Abstract
The introduction of exogenous DNA in human somatic cells results in a frequency of random integration at least 100-fold higher than gene targeting (GT), posing a seemingly insurmountable limitation for gene therapy applications. We previously reported that, in human cells, the stable over-expression of the Saccharomyces cerevisiae Rad52 gene (yRAD52), which plays the major role in yeast homologous recombination (HR), caused an up to 37-fold increase in the frequency of GT, indicating that yRAD52 interacts with the double-strand break repair pathway(s) of human cells favoring homologous integration. In the present study, we tested the effect of the yRad52 protein by delivering it directly to the human cells. To this purpose, we fused the yRAD52 cDNA to the arginine-rich domain of the TAT protein of HIV (tat11) that is known to permeate the cell membranes. We observed that a recombinant yRad52tat11 fusion protein produced in Escherichia coli, which maintains its ability to bind single-stranded DNA (ssDNA), enters the cells and the nuclei, where it is able to increase both intrachromosomal recombination and GT up to 63- and 50-fold, respectively. Moreover, the non-homologous plasmid DNA integration decreased by 4-fold. yRAD52tat11 proteins carrying point mutations in the ssDNA binding domain caused a lower or nil increase in recombination proficiency. Thus, the yRad52tat11 could be instrumental to increase GT in human cells and a ‘protein delivery approach’ offers a new tool for developing novel strategies for genome modification and gene therapy applications.
INTRODUCTION
In mammalian cells, when a linear double-stranded DNA fragment sharing perfect homology with a target chromosomal locus is transfected into the cell, it can be integrated at the specific locus by homologous recombination (HR); this event is referred to as gene targeting (GT; 1). Two important known applications of GT in somatic cells are generation of ‘knockouts’, that determines the loss-of-function of that particular gene, and of ‘knock-ins’, wherein a mutated allele is corrected back to the wild-type sequence or a new mutation is introduced (2). A major limitation to apply GT strategy in human cells is given by the integration of exogenous DNA at random sites by non-homologous recombination (random integration: RI), an occurrence 100–1000 times more frequent than HR (3). This represents a severe drawback in the current gene therapy experimentation.
Several efforts have been made to increase in mammalian cells the frequency of GT over RI, by modifying the length of homology between vector and locus, by inducing a DNA double-strand break at the target locus, or by overexpressing recombination proteins (4–6). Indirect approaches have also been envisaged to enhance GT by inhibiting the expression of proteins involved in non-homologous recombination using RNA interference (7–9).
In the yeast, Saccharomyces cerevisiae, HR events require both RAD51 and RAD52 proteins (10,11). In fact, Rad51 and Rad52 deletion mutants show 4000 and 3000-fold, respectively, reductions in gene conversion of inverted repeats as compared to the wild type (12,13). Although in human cells GT is thought to occur by a strand invasion mechanism that completely depends on the human Rad51 orthologue (14), the hsRad51 overexpression stimulated GT only by 2- to 3-fold, whereas it actually inhibited intrachromosomal recombination, suggesting that in humans Rad51 gives a relatively modest contribution to Rad52 activity in promoting HR via strand annealing and exchange. No appreciable effect on both mitotic and meiotic HR and on GT is observed following hsRad51 overexpression. Conversely, overexpression of hsRad52 in monkey cells enhances spontaneous HR and resistance to ionizing radiation (15). Moreover, the overexpression of hsRad52 in human HT1080 cells inhibits GT while favoring extrachromosomal HR (14).
Rad52 is a multi-domain protein composed of three regions with separate molecular functions. The N-terminal domain is involved in DNA binding and the C-terminal one in the interaction with the Rad51 protein, whereas an RPA binding domain is found in between (9,16,17). The N-terminal domain of yRad52 from amino acid 21 to 159 is conserved from yeast to higher eukaryotes (18). Genetic evidence in yeast indicates that two point mutations at the N-terminus of yRad52 (Y66A and R70A) cause a pronounced increase in the sensitivity to ionizing radiations without significantly affecting spontaneous recombination. A recent report showed that the R70A mutation actually impairs the single-stranded DNA (ssDNA) binding activity of Rad52 affecting the final step of gene conversion (19). Moreover, it was recently reported that the Y66A mutant has a slow DNA recombinational repair process (20).
The human orthologue of Rad52 is smaller than the yeast molecule: 418 amino acids versus 471. At the N-terminus, the human molecule lacks the first 33 amino acids of the yeast counterpart; amino acids 34–198 of yRad52 share 46% homology with the N-terminus of the human protein, whereas the C-termini of the two molecules share little or nil homology. Human and yeast Rad52 proteins display similar physico-chemical properties, such as the spontaneous organization in homo-oligomeric ring structures, affinity for double- and (preferentially) ssDNA and ability to facilitate DNA re-annealing (21).
The significant differences in the structures of the yeast and human Rad52 orthologue are accompanied by a greater proneness of the yeast molecule to favor HR (22,23). This property could therefore be exploited to overcome the inherent limitations of hsRad52 overexpression strategies in gene therapy approaches. Actually, we recently demonstrated that the expression of yRad52 in human cells induces an up to 37-fold increase of the frequency of GT and an up to 4-fold reduction of RI of exogenous DNA. We proposed that yRad52 has a greater affinity for ssDNA than hsRad52 and is consequently more efficient in promoting strand exchange (24). This observation suggested that the yeast molecule could be used as a tool to develop new strategies for gene therapy of single gene disorders. To this purpose, we investigated the possibility of directly introducing the molecule into the nucleus without the mediation of transient or stable transfection procedures.
In this perspective, highly relevant are the recent observations of the successful employment of membrane-penetrating proteins or cell-penetrating peptides (CPPs), containing protein transduction domains, for direct transfer inside the cells of biologically active proteins, peptides and drugs (25–27). These tools are considered very promising for both basic research and development of gene delivery techniques for cancer therapeutics and vaccination (28). Recently, peptides like the cyclic 13-mer Pep42, that specifically binds the glucose-regulated protein 78 (GRP78) and is internalized into cancer cells, have been reported to be excellent vehicles for tumor cell-specific chemotherapy (29). Yet, the most used CPPs in therapeutic approaches consist of short peptides, usually less than 30 residues, corresponding to the membrane-permeating domain of the human immunodeficiency virus (HIV) Tat protein, or to VP22, penetratin, polyarginine and transportan (30). The Tat protein, whose fundamental role is the control of HIV-1 gene expression, possesses the unique property of entering the cells and translocating to the nucleus when present in the extracellular environment (25,31–33). Previous reports showed that the addition of an 11-amino acid stretch from the basic domain of Tat (amino acids 48–58, named tat11) to heterologous molecules, even of a large size, mediates their cellular uptake and can deliver in the nucleus large molecules or particles, including liposomes, plasmid DNAs, phage vectors and nanoparticles (27,34,35). This property is currently widely exploited as a biotechnological tool for protein transduction, including therapeutic applications like protection from apoptosis in brain and in heart, extension of cytotoxic activity of herpes simplex virus-1 thymidine kinase in cancer gene therapy, and enhancement of viral-mediated gene therapy (36–39).
We explored therefore the possibility that the addition of tat11 to the yRAD52 protein could allow its permeation into the nucleus where it could display its enhancing effect in HR events, including GT.
MATERIALS AND METHODS
Plasmids
The plasmid pQE-60-Rad52 (a gift from Rodney Rothstein) containing the Saccharomyces cerevisiae Rad52-His6 DNA sequence has been modified by inserting, at the C-terminus of the yRAD52, the sequence coding the arginine-rich (48–58 aa) domain from HIV-TAT as follows: two complementary ssDNA oligonucleotides carrying the DNA sequence corresponding to the TAT domain were designed with one end sticky to the NcoI restriction site and the other end to the EcoRI site. The two oligonucleotides were annealed and the resulting double strand was inserted at the NcoI–EcoRI sites to produce the pQE60-Rad52-Tat11. The correct frame of TAT domain insertion in RAD52 was confirmed by direct sequence analysis. The site specific point mutations Y66A and R70A were created by using the Quick change II Site-Directed mutagenesis kit (from Stratagene) following the recommended protocol. The sequences of all oligonucleotides are available upon request. The GT plasmid pHyg− carrying the hygromycin resistance gene mutated at the SacII site (named hyg2) was constructed previously by cloning the 2.2 kb BamHI fragment from pTPSN into BamHI site of Litmus 28 (New England Biolab; 24). The plasmid pSRα-bsr-pA, kindly provided by Monica Zoppè, was constructed by cloning the blasticidin resistance gene into pBluescript SK+ (Stratagene).
Purification of yRad52Tat11
Plasmid pQE-Rad52-His6, pQE-Rad52-Tat11-His6 and plasmids coding the mutated Rad52 protein (Y66A and R70A) were introduced into Escherichia coli BL-21 competent cells. Bacterial cultures were grown in Luria broth until OD600= 0.4 and then treated with 1 mM isopropyl β-d thiogalactoside for 4 h at 37°C. All the purification steps were performed at 4°C. The cells were lysed on ice by sonication (3 cycles/3 s) in 10 ml equilibration buffer [20 mM NaH2PO4 pH 8, 400 mM NaCl, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] along with lysozyme 10 mg/ml (Boehringer Mannheim, GmbH) and 5% Triton. After cell lysis, the supernatant was collected by centrifugation. The yRad52tat11 protein was purified by loading the supernatant onto Ni-NTA agarose resin column that was equilibrated with the equilibration buffer prior to use. Later, the column was washed for 4–6 times with 10 ml wash buffer (20 mM NaH2PO4 pH 8, NaCl 400 mM, 20 mM imidazole pH 7.8, 0.1 mM PMSF) and eluted with elution buffer (20 mM NaH2PO4 pH 8, 400 mM NaCl, 150 mM imidazole pH 7.8, 5 mM MgCl2, 10% glycerol, 0.1 mM PMSF). The protein was eluted three times from the column with 800 µl of elution buffer and stored at −80°C. The purity and the concentration of the yRad52 proteins was checked by polyacrylamide gel electrophoresis, loading multiple dilutions of the purified protein and comparing the band intensity to known amounts of bovine serum albumin. The final yield of the protein was quantified using the Bradford method (from BioRad).
DNA mobility shift assay
For this assay, we used the circular ssDNA from ΦX174 virion purchased from New England Biolabs. The mobility shift assay was carried out according to the protocol previously reported (40). The reactions for DNA mobility shift assay contained ssDNA (30 µM nucleotides) co-incubated with different amounts of wt-yRad52, mut-yRad52 or eGFPTat11 (kindly provided by Mauro Giacca) in 10 µl of reaction buffer (35 mM K-MOPS, pH 7.2, 1 mM DTT, 100 µg/ml BSA, 2.5 mM ATP and 3 mM Mgcl2). The reaction mixtures were incubated at 25°C for 10 min and then mixed with loading dye (0.1% Orange G in 30 mM Tris–HCl, pH 7.5, containing 50% glycerol). The mixture was loaded onto 0.9% agarose gel and separated by electrophoresis at 100 mA in TAE buffer at 25°C until the dye front has migrated 4 cm. DNA was stained with 1% ethidium bromide.
Protein delivery and immunoblotting
The yRad52tat11 was added directly to the cell culture medium using a MultiwellTM 24-well plate (Becton Dickinson, NJ, USA). Increasing concentrations of purified protein ranging from 8 to 30 μg/ml were added directly with fresh Dulbecco medium (Eurofin) containing 10% fetal calf serum along with 1 μg/ml of chloroquine diphosphate and incubated from 6 to 48 h (31). The preparations of total and nuclear extracts were carried out as reported previously (41,42). All the steps were carried out at 4°C unless otherwise mentioned. Total cell extracts were prepared by using lysis buffer containing PMSF. The nuclear extracts were prepared by lysing the cells in hypotonic buffer (20 mM HEPES–KOH pH 8, 5 mM KCl, 1.5 mM MgCl2, protease inhibitors cocktail tablet pH 7.9, 5 mM Na butyrate, 0.1 mM DTT). The lysate was then homogenized in a glass Dounce homogenizer and centrifuged at 600g for 15 min at 4°C. The nuclei were washed three times with nuclear extraction buffer (15 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.4 M NaCl, 10% sucrose, protease inhibitors cocktail pH 7.9, 1 mM DTT). The supernatant soluble fraction was collected separately and the pellet insoluble fraction was lysed with buffer containing PMSF. The concentrations of total cellular and nuclear extracts for both soluble and insoluble fraction were determined by BioRad Protein assay (Biorad Laboratories, GmbH). The cell and nuclear extracts were analyzed by SDS–PAGE electrophoresis. Purified wt-yRad52tat11 protein was used as a control. The membrane hybridization was carried out with mouse monoclonal anti-penta-histidine antibody (Qiagen). The secondary antibody used was goat secondary antimouse IgH-HRP antibody (Santa Cruz Biotechnology). The reference antibody for total cell extracts used was α-tubulin mouse monoclonal antibody and goat secondary anti-mouse IgH-HRP antibody (Santa Cruz Biotechnology).
Intrachromosomal recombination and GT assay
To determine the effect of yRAD52 in the intrachromosomal recombination, we used the HeLaG1 cells (from Margherita Bignami) that allow to measure gene conversion events between two differentially mutated hygromycin-resistance (HygR) genes (43). One HygR gene is mutated at the PvuI site (hyg1), the other HygR at the SacII site (hyg2, Figure 1A). An intrachromosomal recombination event leads the restoration of HygR to the wild type; therefore the frequency of intrachromosomal recombination was calculated as total number of HygR clones per 105 viable cells. Twenty four or 48 h after the delivery of the yRad52 protein by directly adding the protein aliquot to the DMEM medium, the cells were trypsinized, counted and seeded in triplicate in 10-cm dishes at cell density of 1 × 105/plate for recombination and in duplicate in p60 mm dishes at cell density of 1 × 102/plate for cell survival. Hygromycin at the concentration of 300 μg/ml was added 24 h later. Selective medium was changed after a week. After 15 days, HygR clones and survivors were stained with crystal violet and counted.
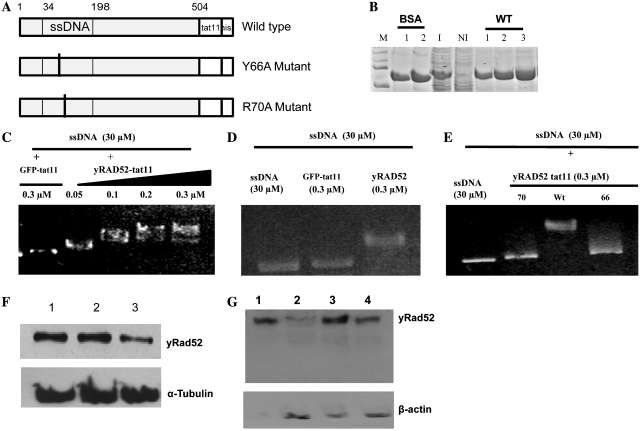
Figure 1.
Purification, ssDNA binding activity and delivery of yRad52tat11. (A) Maps of the yRad52tat11 wild-type fusion protein, the Y66A and the R70A mutant, that for simplicity only show the N-terminal DNA binding domain; the numbers represent amino acid residues in the wild-type yeast molecule; the ssDNA binding domain is shown. Both mutations are located in this domain; the tat11 and the his-tag are located at the C-terminus. (B) Coomassie staining of purified wild-type-yRad52-tat11 protein (about 52 kDa); M-lane, markers; BSA-lanes 1 and 2, 200 and 400 ng of bovine serum albumin; I-lane, 5 µg of total protein extract from IPTG-induced bacteria; NI-lane, 5 µg of total protein extract from not induced bacteria; WT-lanes 1, 2 and 3, wild-type yRad52tat11 purified protein elutions; the yield was 80, 70 and 50 ng/μl for three different elutions. (C) In vitro mobility shift assay with ssDNA (30 µM as nucleotides) and increasing concentrations (0.05, 0.1, 0.2, 0.3 µM) of wt yRad52tat11 protein or GFPtat11 (0.3 µM) as control; the reaction mixtures were loaded in a 0.9% agarose gel, electropheresed and stained with ethidium bromide. (D) In vitro mobility shift assay with ssDNA (30 µM as nucleotides) alone, in presence of 0.3 µM concentration of wild-type yRad52 or GFPtat11. (E) In vitro mobility shift assay with ssDNA (30 µM as nucleotides) alone or in presence of 0.3 µM concentration of wild-type yRad52tat11, R70A or Y66A mutated protein, as indicated. (F) Western blot analysis of total cell extracts; 6 × 105 HeLaG1 cells were incubated with 20 µg/ml wild-type-yRad52tat11 or mutant proteins for 24 h; cell lysis and total cell extracts were carried out as reported in ‘Materials and Methods’ section; 10 µg of total proteins were loaded in each well; the blot was hybridized with anti-penta-6-histidine antibody; lane 1, extract from wild-type yRad52tat11-exposed cells; lane 2, extract from R70A treated cells; lane 3, extract from Y66A treated cells; tubulin was used as loading control. (G) Western blot analysis of nuclear extracts from 3 × 106 total cells treated with 20 µg/ml of wild type or mutated yRad52tat11 protein for 24 h; aliquots corresponding to 10 µg of nuclear protein extract were loaded and electrophoresed; lane 1, 1 µg of purified yRad52tat11protein; lane 2, nuclear extract from Y66A mutant-treated cells; lane3, nuclear extract from wild-type yRad52tat11-treated cells; lane 4, nuclear extract from R70A mutant-treated cells.
The GT experiments were carried out using the in HeLa1B cell line according to the previously reported procedure (24). GT events occur between a HygR mutated gene (hyg1) integrated into the genomic DNA as single copy, and another HygR copy (hyg2) transfected to the cells by electroporation. This event leads to the reversion of the HygR to wild type. The frequency of GT was measured as number of HygR per 105 viable cells. In detail, 3–5 × 106 cells were exposed to 16–30 μg of yRad52 for 6 h. Thereafter, cells were transfected with 10 µg of hyg2 fragment by electroporation (24). After electroporation, cells were seeded in 10-cm dishes at cell density of 1 × 105/plate for recombination and in p60 mm dish at cell density of 1 × 102/plate for cell survival. Hygromycin (300 μg/ml) was added after 24 h. Selective medium was changed after a week. After 15 days, HygR clones and survivors were stained with crystal violet and counted.
To assess whether the GT occurred, the genomic DNA was extracted from six independent HygR clones as previously reported and PCR analysis was performed to amplify the 359-bp fragment of the HygR gene. A total of 10 μl aliquots of PCR products were digested with PvuI as reported (24).
Random integration
The effect of yRAD52 on plasmid integration was determined in conditions allowing only non-homologous recombination (RI). The 3–5 × 106 HeLa1B cells were transfected with 4 µg of pSRα-bsr-pA by electroporation as described previously (24). The cells were seeded in 10-cm dishes at cell density of 1 × 105 cells/plate with 0.2 µg/ml of blasticidin, and at a density of 1 × 102 in p60 mm dish for survival. The culture medium was changed once a week and after 15 days clones were stained and counted. The frequency of random integration was calculated by dividing the number of blasticidin-resistant clones by the number of viable cells.
RESULTS AND DISCUSSION
Production of the yRad52Tat11 protein and mutant derivatives
The tat11 peptide was cloned, with the procedure described under ‘Materials and Methods’ section, at the C-terminus of the yRAD52 gene, to avoid any possible interference with the ssDNA binding capacity that resides in the N-terminus domain. We produced also two mutants in the ssDNA binding domain (Y66A and R70A), corresponding to ionizing radiation-sensitive phenotypes, with a view to assess the importance of the affinity of the molecule for ssDNA, an essential property in the context of strand exchange. Figure 1A shows a map of the constructs utilized in this work. yRad52Tat11 and the two mutant proteins were purified by Ni-NTA affinity column purification system and eluted with the buffer containing 20 mM imidazole and 400 mM NaCl, that was proven to have little cytotoxic effect on HeLaG1 cells (around 60% survival). SDS–PAGE analysis of the different elutions is reported in Figure 1B.
Affinity for ssDNA of the recombinant proteins
In order to determine whether the addition of tat11 and the introduction of the point mutations affect the affinity for ssDNA, we measured by a gel mobility shift assay the ability of yRad52Tat11 and of the two single amino acid substitution mutants (Y66A and R70A) to bind ssDNA in vitro. In the first place, we incubated different concentrations of wild-type yRad52tat11 (the GFPtat11 protein at 0.3 µM was used as control). The highest electrophoretic motility shift was seen when the ssDNA was mixed with 0.3 µM yRad52tat11 (Figure 1C), in agreement with the effect observed with wt yRad52 at the same concentration (Figure 1D). Accordingly, we determined the ssDNA binding activity of the mutant yRad52tat11 proteins by adding 0.3 µM of protein that is the concentration giving the maximum gel retardation with yRad52. The results (Figure 1C) show that the tat11 fusion domain does not affect the ssDNA binding activity, whereas the two mutants had a much lower affinity for ssDNA (Figure 1E). Thus, the tat11 construct with the otherwise wt sequence could be exploited to determine its effect on GT if introduced into the nucleus.
Permeation of the yRad52Tat11 fusion protein into HeLa cells and nuclei
To determine whether the tat11 peptide is actually adequate to deliver a functional yRad52 inside the cell, we measured the intracellular level of the yRad52tat11 and of the two mutant derivatives after incubation of the HeLaG1 cells with the proteins at concentrations ranging between 12 and 24 µg/ml for 24 h. We prepared total cell and nuclear extracts and carried out western blots. The three yRad52 proteins (wild type, Y66A, R70A) were found at similar level in the total cell extract (Figure 1E). On the other hand, the mutant protein Y66A accumulated in the nucleus at lower level than the wt and the R70A mutant (Figure 1F). This indicates that the mutant protein Y66A is probably less stable in intracellular conditions.
The data demonstrate that the tat11 domain fused to Rad52 proteins is indeed able to ‘deliver’ the protein inside the cell by crossing the membrane and reaching the nucleus, even if the mutants do so to a lesser extent.
Enhancement of intrachromosomal recombination
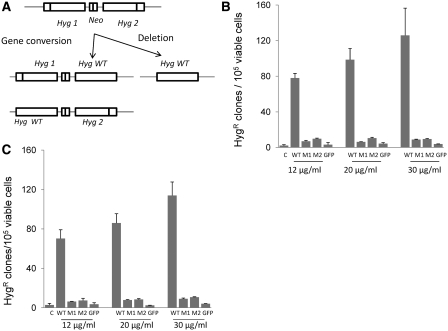
In order to test whether the yRad52tat11 constructs that have maintained to greater or lesser extent the ability to bind ssDNA, when exogenously introduced had the same effect on the recombination processes as the intracellularly produced protein, we first measured the efficiency of intrachromosomal recombination. To this purpose, we used the HeLaG1 cell line which harbors two defective hygR genes oriented as direct repeats separated by a G418 resistance gene (43). A recombination event can occur by gene conversion or intrachromosomal deletion leading to the restoration of HygR gene to wild type (Figure 2A). Hence, the frequency of intrachromosomal recombination is measured as the frequency of HygR clones obtained. If the clones arise from gene conversion they must keep the NeoR gene and be resistant to G418, whereas the HygR reversion by intrachromosomal deletion would determine the loss of the NeoR gene and clones would be G418 sensitive. Wt-yRad52tat11, the mutant Y66A, or the mutant R70A protein were added to cell culture medium at concentrations ranging between 12 and 30 μg/ml and incubated at 37°C for 24 or 48 h. The GFPtat11 was used as negative control. We did not observe a significant cytotoxic effect either after 24- or 48-h incubation with the protein. The survival rate calculated by plating 100 cells and counting the clones after 2 weeks, ranged between 70% and 50% (data not shown). After 24-h incubation, the wt-yRad52tat11 caused a strong increase in intrachromosomal recombination (Figure 2B). The effect was measured as the fold increase over the recombination observed with the negative control. At 12 µg/ml, yRad52 increased recombination 37-fold, while the mutants stimulated recombination only 3- to 4-fold. At the concentration of 20 and 30 μg/ml, the wt yRad52tat11 enhanced recombination by 47- and 60-fold, respectively; at the same concentration the mutant protein did not further increase recombination. After 48-h incubation (Figure 2C), at the same concentration range (12–30 µg/ml), the wt-yRad52tat11 protein enhanced recombination by 24- to 39-fold, while the mutant proteins had only a 2- to 4-fold effect.
Figure 2.
Effect of yRad52tat11 on intrachromosomal recombination. (A) HeLaG1 cells contain two copies of HygR genes inactivated by 10-bp insertions, either at a unique PvuI site (hyg1) or at a unique SacII site (hyg2); the two mutated hyg genes are in direct repeat orientation and are separated by a sequence containing the amino-glycoside phosphotransferase (Neo) gene conferring resistance to G418; an intrachromosomal recombination event occurring by gene conversion between the two hyg sequences results in restoration of one of the mutant hyg genes to wild type; the intrachromosomal deletion of the DNA sequence between the two mutated hyg genes leads to the formation of a HygR wild type (Hyg WT) with loss of intervening sequence; the intrachromosomal recombination was measured after incubating HeLaG1 cells with 12, 20 or 30 µg/ml yRad52tat11 proteins for 24 h (B) or 48 h (C). The frequency of intrachromosomal recombination was determined as total HygR clones per 105 viable cells; the data are reported as mean of three to five independent experiments ± standard deviation; GFPtat11 (12, 20 and 30 µg/ml) was used as negative control (C in B and C); WT, wild-type protein; M1 and M2, Y66A and R70A mutant proteins, respectively.
The enhanced yield of hygromycin-resistant colonies, as mentioned above, could in principle be due to either gene conversion or to intrachromosomal deletion. Analysis of the presence of the NeoR gene showed that nine HygR clones out of ten were G418 resistant, indicating that the 90% of the recombination events are due to gene conversion (43).
Thus, these results demonstrate that yRad52tat11 enhances intrachromosomal recombination by up to 60-fold without affecting cell viability. These results are to be compared with the 37-fold increase in HR previously obtained by overexpression of yRad52 in human cells (24) and with the actual inhibition of intrachromosomal recombination caused by overexpression of human Rad52 in the same cells (14). This agrees with the observed higher efficiency of yRad52 versus hsRad52 in the recombination process.
We can thus conclude that the exogenously introduced yRad52 can indeed significantly enhance intrachromosomal recombination rate. This effect, not surprisingly, requires the maintenance of the affinity for ssDNA, as shown by the severe reduction of recombination enhancement capacity demonstrated by the mutants impaired in ability to bind ssDNA, an ability that is an obvious prerequisite for exerting the probable strand invasion process operated by this molecule. Actually, the property of yRad52 to bind ssDNA is essential for assuring its capacity to stimulate the re-annealing of complementary molecules, an obligatory step for HR.
Enhancement of GT by intranuclear permeation of yRad52 protein
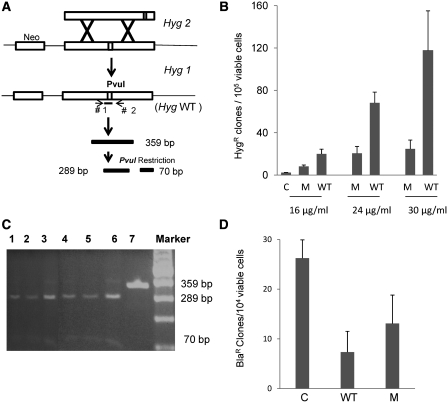
To test whether the demonstrated stimulatory effect of yRad52 on intrachromosmal HR extend also to GT, considering the extensive overlap in the molecules involved in the two processes, we used the HeLa1B cell line, which bears a single mutated copy of hygR gene (hyg1) integrated into the genome (24). The HeLa1B cells were incubated for 6 hours at 37°C in suspension cultures with different concentrations of wt-yRad52tat11 or mutant Y66A protein, chosen since this mutation caused the most pronounced reduction in intrachromosomal recombination (Figure 2B and C). The cells were then transfected by electroporation with the targeting hyg2 fragment. As shown in Figure 3A, a GT event between hyg1 and hyg2 leads to a functional HygR gene and the restoration of a diagnostic PvuI site in the HygR gene. The frequency of GT is measured by the number of HygR clones per 105 viable cells. The wild type yRad52tat11 strongly enhanced GT frequency in HeLa cells (Figure 3B). At 16 μg/ml, the wild-type protein increased GT almost 9-fold over the control; at the highest concentration (30 μg/ml), the enhancement was over 50-fold (Figure 3B). Also in this case, the mutant protein with impaired ssDNA binding ability increased GT at the highest concentration, only by 10-fold (Figure 3B).
Figure 3.
Effect of yRad52tat11 protein on GT and RI. (A) Measure of GT events between the single chromosomal HygR gene utilized in the experiments of Figure 2 (hyg1), and the BamHI fragment transfected by electroporation to the cells, corresponding to mutant hyg2 in the same figure; a GT event leads to the formation of wild-type HygR gene (Hyg WT) with the restoration of the restriction site PvuI. (B) GT experiment in HeLa1B cells carried out transfecting by electroporation 10 μg of BamHI hyg2 DNA fragment purified from agarose gel; cells were pre-incubated for 6 h with 16, 24 or 30 μg/ml yRad52tat11 or Y66A protein (M); GT frequency is expressed as total HygR clones per 105 viable cells and the results are the mean of at least three independent experiments ± standard deviation. (C) The 359 bp Hyg fragment was amplified by PCR from the genomic DNA of HygR clones and digested with PvuI; the digestion gave a 289- and a 70-bp fragment; lanes 1–6, DNA from HygR clones digested with PvuI; lane 7, not restricted DNA. (D) Effect of wild-type yRad52tat11 or Y66A protein (M) on random integration; the frequency of random integration was determined in HeLa1B cells by electroporating 8 µg of plasmid DNA and measured as number of total BlaR clones per 104 viable cells; the results are the mean of four independent experiments ± standard deviation.
In order to check if the HygR clones were produced by an accurate GT event, we isolated the genomic DNA from six independent HygR clones, amplified the hygR fragment and restricted with PvuI as previously reported (24). The results showed that all clones were due to GT because all the 359-bp fragments were digested by PvuI giving the expected 289- and 70 bp band (Figure 3C).
Thus, we can conclude that, whereas permanent expression of yRad52 in human cells caused an ∼37-fold enhancement of GT, the exogenously introduced molecule caused an up to 50-fold enhancement of the process.
Effect of intranuclear permeation of yRad52 on RI
Considering that in the case of endogenous expression the yRad52tat11 actually decreases the frequency of RI events by a factor of three, we investigated whether when introduced into the cell, it affects exogenous DNA integration by non homologous recombination. To this purpose, we electroporated in HeLa cells a plasmid DNA bearing the gene for blasticidin resistance, which shares no homology with the chromosomal DNA, in presence of yRad52tat11 (30 μg/ml). This treatment caused a 4.4 fold reduction in random integration frequency, whereas the mutant protein caused a 2.2-fold reduction (Figure 3D). This effect can be interpreted in the framework of a competition between the pathways for HR and non-homologous end-joining: the yRad52 appears to displace the equilibrium towards HR, while also the mutated proteins displace the equilibrium in the same direction, but to a much lower extent. Also these data indicate that the relatively higher efficiency of yRad52, versus its human counterpart, in channeling exogenous DNA towards GT, is maintained in the exogenously added protein.
CONCLUDING REMARKS
The results here described show that a strategy based on the intranuclear permeation of proteins affecting DNA transaction is potentially adequate to develop conditions greatly, if not exclusively, favoring GT. Without such intervention, the integration by non-homologous recombination is at least 100-fold more frequent than HR. Here, we exploited tat11 promoted permeation and the properties of the yRad52 proteins, in the hypothesis that this molecule, in human cells, is efficiently involved in the search for homology and promotes strand invasion thereby suppressing RI events. With this strategy we obtained an enhancement of GT by 50-fold and a parallel reduction of RI by 4.4-fold. In absolute terms, therefore, the GT frequency was twice higher than RI, to be compared to a normal situation of less than one GT event per 100 RI ones. We propose therefore the yRad52tat11 as a novel tool to increase GT in human cells, to be exploited in both basic and applied research. It is conceivable that the utilization of this strategy, possibly exploiting also other proteins involved in DNA transactions, may bring to the development of effective methodologies for correcting in vitro (or ex vivo) single mutations involved in monogenic disorders.
FUNDING
Associazione Italiana per la Ricerca sul Cancro; Istituto Toscano Tumori (to A.F.). Funding for open access charge: Istituto di Fisiologia Clinica CNR, Pisa Italy.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Rodney Rothstein, Uffe Mortensen, Patrick Sung, Monica Zoppè and Mauro Giacca for plasmids and eGFPTat11 protein. We are grateful to Margherita Bignami for the HeLaG1 cell line.
REFERENCES
- 1.Smith K. Theoretical mechanisms in targeted and random integration of transgene DNA. Reprod. Nutr. Dev. 2001;41:465–485. doi: 10.1051/rnd:2001102. [DOI] [PubMed] [Google Scholar]
- 2.Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jasin M, Berg P. Homologous integration in mammalian cells without target gene selection. Genes Dev. 1988;2:1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- 4.Thomas KR, Deng C, Capecchi MR. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol. Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vispe S, Cazaux C, Lesca C, Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia SJ, Shammas MA, Shmookler Reis RJ. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol. Cell Biol. 1997;17:7151–7158. doi: 10.1128/mcb.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 8.Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl Acad. Sci. USA. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Secretan MB, Scuric Z, Oshima J, Bishop AJ, Howlett NG, Yau D, Schiestl RH. Effect of Ku86 and DNA-PKcs deficiency on non-homologous end-joining and homologous recombination using a transient transfection assay. Mutat. Res. 2004;554:351–364. doi: 10.1016/j.mrfmmm.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr. Genet. 1995;27:298–305. doi: 10.1007/BF00352096. [DOI] [PubMed] [Google Scholar]
- 11.Galli A, Cervelli T, Schiestl RH. Characterization of the hyperrecombination phenotype of the pol3-t mutation of Saccharomyces cerevisiae. Genetics. 2003;164:65–79. doi: 10.1093/genetics/164.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne GT, Weaver DT. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- 13.Bai Y, Davis AP, Symington LS. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanez RJ, Porter AC. Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic Acids Res. 2002;30:740–748. doi: 10.1093/nar/30.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park MS. Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem. 1995;270:15467–15470. doi: 10.1074/jbc.270.26.15467. [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen UH, Erdeniz N, Feng Q, Rothstein R. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics. 2002;161:549–562. doi: 10.1093/genetics/161.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi I, Hallwyl SC, Seong C, Mortensen U, Rothstein R, Sung P. Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem. 2009;284:33275–33284. doi: 10.1074/jbc.M109.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Mayolo AA, Sunjevaric I, Reid R, Mortensen UH, Rothstein R, Lisby M. The rad52-Y66A allele alters the choice of donor template during spontaneous chromosomal recombination. DNA Repair. 2010;9:23–32. doi: 10.1016/j.dnarep.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dyck E, Stasiak AZ, Stasiak A, West SC. Visualization of recombination intermediates produced by RAD52-mediated single-strand annealing. EMBO Rep. 2001;2:905–909. doi: 10.1093/embo-reports/kve201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohl TJ, Nickoloff JA. Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol. Cell Biol. 2008;28:897–906. doi: 10.1128/MCB.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haber JE, Hearn M. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Primio C, Galli A, Cervelli T, Zoppe M, Rainaldi G. Potentiation of gene targeting in human cells by expression of Saccharomyces cerevisiae Rad52. Nucleic Acids Res. 2005;33:4639–4648. doi: 10.1093/nar/gki778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 26.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 27.Sonnemann KJ, Heun-Johnson H, Turner AJ, Baltgalvis KA, Lowe DA, Ervasti JM. Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med. 2009;6:e1000083. doi: 10.1371/journal.pmed.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, Fujita S, Hayakawa T, Takeda K, Hasegawa M, et al. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J. Biol. Chem. 2001;276:26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- 29.Yoneda Y, Steiniger SC, Capkova K, Mee JM, Liu Y, Kaufmann GF, Janda KD. A cell-penetrating peptidic GRP78 ligand for tumor cell-specific prodrug therapy. Bioorg. Med. Chem. Lett. 2008;18:1632–1636. doi: 10.1016/j.bmcl.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris MC, Deshayes S, Heitz F, Divita G. Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol. Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- 31.Fittipaldi A, Ferrari A, Zoppe M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 32.Cardarelli F, Serresi M, Bizzarri R, Giacca M, Beltram F. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol. Ther. 2007;15:1313–1322. doi: 10.1038/sj.mt.6300172. [DOI] [PubMed] [Google Scholar]
- 33.Gustafsson AB, Sayen MR, Williams SD, Crow MT, Gottlieb RA. TAT protein transduction into isolated perfused hearts: TAT-apoptosis repressor with caspase recruitment domain is cardioprotective. Circulation. 2002;106:735–739. doi: 10.1161/01.cir.0000023943.50821.f7. [DOI] [PubMed] [Google Scholar]
- 34.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 35.Brooks H, Lebleu B, Vives E. Tat peptide-mediated cellular delivery: back to basics. Adv. Drug Deliv. Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Cao G, Pei W, Ge H, Liang Q, Luo Y, Sharp FR, Lu A, Ran R, Graham SH, Chen J. In vivo delivery of a Bcl-xL fusion protein containing the TAT protein transduction domain protects against ischemic brain injury and neuronal apoptosis. J. Neurosci. 2002;22:5423–5431. doi: 10.1523/JNEUROSCI.22-13-05423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tasciotti E, Zoppe M, Giacca M. Transcellular transfer of active HSV-1 thymidine kinase mediated by an 11-amino-acid peptide from HIV-1 Tat. Cancer Gene Ther. 2003;10:64–74. doi: 10.1038/sj.cgt.7700526. [DOI] [PubMed] [Google Scholar]
- 38.Sandgren S, Cheng F, Belting M. Nuclear targeting of macromolecular polyanions by an HIV-Tat derived peptide Role for cell-surface proteoglycans. J. Biol. Chem. 2002;277:38877–38883. doi: 10.1074/jbc.M205395200. [DOI] [PubMed] [Google Scholar]
- 39.Kabouridis PS, Hasan M, Newson J, Gilroy DW, Lawrence T. Inhibition of NF-kappa B activity by a membrane-transducing mutant of I kappa B alpha. J. Immunol. 2002;169:2587–2593. doi: 10.4049/jimmunol.169.5.2587. [DOI] [PubMed] [Google Scholar]
- 40.Song B, Sung P. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- 41.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haglund RE, Rothblum LI. Isolation, fractionation and reconstitution of a nuclear extract capable of transcribing ribosomal DNA. Mol. Cell Biochem. 1987;73:11–20. doi: 10.1007/BF00229371. [DOI] [PubMed] [Google Scholar]
- 43.Ciotta C, Ceccotti S, Aquilina G, Humbert O, Palombo F, Jiricny J, Bignami M. Increased somatic recombination in methylation tolerant human cells with defective DNA mismatch repair. J. Mol. Biol. 1998;276:705–719. doi: 10.1006/jmbi.1997.1559. [DOI] [PubMed] [Google Scholar]