution of quantitative real-time PCR. Nat. Methods. 2005;2:313–316. [Google Scholar]
- 18.Grünweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 19.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 20.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wengel J, Nielsen P. Oligonucleotide analogues. 2002 Patent US 2002/0068708 A1. [Google Scholar]
- 22.Wurm R, Neusser T, Wagner R. 6S RNA-dependent inhibition of RNA polymerase is released by RNA-dependent synthesis of small de novo products. Biol. Chem. 2010;391:187–196. doi: 10.1515/bc.2010.018. [DOI] [PubMed] [Google Scholar]