Abstract
The use of next-generation sequencing technologies to produce genomic copy number data has recently been described. Most approaches, however, reply on optimal starting DNA, and are therefore unsuitable for the analysis of formalin-fixed paraffin-embedded (FFPE) samples, which largely precludes the analysis of many tumour series. We have sought to challenge the limits of this technique with regards to quality and quantity of starting material and the depth of sequencing required. We confirm that the technique can be used to interrogate DNA from cell lines, fresh frozen material and FFPE samples to assess copy number variation. We show that as little as 5 ng of DNA is needed to generate a copy number karyogram, and follow this up with data from a series of FFPE biopsies and surgical samples. We have used various levels of sample multiplexing to demonstrate the adjustable resolution of the methodology, depending on the number of samples and available resources. We also demonstrate reproducibility by use of replicate samples and comparison with microarray-based comparative genomic hybridization (aCGH) and digital PCR. This technique can be valuable in both the analysis of routine diagnostic samples and in examining large repositories of fixed archival material.
INTRODUCTION
Systems medicine is expected to link complex molecular data underlying disease phenotypes to patient outcomes (1). As technology for data generation, such as next-generation (NG) sequencing, and methodology for computational analysis develop rapidly, the accessibility of clinical material of sufficient quality and quantity is likely to be rate limiting in the discovery process. For cancer patients, diagnosis is virtually always based on the evaluation of a tumour sample, ideally a biopsy or surgical specimen. Left-over tissue, stored in pathology archives and no longer required following diagnosis or clinical management, proves an immense resource for molecular analysis, as each cancer patient is a potential donor. The fixation procedures, necessary for tissue preservation in the diagnostic setting, may however compromise the quality of the material with regard to some applications. One of the applications described for NG sequencing is the production of copy number variation (CNV) data by analysing the distribution of aligned reads to a reference genome (2–5). These initial studies have used a depth of sequencing that would preclude the routine analysis of large numbers of samples. They have also used optimal starting conditions in terms of DNA quality and quantity, which are not likely with clinical samples. However, Schweiger et al. (6) recently showed that DNA isolated from formalin-fixed paraffin-embedded (FFPE) tumours could be used for sequencing-based CNV analysis. Here, we report how we have modified the current methods for sequencing-based CNV analysis and overcome their limitations, to allow the high-throughput copy number analysis of DNA from suboptimal quality material, such as FFPE tissue. Specifically, we have investigated the feasibility of undertaking sequence analysis of samples in pools using unique oligonucleotide tags to distinguish individual patients. We demonstrate that up to 10 patient samples can be pooled in one sequencing lane of an Illumina Genome Analyzer II (GAII; Illumina Inc, San Diego, CA, USA). We also show that as little as 5 ng (compared to Illumina’s recommended 1 µg) of template DNA from FFPE specimens is sufficient to generate a library of fragments for sequence analysis, resulting in a copy number karyogram that is indistinguishable from a karyogram generated from 1 µg of template DNA or from DNA isolated from the same fresh-frozen tumours without prior fixation. These modifications will enable exploitation of the vast archives of FFPE material to supply the systems medicine research pipeline, and will bring this methodology within the reach of routine clinical analysis, while preserving the majority of the tissue block for other investigations.
To illustrate the value of this approach, we have obtained FFPE samples of repeat biopsies and resection specimens from the same patient whose oral cancer recurred over a 2-year period. We show that all samples share certain copy number aberrations indicating a likely common progenitor, but that the most recent samples display a new region of chromosomal amplification indicating that an initial clonal population of tumour cells has continued to mutate.
MATERIALS AND METHODS
Sample selection
Cell lines
LUDLU-1 cell line was established from a lung squamous cell carcinoma (7). AGLCL cell line was established following EBV infection of normal B cells from the same patient. Both cell lines were cultured in RPMI 1640 medium supplemented with 2 mM glutamine, 50 U/ml penicillin, 50 µg/ml streptomycin and 10% foetal bovine serum using standard cell culture techniques. The HONE-1 cell line is an epithelial cell line derived from a poorly differentiated nasopharyngeal squamous cell carcinoma (8). It was cultured in Iscove’s modified Dulbecco’s medium, supplemented with 2 mM glutamine, 50 U/ml penicillin, 50 µg/ml streptomycin and 10% foetal bovine serum using standard cell culture techniques.
Clinical samples
Surgical resection specimens of lung tumours and corresponding normal lung tissue were prospectively collected at the local Department of Thoracic Surgery, snap frozen and stored in aliquots at −80°C. FFPE blocks of lung tumours, oral tumours and dysplastic lesions were retrieved from the local pathology archive. Approval was obtained from the local ethics committee and written informed consent for the use of their tissue for research was available for all patients.
DNA isolation
Snap-frozen tissues and cell lines
Genomic DNA from clinical samples and cell lines was prepared by sequential phenol/chloroform extraction followed by ethanol precipitation, adapted from a previously described method (9). Briefly, frozen tissue was ground using pestle and mortar and the resulting powder suspended in 0.4% LiDS lysis buffer; cell lines were harvested from the culture flask, pelletted by centrifugation and resuspended in 0.4% LiDS lysis buffer. Proteinase K was then added to the cell lysate to a final concentration of 100 μg/ml and incubated overnight at 55°C. Following three phenol extractions and one with chloroform, the final aqueous solution was added with 0.3 M sodium acetate pH 5.2 (1:10 vol:vol) and DNA precipitated by the addition of two volumes of 95% ethanol. The DNA precipitate was then collected by centrifugation, washed with 70% ethanol, air dried and finally dissolved in sterile water.
FFPE tissue sections
Areas of dysplasia or tumour to be dissected were identified and marked by a pathologist on an haematoxylin and eosin (H+E) stained slide cut from each FFPE block to be sampled. Ten 7 μm sections were then cut and the dysplastic or tumour tissue was macro-dissected with a scalpel blade using the marked H+E slide as a guide. A further section was cut and then H+E stained to confirm persistence of histology throughout the areas sampled. DNA extraction was performed using the QIAamp DNA micro kit (Qiagen, Sussex, UK) according to the manufacturer’s instructions and DNA was eluted in 25 μl of AE Buffer.
DNA concentration and purity was determined using the Nanodrop-8000 (Fisher Scientific UK Ltd, Leicestershire, UK) and the Quant-iT PicoGreen dsDNA BR assay (Invitrogen, Paisley, UK).
Array-CGH
DNA from the LUDLU-1 and AGLCL cell lines (450 ng each) was labelled in the presence of Cy3-or Cy5-labelled nucleotides as previously described (10) but purified using a PureLink PCR Purification Kit (Invitrogen) according to the manufacturer’s instructions. Labelled material was combined, hybridized to Agilent Human Genome 244K CGH microarrays (Agilent Technologies, Santa Clara, CA, USA), washed and scanned according to Agilent’s Oligonucleotide Array-based CGH for Genomic DNA Analysis protocol. Data were extracted from the scanned images using Feature Extraction software (v7.1; Agilent Technologies) and analysed using Agilent’s DNA Analytics package.
Illumina Genome Analyser library preparation
Between 5 ng and 1 μg genomic DNA were used to prepare the DNA libraries for sequencing, following standard Illumina protocols. DNA was sheared on a Covaris S2 Sample Preparation System (Covaris Inc., Woburn, MA, USA) and checked for appropriate size distribution on an Agilent Bioanalyser DNA 1000 LabChip. End repair was performed by using the End-It DNA End Repair Kit (Epicentre Biotechnologies, Madison, WI, USA) and Klenow DNA polymerase, followed by ligation of 6 bp unique tag adapter oligonucleotides, using previously established methods (11). Tags were chosen so as to avoid over-representation of any one base at each position, which can interfere with cluster recognition. Fragments were size selected to 200 bp using a gel cut step. Samples were enriched using a 12-cycle enrichment PCR. For low concentration DNA samples, an 18-cycle enrichment PCR was performed before the gel cut stage rather than afterwards. Libraries were then examined using an Agilent Bioanalyser DNA 1000 LabChip and Invitrogen’s Quant-iT Picogreen dsDNA BR assay kit to assess for DNA quality and concentration, respectively. This information was used to pool equal amounts of each sample library for cluster amplification and either 51 or 76 cycles of Illumina sequencing by synthesis, resulting in 45/70 bp of genomic DNA sequence and 6 bp of tagged adapter. Sequencing was initially done with 51-bp reads but the move was made to 76-bp reads as machine and analysis package upgrades resulted in better base calling for longer reads.
Data analysis
Image analysis and base calling were performed using Illumina’s CASAVA pipeline. Reads were trimmed of their 6 bp tags with the USE_BASES option and uniquely aligned to the human genome (UCSC version hg19) using the alignment algorithm Eland (12). Python scripts were used to first split the reads into files according to tag, and then to make pairwise comparisons of each tumour and normal sample by splitting the genome into non-overlapping windows of equal numbers of normal reads, typically 400, and counting the number of tumour reads which fell into each window.
Copy number analysis was done in R, first by normalizing the number of tumour and normal counts across the genome and calculating the log2 ratio of normalized tumour:normal read counts for each window. Second, segments of equal copy number were called using the Bioconductor DNAcopy package (13). These segments were converted into bedgraphs suitable for uploading onto the UCSC genome browser (http://genome.ucsc.edu/) (14) as well as being plotted over graphs of tumour read counts. A summary of the number of aligned reads and subsequently detected CNVs for each sample is listed in Table 1.
Table 1.
Summary statistics for each sample
| Sample | Aligned tumour reads | Fraction reads aligning (percent) | Aligned normal reads | Mean window size (kb) | Number of CNVs | Mean CNV size (Mb) | Minimum CNV size (Mb) |
|---|---|---|---|---|---|---|---|
| LUDLU1 | 2 551 569 | 76 | 4 973 483 | 498 | 163 | 18.3 | 0.56 |
| LS010 0.035X | 2 330 765 | 75 | 1 894 107 | 653 | 101 | 29.2 | 1.15 |
| LS010 0.016X | 1 091 212 | 75 | 888 169 | 1224 | 78 | 38.2 | 1.59 |
| LS010 0.002X | 136 405 | 75 | 882 800 | 1388 | 47 | 62.9 | 6.20 |
| LS035 | 931 088 | 74 | 809 154 | 1524 | 53 | 57.1 | 1.35 |
| LS035 FFPE | 990 577 | 74 | 809 154 | 1524 | 65 | 45.6 | 1.84 |
| LS041 | 888 436 | 70 | 824 869 | 1417 | 74 | 40.2 | 1.02 |
| LS041 FFPE | 169 202 | 64 | 824 869 | 1417 | 55 | 53.7 | 2.65 |
| LS041 50 ng | 939 186 | 58 | 824 869 | 1417 | 105 | 27.7 | 1.43 |
| LS041 10 ng | 181264 | 21 | 824 869 | 1417 | 94 | 31 | 2.39 |
| LS041 5 ng | 136 350 | 20 | 824 869 | 1417 | 107 | 27.1 | 2.39 |
| LS043 | 1 002 978 | 60 | 1 259 862 | 895 | 110 | 27.3 | 1.18 |
| LS043 FFPE | 511 550 | 76 | 1 259 862 | 895 | 72 | 41.2 | 0.91 |
| LS043 FFPE 50 ng | 4 697 581 | 60 | 2 642 990 | 468 | 151 | 19.9 | 0.47 |
| LS043 FFPE 10 ng | 2 296 802 | 61 | 2 642 990 | 468 | 194 | 15.3 | 0.76 |
| LS043 FFPE 5 ng | 3 570 091 | 62 | 2 642 990 | 468 | 144 | 20.9 | 0.85 |
| PG019-1 | 879 715 | 76 | 1 894 107 | 653 | 64 | 47 | 1.11 |
| PG019-4 | 564 178 | 74 | 1 894 107 | 653 | 105 | 28.2 | 1.10 |
| PG019-5 | 520 740 | 71 | 1 894 107 | 653 | 70 | 43 | 2.23 |
| PG019-6 | 844 130 | 76 | 1 894 107 | 653 | 64 | 47 | 0.77 |
| PG019-7 | 303 584 | 57 | 1 894 107 | 653 | 36 | 84 | 2.39 |
| PG019-8 | 2 249 010 | 75 | 1 894 107 | 653 | 37 | 81.8 | 3.89 |
| PG019-9 | 1 305 492 | 77 | 1 894 107 | 653 | 41 | 73.7 | 3.29 |
The number of aligned reads, fraction of reads aligning, number of reference reads, mean window size and size and number of detected CNVs are shown
A Pearson correlation was calculated between the array-CGH (aCGH) and sequencing data by calculating an average aCGH generated copy number for every genomic window of sequencing reads. This made it possible to perform a pairwise comparison of two files of equal length.
A similar method was used to calculate the correlation between molecular copy number counting (MCC) and sequencing data, sampling the sequence based copy number for every MCC data point.
RESULTS
Reproducibility and validation of NG sequencing for CNV analysis
We began by analysing the copy number profile of genomic DNA from the lung squamous cell carcinoma cell line LUDLU-1 and the DNA from the paired normal B cell line AGLCL. We confirmed that the tumour karyogram demonstrated copy number gain and loss features predicted for a squamous cell lung carcinoma such as 3p loss and distal 3q and 8q amplification (15–17) (Supplementary Figure S1). We confirmed by comparing the normal DNA from five individuals that comparison of normal DNA against normal DNA resulted in a featureless karyogram with no detectable gain or loss (result not shown).
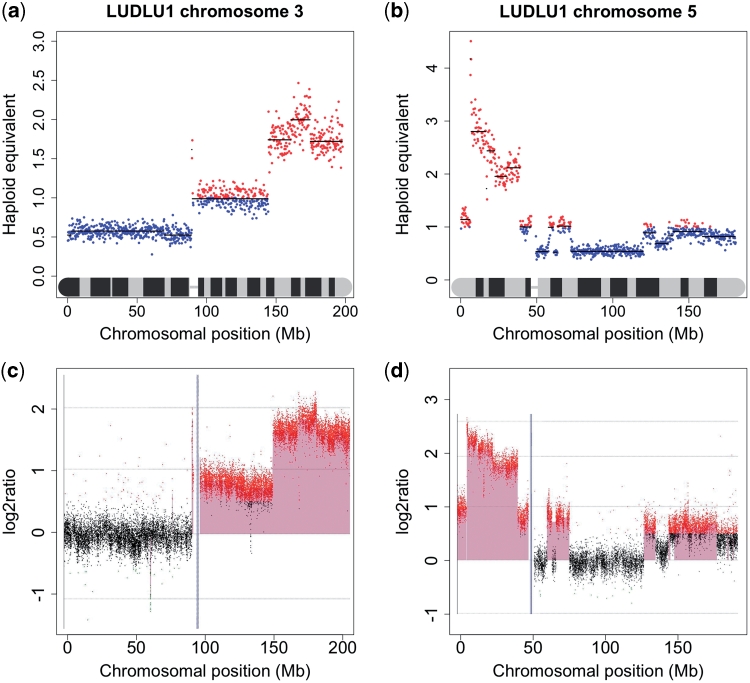
Further validation was achieved by comparing the copy number profile generated by NG sequencing with that obtained by aCGH. High-resolution aCGH was performed using an Agilent 244K array using DNA from LUDLU-1 and AGLCL cells. The aCGH-generated copy number profiles appeared almost identical to those obtained from sequence analysis. Every chromosome showed the same pattern of gain and loss. Even smaller features such as small spikes of gain or loss were replicated. A Pearson correlation of 0.9362277 was calculated between the two data sets. Examples from the aCGH profile compared to a copy number karyogram generated from NG sequencing data is shown in Figure 1.
Figure 1.
Examples of a comparison of copy number analysis by sequencing and aCGH. (a) Chromosome 3 analysed by NG sequencing; (b) chromosome 5 analysed by NG sequencing; (c) chromosome 3 analysed by aGCH; (d) chromosome 5 analysed by aCGH.
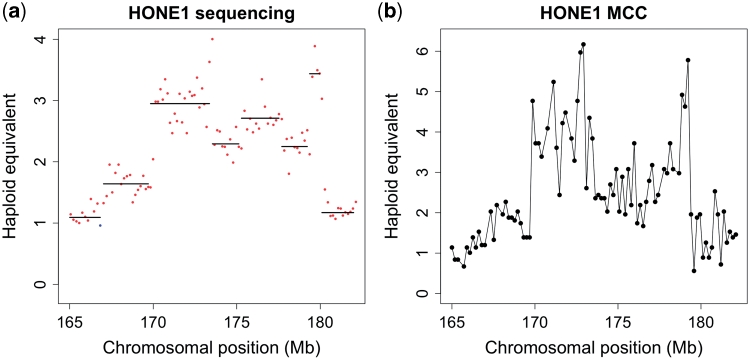
To determine the validity of copy number annotation by sequencing even at high resolution, we compared its performance to another method that also generates copy number data in a digital format, MCC (18,19). We compared the copy number profile generated by NG sequencing with the MCC data for the 17 Mb amplicon in the distal part of chromosome 3q in the HONE-1 cell line and found the profiles to be very similar. Gains and losses were seen in the same places, although the MCC data were noisier and suggested a slightly higher copy number in places. A Pearson correlation of 0.8126372 was calculated between the two data sets (Figure 2).
Figure 2.
Comparison of copy number analysis by (a) sequencing and (b) a digital PCR method (MCC). The 88 MCC markers between 165 and 182 Mb of chromosome 3 of the HONE-1 cell line were analysed and compared to the sequence-based copy number data from the same region.
The reproducibility of copy number data produced from NG sequencing was confirmed by demonstrating virtually identical copy number karyograms on analysis of duplicates of four sets of DNA from tumour:normal pairs of fresh frozen lung squamous cell carcinomas (Supplementary Figure S2).
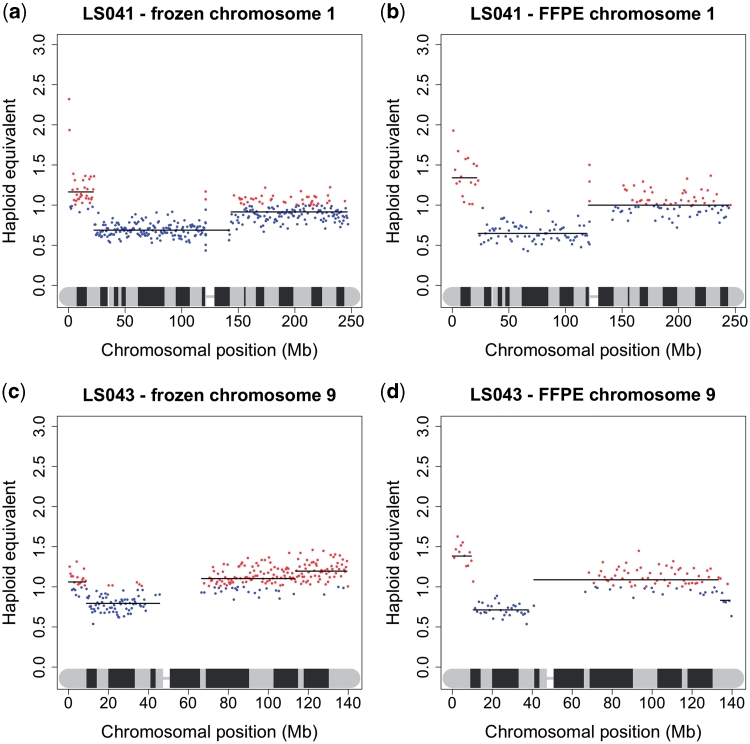
Finally, we compared the copy number karyograms for DNA extracted from snap frozen versus FFPE material from the same lung squamous cell carcinomas (Figure 3). As previously shown by Schweiger et al. the matching fresh and fixed copy number karyograms for some of the samples were almost identical even when examined at high resolution. For other samples, the positions of regions of CNV in the fixed samples were identical to the frozen samples, but the magnitude of variation was greater. For example, in the fresh-frozen sample, LS043, shown in Figure 3, the distal 9 Mb of 9p has a tumour:normal ratio of 1.1:1, while the rest of the 9p arm has a ratio of 0.82:1. In the FFPE sample the two regions are in the same position, but have ratios of 1.51:1 and 0.75:1, respectively. This difference is probably due to the removal of non-cancerous cells (i.e. stromal cells, inflammatory cells and endothelial cells) by macrodissection in the fixed samples.
Figure 3.
Copy number karyograms of DNA extracted from snap frozen and FFPE tissue. Representative individual chromosome plots for two lung squamous cell carcinoma patients (LS041 and LS043) are shown. (a) LS041 snap frozen chromosome 1; (b) LS041 FFPE chromosome 1; (c) LS043 snap-frozen chromosome 9; (d) LS043 FFPE chromosome 9.
The effect of using decremental amounts of starting DNA
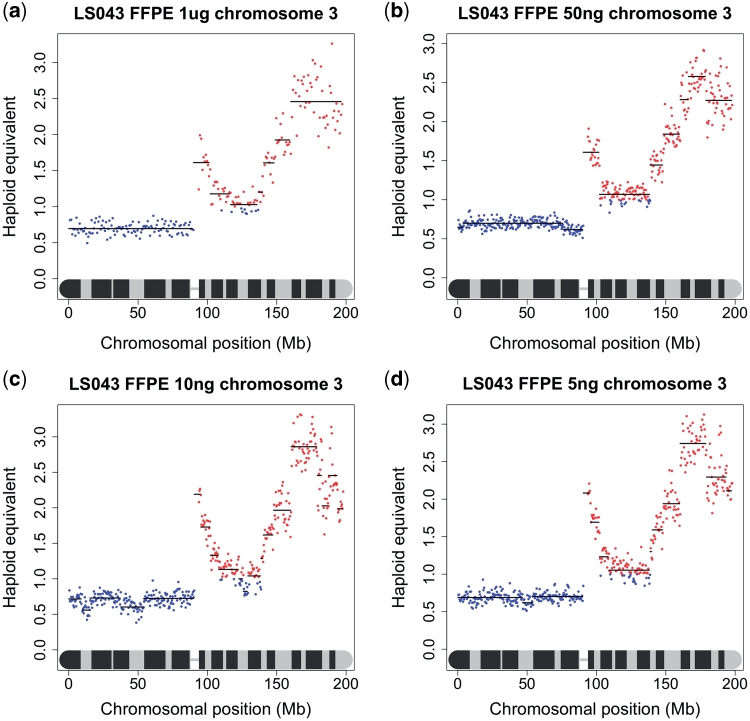
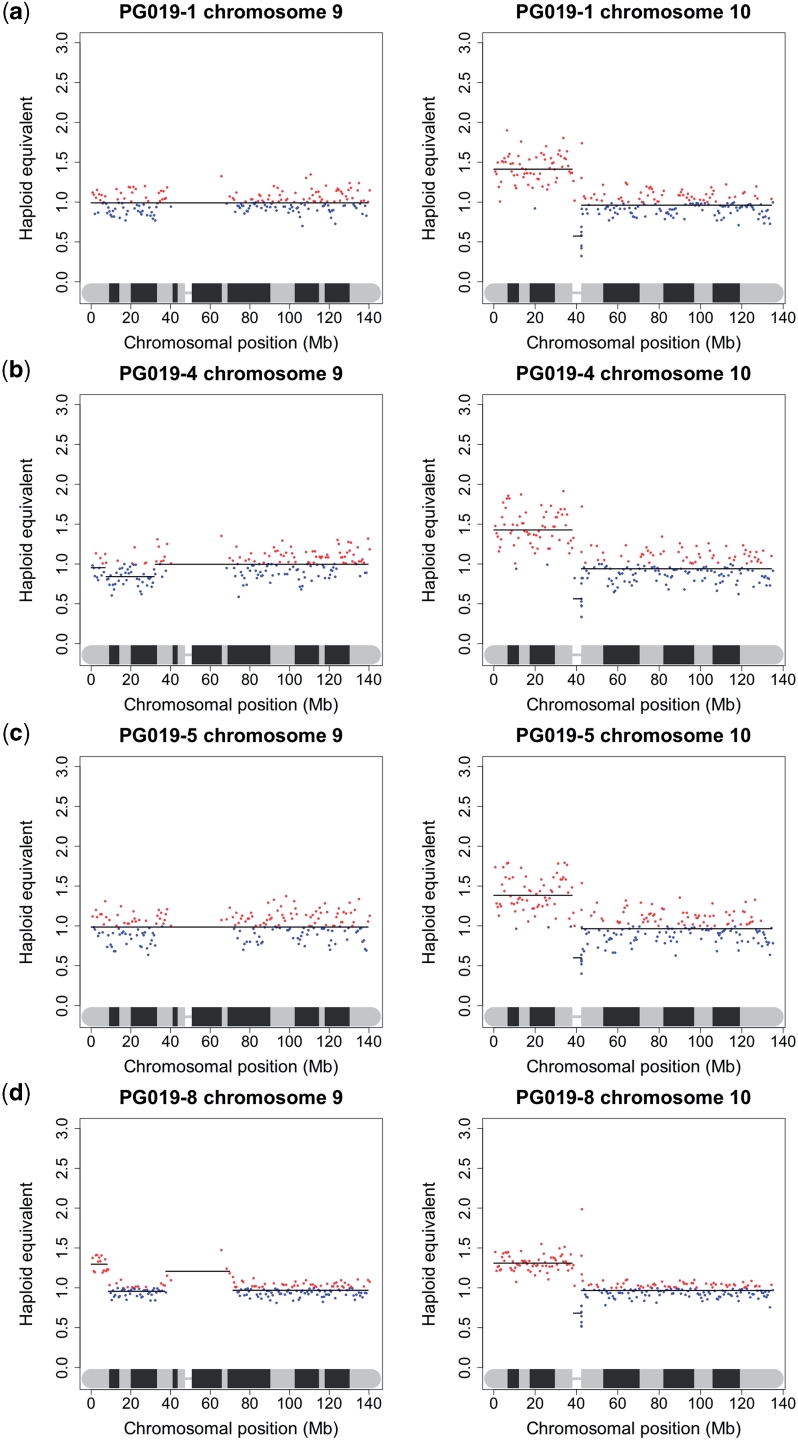
Due to the difficulty in obtaining large amounts of good quality DNA from many clinical samples, especially fixed archival surgical and biopsy specimens, we undertook to investigate the minimum amount of DNA that is needed to produce acceptable sequence data for copy number analysis. We performed a series of dilutions of DNA from one of the frozen tumour samples and one of the FFPE samples, so that sequencing libraries were generated from 400 ng, 200 ng, 100 ng, 50 ng, 10 ng and 5 ng of starting DNA, compared to Illumina’s recommended 1 µg. As all the libraries appeared to be within normal parameters as judged by Agilent readings, only those prepared from 50 ng, 10 ng and 5 ng were sequenced. While it was difficult to accurately titrate the lower concentration libraries to give a consistent number of sequencing reads, and some samples gave a low percentage of alignable reads, the karyograms produced were almost identical to those made under Illumina’s recommended conditions suggesting that copy number data could be obtained from nanogram quantities of DNA isolated from tissue sections of a FFPE block (Figure 4). To confirm this in a real sample, we obtained FFPE blocks of sequential biopsies and surgical specimens from a patient with multifocal oral cancer who has been under the care of the local maxillo-facial unit for several years. Specifically, we obtained blocks for (i) a biopsy of a tumour in the left tongue in May 2007, (ii) a biopsy of dysplasia in the right floor of the mouth in June 2008, (iii) a wide excision specimen of the same dysplasia in July 2008 and (iv) four distinct specimens of tumour-associated dysplasia obtained from a further anterior floor of mouth surgical resection in August 2009. DNA was isolated from macrodissected dysplastic and tumour tissue and libraries were prepared from template DNA ranging from 55 to 270 ng. The karyograms were mostly normal (results not shown): the most notable feature was gain of 10p, which was common to all lesions. The synchronous biopsies obtained in the surgical field in August 2009 carried an 8Mb region of amplification of distal 9p in addition to gain of 10p (Figure 5).
Figure 4.
NG sequencing-based copy number karyograms using decremental amounts of starting DNA, i.e. (a) 1 µg, (b) 50 ng, (c) 10 ng and (d) 5 ng. The copy number profiles for chromosome 3 using genomic DNA from the FFPE squamous cell carcinoma LS043 are shown here. Similar results were obtained for the other chromosomes and also using the fresh frozen sample LS010 (not shown).
Figure 5.
Generation of copy number karyograms from small clinical samples. Chromosomes 9 and 10 copy number profiles of DNA from four sequential FFPE specimens obtained from different areas of the oral cavity of the same patient (PG019) over a two year period. The samples shown are (a) PG019-1, a biopsy from May 2007; (b) PG019-4, a biopsy from June 2008; (c) PG019-5, wide local excision from July 2008; (d) specimen from surgical resection from August 2009. All samples show a similar gain of the short arm of chromosome 10. PG019-8 shows a discreet amplification in 9p: this amplification was detected in three additional samples from the August 2009 surgical resection (data not shown).
Feasibility of multiplexing
We investigated sequencing various numbers of pooled libraries together in order to gain an understanding of the quality of data obtainable from different levels of reagent and machine capacity investment. The initial experiment using the LUDLU-1:AGLCL cell line pair was performed using one eighth of an Illumina GAII run and produced 7 897 570 uniquely aligning reads. Of these, tagging indicated that 2 547 384 were from LUDLU1. A total of 2 500 000 reads of 45 bp (51 bp – 6 bp tag) represents 113 Mb, or ∼0.04× coverage. The 206 CNV regions were detected with mean size of 15 Mb and a smallest detectable size of 15 Kb.
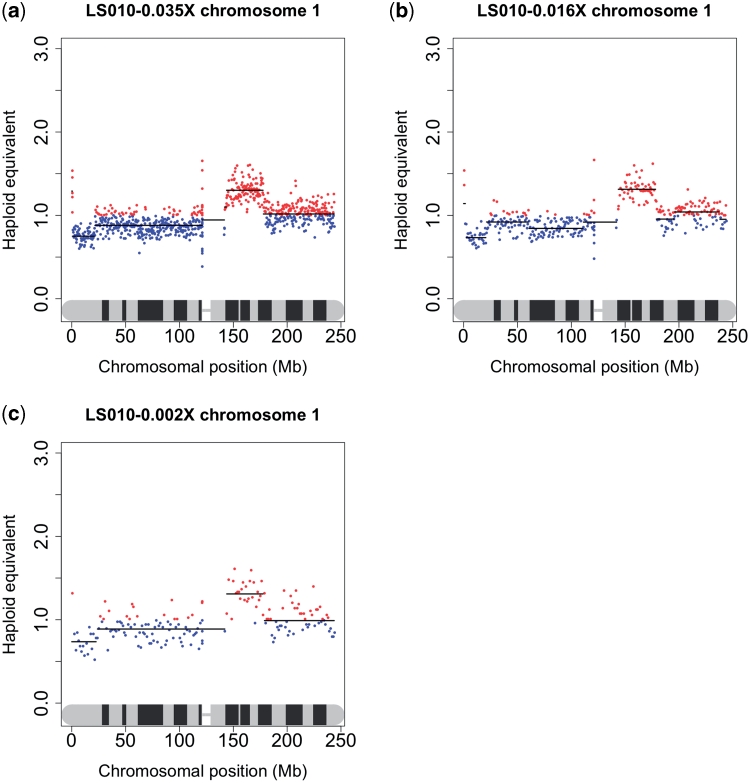
For the subsequent experiment using DNA from snap-frozen tumour:normal pairs, the level of multiplexing was increased so that 10 samples (5 pairs) were pooled and analysed together on one eighth of an Illumina GAII run. Between 847 576 and 1 321 486 reads per sample were obtained. Between 57 and 115 copy number variation regions were detected, averaging between 26 and 58 Mb in size, with a smallest detectable region between 0.9 and 1.5 Mb. This experiment was duplicated and the reads from each sequencing run combined to give between 1 873 100 and 2 328 268 reads per sample (0.028–0.035× coverage). The overall CNV pattern of each sample remained unchanged (Figure 6a and b), but the DNAcopy algorithm was able to detect smaller regions of copy number variation, between 89 and 200 Kb. As a result, the number of observed regions increased to between 81 and 187.
Figure 6.
The effect of increased multiplexing. Sequencing was performed using 5 and 10 samples per lane of an Illumina GAII. Shown here is chromosome 1 from sample LS010 at the level of (a) five samples per lane generating 2 330 765 sequence reads for this sample or 0.035× genomic coverage; (b) 10 samples per lane generating 1 091 212 reads or 0.016× coverage and (c) an in silico simulation of 80 samples per lane generating 136 405 reads or 0.002× coverage.
To determine how little data would give reproducible results, 90% of the reads from one of the frozen samples (LS010) were randomly removed in silico, leaving 136 405 tumour reads or 0.002× coverage. This is an approximate simulation of running 80 tagged samples on one lane, or 0.15% of the sequencer’s total capacity. Currently, this is not technically feasible, but may be achievable in the near future as methods such as DNA Sudoku mature (20). Unsurprisingly, all fine scale data were lost, the smallest region of variation being detected being 5 Mb. It was still perfectly possible to see large-scale aberrations such as the gain or loss of whole chromosomal arms (Figure 6c), suggesting that this methodology could still provide useful data even when highly multiplexed.
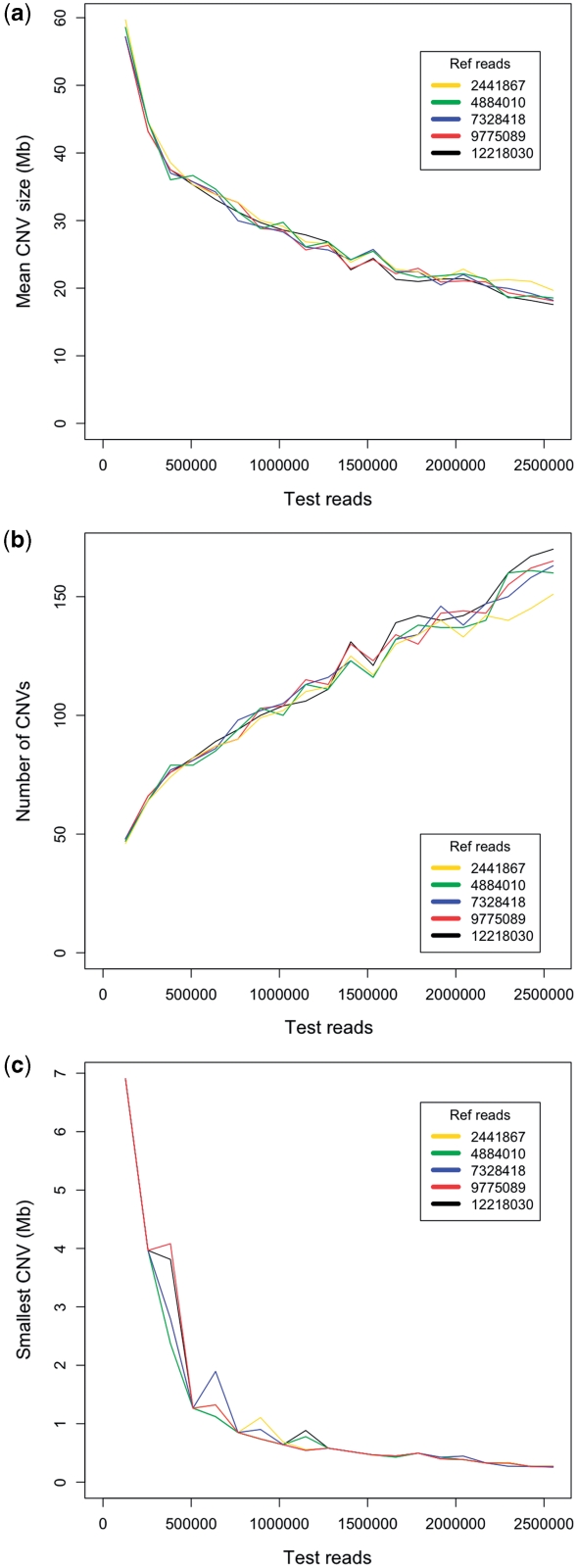
To further explore the theoretical limits of multiplexing, a series of simulations were performed using the LUDLU-1:AGLCL cell line pair. An additional 7.4 million AGLCL reads were sequenced, giving a total of 12 218 030 reads. Reads were randomly stripped away in silico from both samples, resulting in five files of AGLCL reads ranging between 2 441 867 and 12 218 030 and 20 files of LUDLU-1 reads ranging between 127 421 and 2 551 569. 127 421 reads is <1% of the standard output from one lane of an Illumina GAII during our experiments. Every combination of these two samples was then analysed, using LUDLU-1 as the test and AGLCL as the reference sample, and keeping the window sizes equivalent to 200 LUDLU-1 reads. The results are shown in Figure 7. The number of reference reads appeared to have almost no effect on either the number of CNVs, mean size or smallest detectable CNV. Not surprisingly, the number of LUDLU-1 reads, and hence the size of the windows used had a much greater effect. Resolution gradually decreased alongside read number, but with a sudden decrease once read numbers dropped below 500 000. It may be that this is the point at which a window size of 200 reads is bigger than many of the actual CNVs from this sample.
Figure 7.
Simulated data showing the effect of increased multiplexing. Twenty different numbers of LUDLU-1 reads between 127 421 and 2 551 569 were compared to five different numbers of AGLCL reads between 2 441 867 and 12 218 030. Shown here are: (a) the mean CNV size; (b) the number of CNVs detected; (c) the smallest detectable CNV.
DISCUSSION
We have demonstrated that NG sequencing platforms can be used in a high-throughput, cost-effective manner to elucidate copy number information from a variety of DNA sources, including cell lines, frozen tumour samples and FFPE material. We have shown that good quality data can be obtained when multiplexing up to 10 samples on one lane of an Illumina GAII. We have shown that the method is reproducible, and that the resolution is highly flexible and adjustable. The resolution has been shown to be comparable with aCGH when performed at low levels of multiplexing and has a high degree of correlation. NG sequencing also produces much more data than a comparable investment in a PCR-based method such as loss of heterozygosity (LOH) analysis or MCC when performed at higher levels of multiplexing. Sequence data and MCC does show a strong correlation, but not as strong as that between sequencing and aCGH. We have concentrated on tumour samples, but the method is equally applicable to studies of CNV in constitutional DNA. In fact, the data are much easier to interpret when all the cells in a sample have the same genotype.
Currently, the leading technology for investigating CNV is aCGH. However, limitations remain for certain applications. aCGH has proved difficult to use with DNA from FFPE samples. Typically, researchers have had to devise ingenious upstream methods in order to study archival material (21). Also, aCGH typically requires microgram quantities of DNA. When smaller samples are studied, a whole-genome amplification step is generally incorporated (22). Our method requires almost no extra fine tuning as we moved from cell line DNA to nanogram amounts of DNA from archival FFPE material, showing that neither a large amount of template or a whole genome amplification step are required. In addition, although we have only demonstrated the use of these data for copy number analysis, each lane of sequencing generates in the region of 700 Mb of sequence data, which can be analysed for other purposes such as searching for genetic variants or viral infection.
Previous studies have used read depth of sequence to examine copy number but with ideal reaction conditions and greater depth of coverage (2–5). Other studies have explored the limits of this technology further, by use of multiplexing (23) or by using DNA from FFPE samples (6). We have sought to combine both approaches and to further push the limits as to the minimum amounts of starting DNA required. The multiplexing aspect is important because it allows researchers to tailor their study design according to the number of samples, desired resolution and available resources. The same sequencing library can be used for a 1 Kb or a 10 Mb resolution experiment. The only difference is the amount of sequencing required. Libraries can be aliquoted for use a number of times so that samples can be initially screened cheaply at low resolution and then further examined at high resolution at a later date and with no additional preparation. Data from two duplicate experiments on the same sample can be merged to double the coverage.
We have demonstrated that sequencing libraries can be constructed starting with nanogram quantities of DNA and, by combining individually tagged libraries, copy number karoygrams can be generated from 10 samples in a single Illumina sequencing run, with the theoretical possibility of extending this number up to at least 80. Besides demonstrating this high-throughput potential, we have also confirmed the observations of Schweiger et al. (6) that DNA isolated from FFPE material can serve as a template to create sequencing libraries for NG sequencing. We have substantially extended the range of samples that can be studied by reducing the amount of template required for library construction from 1.5 µg to less than 100 ng. This is an important practical consideration because it allows minute tissue samples such as biopsies to be analysed. Not only are biopsies small but also the way they are obtained from patients often incorporates underlying stroma, requiring micro-dissection to increase the proportion of abnormal epithelial cells, decreasing the amount of DNA template still further. However these difficulties must be overcome, as analysis of biopsies is essential for both basic and translational cancer research.
In attempting to identify the genomic drivers of malignant tumour initiation and progression of upper aerodigestive cancer, we have used a number of molecular genetic techniques to compare candidate regions, genes and chromosomes of pre-invasive lesions, obtained as biopsies, and the subsequent tumours that develop at the site of the earlier pre-invasive lesions (24–26). In this study, we have used NG sequencing to generate whole genome copy number karyotypes of biopsies and tumours obtained in a chronologic series from a patient with oral cancer. The patient’s cancer has had a relatively indolent disease course reflected by several invasive tumours and dysplastic lesions but no metastatic disease. The karyotypes of the tumour and dysplastic lesions obtained carry relatively few gains and losses compared other the other carcinoma karyograms we examined. This may be a reflection of the tumour’s histology, a verrucous squamous cell carcinoma, whose benign growth pattern may in turn be a consequence of a relatively normal genotype. However, all specimens examined carry gain of the short arm of chromosome 10 indicating a clonal origin. Those obtained from the most recent surgery demonstrated genomic progression, having acquired amplification of distal 9p, an 8 Mb region encompassing 63 predicted genes (27). 9p loss has been associated with tumour progression (28,29), but gain in this region so far has not.
Besides the analysis of biopsies for laboratory studies of clonality and the genomics of tumour progression, the genomic assessment of tumours via the preliminary diagnostic biopsy may a be a useful supplement to routine histopathology, as knowledge of a tumour’s genotype can provide information for prognosis and treatment response (30). The international initiatives to document cancer genomes (http://www.icgc.org/, http://cancergenome.nih.gov/about/index.asp) will substantially augment our current knowledge. We have used fixed archival specimens in this study, but the current samples of each new patient are also fixed as part of routine clinical practice, and the ability to generate whole-genome information from the fixed samples obtained as demonstrated here, indicates how cataloguing genome architecture could be become part of the repertoire of diagnostic tests for cancer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Yorkshire Cancer Research (L341PG); Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme (219540 to O.B.); Cancer Research UK and the Wellcome Trust (to D.J.A.). Funding for open access charge: Yorkshire Cancer Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Maria Kost-Alimova, Karolinska Institute, Stockholm, Sweden, for kindly providing the HONE-1 cell line used in this study.
REFERENCES
- 1.Auffray C, Chen Z, Hood L. Systems medicine: the future of medical genomics and healthcare. Genome Med. 2009;1:2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, Zatloukal K, Lehrach H. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS ONE. 2009;4:e5548. doi: 10.1371/journal.pone.0005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabbitts P, Douglas J, Daly M, Sundaresan V, Fox B, Haselton P, Wells F, Albertson D, Waters J, Bergh J. Frequency and extent of allelic loss in the short arm of chromosome 3 in nonsmall-cell lung cancer. Genes Chromosomes Cancer. 1989;1:95–105. doi: 10.1002/gcc.2870010115. [DOI] [PubMed] [Google Scholar]
- 8.Glaser R, Zhang HY, Yao KT, Zhu HC, Wang FX, Li GY, Wen DS, Li YP. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc. Natl Acad. Sci. USA. 1989;86:9524–9528. doi: 10.1073/pnas.86.23.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis LG, Kuehl WM, Battey JF. Basic Methods in Molecular Biology. 2nd edn. Norwalk: Appleton & Lange; 1994. pp. 16–21. [Google Scholar]
- 10.Fiegler H, Redon R, Carter NP. Construction and use of spotted large-insert clone DNA microarrays for the detection of genomic copy number changes. Nat. Protocol. 2007;2:577–587. doi: 10.1038/nprot.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 14.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjorkqvist AM, Husgafvel-Pursiainen K, Anttila S, Karjalainen A, Tammilehto L, Mattson K, Vainio H, Knuutila S. DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma. Genes Chromosomes Cancer. 1998;22:79–82. [PubMed] [Google Scholar]
- 16.Fong KM, Biesterveld EJ, Virmani A, Wistuba I, Sekido Y, Bader SA, Ahmadian M, Ong ST, Rassool FV, Zimmerman PV, et al. FHIT and FRA3B 3p14.2 allele loss are common in lung cancer and preneoplastic bronchial lesions and are associated with cancer-related FHIT cDNA splicing aberrations. Cancer Res. 1997;57:2256–2267. [PubMed] [Google Scholar]
- 17.Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res. 1997;57:2116–2120. [PubMed] [Google Scholar]
- 18.Daser A, Thangavelu M, Pannell R, Forster A, Sparrow L, Chung G, Dear PH, Rabbitts TH. Interrogation of genomes by molecular copy-number counting (MCC) Nat. Methods. 2006;3:447–453. doi: 10.1038/nmeth880. [DOI] [PubMed] [Google Scholar]
- 19.McCaughan F, Darai-Ramqvist E, Bankier AT, Konfortov BA, Foster N, George PJ, Rabbitts TH, Kost-Alimova M, Rabbitts PH, Dear PH. Microdissection molecular copy-number counting (microMCC)–unlocking cancer archives with digital PCR. J. Pathol. 2008;216:307–316. doi: 10.1002/path.2413. [DOI] [PubMed] [Google Scholar]
- 20.Erlich Y, Chang K, Gordon A, Ronen R, Navon O, Rooks M, Hannon GJ. DNA Sudoku–harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 2009;19:1243–1253. doi: 10.1101/gr.092957.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hostetter G, Kim SY, Savage S, Gooden GC, Barrett M, Zhang J, Alla L, Watanabe A, Einspahr J, Prasad A, et al. Random DNA fragmentation allows detection of single-copy, single-exon alterations of copy number by oligonucleotide array CGH in clinical FFPE samples. Nucleic Acids Res. 2010;38:e9. doi: 10.1093/nar/gkp881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little SE, Vuononvirta R, Reis-Filho JS, Natrajan R, Iravani M, Fenwick K, Mackay A, Ashworth A, Pritchard-Jones K, Jones C. Array CGH using whole genome amplification of fresh-frozen and formalin-fixed, paraffin-embedded tumor DNA. Genomics. 2006;87:298–306. doi: 10.1016/j.ygeno.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Daines B, Wang H, Li Y, Han Y, Gibbs R, Chen R. High-throughput multiplex sequencing to discover copy number variants in Drosophila. Genetics. 2009;182:935–941. doi: 10.1534/genetics.109.103218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster NA, Banerjee AK, Xian J, Roberts I, Pezzella F, Coleman N, Nicholson AG, Goldstraw P, George JP, Rabbitts PH. Somatic genetic changes accompanying lung tumor development. Genes Chromosomes Cancer. 2005;44:65–75. doi: 10.1002/gcc.20223. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee AK, Read CA, Griffiths MH, George PJ, Rabbitts PH. Clonal divergence in lung cancer development is associated with allelic loss on chromosome 4. Genes Chromosomes Cancer. 2007;46:852–860. doi: 10.1002/gcc.20472. [DOI] [PubMed] [Google Scholar]
- 26.McCaughan F, Pole JC, Bankier AT, Konfortov BA, Carroll B, Falzon M, Rabbitts TH, George PJ, Dear PH, Rabbitts PH. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am. J. Respir. Crit. Care Med. doi: 10.1164/rccm.201001-0005OC. doi:10.1164/rccm.201001-0005OC [18 March 2010, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosin MP, Lam WL, Poh C, Le ND, Li RJ, Zeng T, Priddy R, Zhang L. 3p14 and 9p21 loss is a simple tool for predicting second oral malignancy at previously treated oral cancer sites. Cancer Res. 2002;62:6447–6450. [PubMed] [Google Scholar]
- 29.Martin CL, Reshmi SC, Ried T, Gottberg W, Wilson JW, Reddy JK, Khanna P, Johnson JT, Myers EN, Gollin SM. Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature. Oral Oncol. 2008;44:369–382. doi: 10.1016/j.oraloncology.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, Liao ML, Bischoff H, Reck M, Sellers MV, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J. Clin. Oncol. 2010;28:744–752. doi: 10.1200/JCO.2009.24.3030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.