Abstract
Adaptive immunity is mediated through numerous genetic and cellular processes that generate favourable somatic variants of antigen-binding receptors under evolutionary selection pressure by pathogens and other factors. Advances in our understanding of immunity in mammals and other model organisms are revealing the underlying basis and complexity of this remarkable system. Although the evolution of adaptive immunity has been considered to occur by acquisition of novel molecular capabilities, an increasing amount of information from new model systems suggest that co-option and redirection of preexisting systems are the major source of innovation. We combine evidence from a wide range of organisms to obtain an integrated view of the origins and patterns of divergence in adaptive immunity.
With our increased understanding of mammalian adaptive immunity over the past decade, we have come to recognize the remarkable complexity of its underlying mechanisms. The core elements of this system are now mechanistically understood, such as DNA rearrangement, the generation of immune recognition diversity and the supporting cellular complexity that selects and expands cell populations expressing favourable antigen-binding receptor variants. General features of mammalian adaptive immunity — such as clonal selection, compartmental differentiation of lymphocytes, somatic hypermutation (SHM), allelic exclusion and a form of immunological memory — appeared before the emergence of the modern jawed vertebrates. Over the past several years, studies of immune receptors and immunity in a wide range of vertebrate and invertebrate species have revealed several similarities to present-day mammalian immunity and have provided insights into the evolutionary acquisition of immunological complexity1,2. We are within reach of important breakthroughs in our understanding of how adaptive immunity evolved in the context of an innate immune system and how these molecularly disparate systems are related and remain interdependent3. What has become increasingly clear is that the evolution of adaptive immunity requires the study of a large range of molecular systems and that it cannot be understood from studies that are restricted to mice and humans or even from studies that use alternative vertebrate models, such as bony fish and sharks. Furthermore, we recognize that the complex set of processes that constitutes adaptive immunity can be addressed most effectively by examining its constituent steps; these include (not necessarily in order of evolutionary emergence or of equivalent complexity) the appearance of lymphocytes, the acquisition of antigen-binding receptor diversification mechanisms, the structural basis for recognition specificity, the evolution of mechanisms for receptor selection and the regulatory processes that target and attenuate immune responses. We are now in a better position to understand these essential steps in the evolutionary acquisition of adaptive immune function and the many unique forms of somatic specialization and selection that are associated with it.
Adaptive immunity
Conventional adaptive immunity
Adaptive immunity in all investigated jawed vertebrates is mediated by immunoglobulins and T cell receptors (TCRs), which are generated through the recombination of variable (V), diversity (D) and joining (J) gene segments4. The V(D)J recombination process depends on the recognition of recombination signal sequences (RSSs), which flank the segmental elements and creates extensive variation in the receptor structure at junctional (joining) interfaces (FIG. 1). The V(D)J rearrangement form of somatic recombination occurs in the progenitors of B and T cells and is mediated by recombination-activating gene 1 (RAG1) and RAG2, which function in a lymphocyte- and site-specific recombinase complex (see below) and are supported by ubiquitous DNA repair factors5.
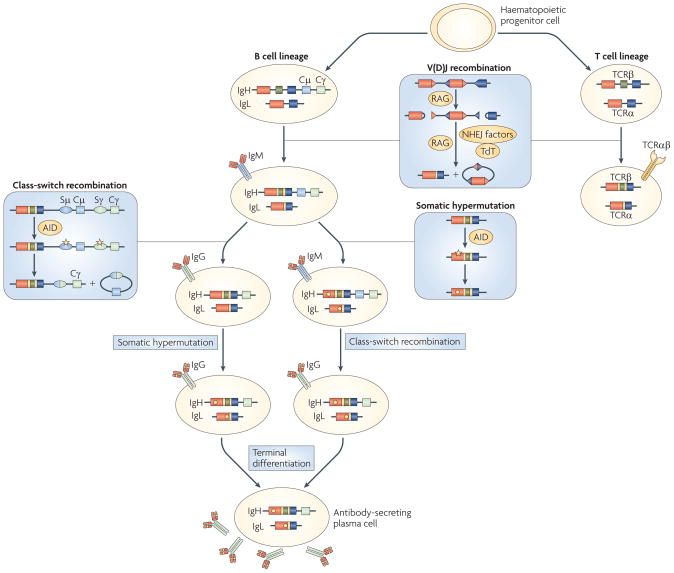
Figure 1. Lymphocyte development and antigen receptor diversification in jawed vertebrates.
A haematopoietic progenitor cell gives rise to distinct B and T cell lineages. Transcriptional networks (not depicted) are crucial for the differentiation and maintenance of cellular identity. Three unique processes — variable, diversity and joining region (V(D)J) recombination, somatic hypermutation and class-switch recombination — diversify antigen receptor genes. For clarity, some details are simplified or omitted. V (red boxes), D (green boxes) and J (dark blue boxes) segments for representative T cells (T cell receptor α-chain (TCRα) and TCRβ)) and B cells (immunoglobulin heavy chain (IgH) and immunoglobulin light chain (IgL)) are shown. The constant region for the Igμ isotype (Cμ) and a single representative downstream Cγ exon within the IgH locus are depicted. Key factors that facilitate each diversification step are shown in yellow ovals. During V(D)J recombination, recombination signal sequences (RSSs; blue and red triangles) direct the recombination-activating gene 1 (RAG1)–RAG2 recombinase complex to individual gene segments (red and blue boxes). The recombinase introduces two double-strand DNA breaks with blunt signal ends and hairpin-sealed coding ends. In the subsequent joining phase, terminal deoxynucleotidyltransferase (TdT), a template-independent DNA polymerase, adds random nucleotides to the junction of the gene elements, thereby increasing repertoire diversity dramatically; the RSSs are joined without further end processing and form excision circles. Once functional DNA rearrangements occur, TCR sequences are unaltered. After encounter with antigen, B cells further recombine the receptor by somatic hypermutation and class-switch recombination. Somatic hypermutation is initiated by activation-induced cytidine deaminase (AID), which deaminates individual cytidines within the V(D)J exon of the immunoglobulin gene, leading to U:G mismatches (yellow star). Subsequent error-prone repair results in individual point mutations (yellow dot in the gene and yellow bar in the immunoglobulin molecules), and B cells with higher affinity for the original antigen are selected. During class-switch recombination, AID creates U:G mismatches in the highly repetitive switch (S) regions (blue and green ovals) that are upstream of the exons encoding the constant regions of different isotypes. Error-prone repair leads to the generation of double-strand DNA breaks, excision of the intervening DNA (containing the Cμ exons) and joining of the remains of the switch regions. The recombined, somatically mutated V(D)J region is then associated with Cγ (green box), instead of Cμ. The precise order in which somatic hypermutation and class-switch recombination occur is unclear. Class-switch recombination has occurred in almost all high-affinity immunoglobulin-expressing B cells. In the course of a humoral immune response, B cells undergo terminal differentiation into plasma cells, which secrete large amounts of soluble immunoglobulins. γδ T cells — in which γδ TCR genes, instead of their αβ TCR genes, have been rearranged — and immunoglobulin gene conversion, which has been demonstrated in differentiating B cells of birds and rabbits, are not shown.
Immunoglobulins function first as membrane-bound receptors on B cells and their precursor cells, and they are selected for both antigen-binding specificity and affinity. A change in RNA splicing converts the membrane-bound receptor to a soluble product and is associated with the differentiation from receptor-expressing B cells to immunoglobulin-secreting plasma cells. Further modification of the primary function of immunoglobulins (that is, antigen recognition) is achieved through SHM or in some species by gene conversion. In the more recently derived tetrapod jawed vertebrates, secondary biological functions of immunoglobulins (such as binding to cell surface receptors and interaction with complement) are imparted through heavy chain class-switch recombination (CSR). Activation-induced cytidine deaminase (AID) mediates SHM, gene conversion and CSR.
The nature of immunoglobulins as diversified multigene families has been addressed from a broad phylogenetic perspective, which has revealed a high degree of variation in both the numbers and organization of the segmental elements1,2,6, as well as a range of mechanisms, including prejoining of individual immunoglobulin gene elements to form functional receptor genes in the germline7,8. A high degree of specialization in the form and function of the V and C (constant) regions of immunoglobulins has been recognized9–11. Prior to the emergence of CSR in the ancestors of modern tetrapods, differences in immunoglobulin class may have been encoded by several independently functioning gene loci, as occurs in modern species of cartilaginous fish (sharks and skates); secondary biological functions of immunoglobulins arose early in phylogeny and show extensive functional variation6,9–11. Although remarkable variation is seen in the organization of immunoglobulin genes and the cellular basis for humoral immunity, their primary mechanisms of rearrangement are conserved across the diverse phylogenetic groups that comprise the jawed vertebrates1,6.
TCRs also undergo V(D)J-type somatic rearrangement but they are exclusively membrane bound. Similar to immunoglobulins, both classes of TCRs (αβ TCRs and γδ TCRs) are heterodimers in which a D segment is a rearranging component of one unit of the receptor heterodimer. This shared characteristic suggests that all three receptor types diverged from a common ancestral heterodimeric receptor. The function of αβ TCRs relies on the polymorphic MHC class I and class II molecules expressed by antigen-presenting cells. By contrast, γδ TCRs function independently of MHC class I and class II molecules and it has been proposed that a forerunner of the rearranging antigen-binding receptors might have been a γδ TCR-like receptor12.
All four TCR chains (α, β, γ and δ) are present in the most divergent jawed vertebrates13, and neither αβ TCRs nor γδ TCRs undergo SHM in mammals. Furthermore, lymphocytes, plasma cells and other cell types of the immune system, as well as the main classes of rearranging antigen-binding receptors, are present in all jawed vertebrates. However, organ- and tissue-specific differences in the distributions of lymphocytes have been noted in various vertebrate species6,14,15.
Unconventional adaptive immunity
Jawless vertebrates (of which the only extant representatives are the agnathans lampreys and hagfish) mount specific responses to bacterial and blood cell antigens16–19, elicit allograft recognition and display other general immune-type responses that are characteristic of ‘cellular’ immunity20, but they do not use a V(D)J recombination-mediated form of adaptive immunity. Molecular genetic analyses and database searches for immunoglobulin domain-containing molecular mediators of specific humoral immunity were unsuccessful21. However, organs and tissues with large numbers of lymphocyte-like cells have been identified in jawless vertebrates and include the thymus-like pharyngeal margins in lampreys20,22,23.
It is now known that humoral (and potentially cellular) mediators of unconventional adaptive immunity in jawless vertebrates are somatically derived variants of leucine-rich repeats (LRRs), termed variable lymphocyte receptors (VLRs). VLRs are assembled from individual LRR segments that are inserted between the common amino-terminal and carboxy-terminal LRR segments, which comprise the non-functional germline VLR gene24,25. Once assembled, the LRR domain forms the VLR ligand-binding surface26,27. The LRR segments upstream of the primary VLR loci contribute sequence information to the functional genes; however, it is unknown whether this information is simply copied into the germline VLR gene using upstream LRR segments as DNA polymerase templates or whether these upstream LRR segments are physically moved by DNA rearrangements. DNA reorganization is a general ontogenetic feature of the lamprey genome28, but the molecular mechanism remains unknown. It is unclear whether AID-like cytidine deaminases might be involved in this process, and it is possible that rearrangement of the VLR locus is an example of this general genome characteristic rather than being an entirely distinct adaptation.
The mechanism of VLR assembly seems to be driven by a copy choice mechanism of recombination that is based on sequence similarities of individual LRR segments rather than by specific recombination elements29. The AID-like deaminases (see below) CDA1 and CDA2 have been implicated in this process30; however, their function in primary or secondary diversification of VLRs is not clear. The assembly (and diversification) of VLRs resembles more closely the region-specific AID-mediated secondary processes — SHM, gene conversion and CSR — that accompany antibody responses of jawed vertebrates than the non-templated somatic variation that occurs during V(D)J rearrangement of immunoglobulin and TCR loci. A process similar to allelic exclusion, which traditionally is defined as the suppression of rearrangement on the opposite allele that occurs after in-frame V(D)J joining, is associated with VLRs24,29,31. VLR assembly is probably as efficient as V(D)J recombination and generates an optimal size repertoire within the limited number of lymphocyte-like cells present in the larvae of jawless vertebrates. Although there is no direct proof, VLRs might undergo affinity maturation, placing this aspect of receptor somatic ‘refinement’ at an early stage of vertebrate evolution32. Although VLRs and immunoglobulins and TCRs are structurally unrelated and generate somatic variation through unrelated mechanisms, they develop clonal specificity through somatic recombination, show evidence for cell lineage-specific compartmentalization of receptor expression (see below) and might share some common aspects of immune receptor regulation. Therefore, the VLRs are important in our understanding of the origins of vertebrate adaptive immunity.
A case for transposon-mediated change
For over a decade, much of our thinking about the evolutionary emergence of adaptive immunity in jawed vertebrates was centred on the ‘big bang’ theory33,34 and related theories. The basis of this hypothesis is that a large number of genes, including those encoding immunoglobulins, αβ TCRs, γδ TCRs and MHC class I and II molecules, as well as cellular processes and organs that mediate adaptive immunity, were acquired over a short evolutionary period in a common ancestor of the modern jawed vertebrates. Therefore, rearranging forms of immunoglobulins and TCRs would not be expected to be present in jawless vertebrates or invertebrates, and the chance of detecting distant orthologues would be remote. Indeed, genome-wide analyses of vertebrates, invertebrate chordates and other deuterostomes has, in recent years, revealed no evidence for clear functional homologues of either the rearranging antigen-binding receptors or the MHC class I and II molecules beyond jawed vertebrates35–37. However, many functional modules that previously were thought to be markers of conventional adaptive immunity existed before the emergence of jawed vertebrates; some of these components probably had active roles in innate immunity, whereas others may have had non-immune functions (BOX 1). As we accumulate more information from deuterostome invertebrates and agnathans, the big bang theory of adaptive immunity seems to be more a set of observations made from a limited phylogenetic sampling and mechanistic focus than a true representation of rapid evolution.
Box 1. Diverse innate systems as a link to adaptive immunity?
Recent genome sequencing projects have revealed new forms of immunity that share characteristics with both conventional innate and adaptive immune systems. Innate receptors are encoded with fully defined specificities, susceptible to change only on an evolutionary scale86. The dominant paradigm describing the function of these receptor systems is centred on their recognition of immutable and defined microbial signature molecules. The receptors in turn are encoded in small gene families that collectively enable the recognition of a large portion of the microbial universe. Relative to many non-self recognition proteins, the genes encoding these innate receptors tend to be evolutionarily stable in both insects and vertebrates. However, large families of innate receptors have been characterized in several invertebrate species, including the purple sea urchin36,82, amphioxus35,84,87 and an annelid88. In these species, large, diversified families of innate immune receptors seem to have evolved quickly. Their patterns of incremental diversity are consistent with the recognition of close variants of evolving targets. The most diverse repertoire of innate receptors is found in the sea urchin genome, which encodes more than 200 Toll-like receptors (TLRs), approximately 300 nucleotide-binding oligomerization domain (NOD)-like receptors and 180 scavenger receptors with several scavenger receptor cysteine-rich (SRCR) regions. Diversity among the sea urchin TLR genes is most evident in the leucine-rich repeat (LRR) elements that are thought to recognize non-self molecules, and mutational changes are consistent with positive evolutionary selection89. Phylogenetic analysis suggests that they are pushed towards a highly diversified repertoire, a feature of adaptive immune system components. Whereas the apparent diversity even in these large innate immune systems is much less than the diversity generated in the adaptive immune systems of vertebrates, the possibility of further somatic or combinatorial variation remains to be explored. As these gene systems become more complex, the distinction between innate and adaptive immune systems begins to become less clear1,73,82,90. The phylogenetic distribution of these systems at the base of the chordate tree, coupled with molecules such as the variable region-containing chitin-binding proteins (VCBPs) suggests that the regulatory systems that control deployment of this innate diversity may have been available as a basis for selection systems in early vertebrate adaptive immune systems.
Despite the integral role of MHC class I and II molecules in adaptive immunity, it is the process of somatic rearrangement of immunoglobulin and TCR genes that is the common element of conventional adaptive immunity. V(D)J recombination involves the RAG1–RAG2 recombinase complex and DNA repair factors (BOX 2). The DNA repair factors are not specialized for the generation of immunoglobulin and TCR diversity but are integral components in the somatic process of segmental rearrangement. The RAG1–RAG2 gene pair is thought to be a remnant of an ancient RAG-like transposon (encoding RAG1 and RAG2 as a transposase complex) that entered the genome of a common ancestor of all jawed vertebrates33 and integrated into an ancestral immunoglobulin-domain gene locus, of which the nature is unclear, but it possibly served as an innate immune receptor (see below). This hypothetical event is a crucial, if not the most important, component of the big bang theory in which the generation of a functional immune receptor depends on the excision of the transposon (or remnants thereof), followed by an imprecise (untemplated) sealing of the genomic break, which gives rise to clonally diverse specificities. Subsequent recombination and gene duplication events then transferred the RAG1 and RAG2 genes to their current location outside of the immunoglobulin and TCR gene loci.
Box 2. DNA repair in V(D)J recombination.
The fundamental basis for conventional adaptive immunity involves the breakage and repair of DNA. During variable, diversity and joining region (V(D)J) recombination, the recombination-activating gene 1 (RAG1)–RAG2 recombinase complex generates double-strand DNA breaks at the border of the gene segments to be joined and their flanking recombination signal sequences (RSS). To join the four DNA ends, the RAG complex cooperates with ubiquitously expressed DNA repair proteins essential for non-homologous end-joining (NHEJ) (reviewed in REF. 91). The core components of NHEJ are Ku70, Ku80, cernunnos (also known as NHEJ1 and XLF1), X-ray-repair cross-complementing protein 4 (XRCC4) and DNA ligase IV. Loss of any one of these components abolishes V(D)J recombination.
The repair features are not unique to vertebrates but can be found in species as phylogenetically removed from vertebrates as yeast. Ku homologues are present in bacteria92. V(D)J recombination also depends on both artemis and catalytic subunit of DNA-dependent protein kinases (DNA-PKcs) 93, both of which are less well conserved in evolution. However, their homologues have been identified in the sea urchin genome, suggesting they were present in the common ancestor of all living deuterostomes. Of the lymphocyte-specific repair factors, only the template-independent DNA polymerase terminal deoxynucleotidyl transferase (TdT) acts exclusively in V(D)J recombination. TdT adds non-templated (N-) nucleotides to the DNA ends before the action of the end-joining factors; it is a close relative of DNA polymerase-μ. Only one such polymerase, which is related equally to TdT and polymerase-μ, is present in the sea urchin genome. It is important to note that polymerase-μ has been recently found to contribute to N-nucleotide diversity independently of TdT in the context of V(D)J recombination in embryos94. Therefore, the DNA repair module that operates in the joining phase of V(D)J recombination probably was available before the appearance of the RAG transposon and subsequently was co-opted for adaptive immune function.
The next stages in the evolution of the conventional adaptive immune response of jawed vertebrates involved locus duplication, tandem element duplication and subsequent differentiation that gave rise to gene segments in the immunoglobulin and TCR gene loci. Thus, an ancestral single-specificity antigen receptor became a highly variable receptor, which might have functioned in innate immunity and given rise to the heterodimeric immunoglobulin and TCR systems found in the jawed vertebrates. Transcriptional and translational control mechanisms, as well as other cellular adaptations, such as those that restrict expression of a single receptor specificity to an individual clonal immune cell, are seen in lymphocytes of both jawed38 and jawless24,29,31 vertebrates and might have been features of the predecessor of the complex system that regulates the expression of these diverse receptors rather than core elements of the big bang theory.
The identification of the RAG1- and RAG2-like gene cluster in the echinoderm purple sea urchin (Strongylocentrotus purpuratus)39 (FIG. 2) and sequence elements with similarity to RAG1 in amphioxus (Branchiostoma floridae), a member of the subphylum Cephalochordata, and other invertebrates35, as well as the discovery of Transib transposons40 encoding a transposase that has sequence homology to the RAG1 core region (but not to RAG2) flanked by RSS-like elements, provide new insights into the origins of segmental recombination. These observations raise two plausible explanations for the origins of the Deuterostome RAG1- and RAG2-like gene clusters: the first possibility is that an ancestral RAG1-like transposon entered the genome of a common Deuterostome ancestor at the site of an ancestral endogenous RAG2-like gene far earlier than anticipated. The second possibility is that an as yet unidentified RAG1- and RAG2-containing transposon was integrated independently into the lineage leading to the sea urchin and an early jawed vertebrate ancestor. A third possibility, that a RAG1 transposon integrated independently near an endogenous RAG2-like gene in each lineage, is unlikely41 given the close linkage of RAG1- and RAG2-like genes in sea urchins.
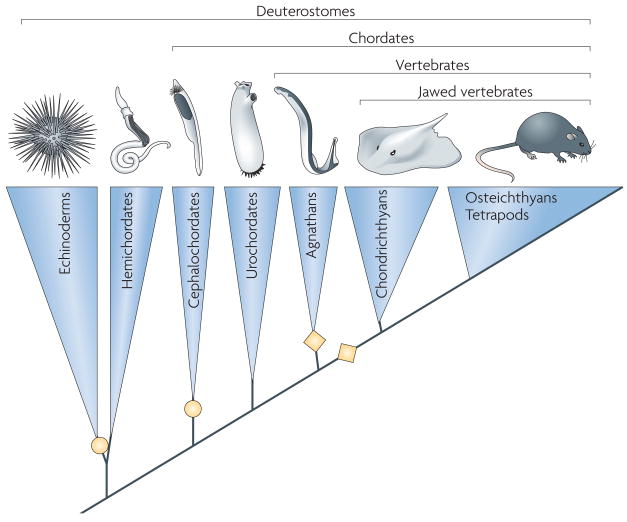
Figure 2. Relationships among animal phyla and subphyla with direct bearing on the origins of V(D)J recombination in jawed vertebrates.
Major groups from the deuterostome superphylum are shown (a small phylum Xenoturbellida is omitted). Recent genome sequences from each of these clades are rapidly increasing our understanding of the origins of jawed vertebrate adaptive immunity. Hemichordates have features of chordates, with which they were once placed, but now are classed as a sister group to the echinoderms. Yellow diamonds indicate placement of two forms of adaptive immunity in jawed and jawless vertebrates; yellow circles indicate clades with complex multigene families encoding innate immune receptors.
Although RAG1 seems to have a longer phylogenetic history, the origins of RAG2 are of equal importance. RAG2 shares no transposase features with RAG1 and might not have been part of the original RAG transposon. RAG2 has a plant homeodomain, a feature shared with many chromatin-associated factors, and interacts with chromatin, particularly at histone H3 that is dimethylated or trimethylated at lysine 4 (H3K4me2 or H3K4me3, respectively)42–44. In developing lymphocytes, RAG2 binds a large number of regions of the genome with H3K4me3, whereas RAG1 specifically binds the immunoglobulin and TCR gene loci45. Therefore, it is RAG1, and not RAG2, that confers the sequence specificity for V(D)J recombination. The chromatin-binding specificity of RAG2 suggests that the ancestor of RAG2 may have functioned in regulating the transcription and/or chromatin structure necessary for the recombination process rather than as a direct mediator of recombination.
The introduction of RAG activity to ubiquitous DNA repair processes, which are necessary for general cell survival, would introduce the capacity for limited somatic variation in an ancestral immune cell and was probably the first of many evolutionary acquisitions that do not have a direct genetic relationship to the generation of immunological diversity. Alternatively, this process could have ‘piggybacked’ on previously existing receptor gene diversification processes that relied on AID-related DNA deaminases (see below)46–48. The identification and functional characterization of AID-like deaminases in jawless vertebrates30 and their circumstantial association with VLR diversification could reflect a primary role of deaminase-mediated gene diversification in the emergence of vertebrate adaptive immunity. As AID-dependent processes involve DNA strand breaks, they could have rendered these receptor genes prime targets for the acquisition of mobile elements. Collectively, these observations spread the genetic innovations that are associated with V(D)J recombination over a longer period of animal evolution than was previously thought. New observations in jawless vertebrates and invertebrates have the potential to provide key insights into the origins of the V(D)J rearrangement mechanism and the cellular innovations that allow efficient selection and regulation of non-self binding potential. A better understanding of the evolution of these systems can be gained by reducing this elaborate process to a series of acquisitions, integrations and adaptations that occurred over a protracted period of phylogenetic time in the lineage leading to the jawed vertebrates, as well as in specific branches of vertebrate evolution as represented in modern species (FIG. 3).
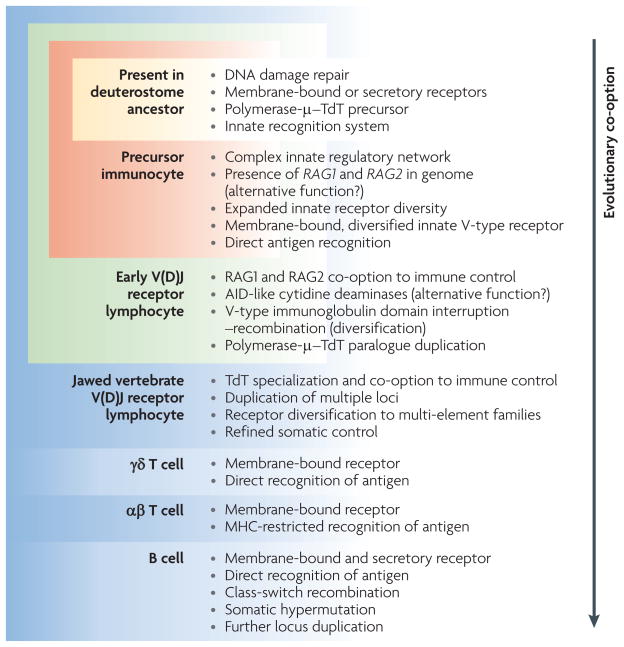
Figure 3. Evolutionary co-option of ancient biological systems into lymphocytes that express V(D)J receptors.
The cumulative acquisition of jawed vertebrate lymphoid properties within the lineage from an invertebrate ancestor to a modern jawed vertebrate is illustrated. The deuterostome ancestor probably had a complex form of innate immunity and the capacity to carry out unrelated, general cellular processes that are integral to modern jawed vertebrate immune cell (lymphocyte) function. The precursor immunocyte represents a collection of features inferred from genome sequence analyses and functional biochemistry of invertebrate deuterostomes, including a single homologue (polymerase-μ–TdT) that possesses features of both TdT (terminal deoxynucleotidyl transferase) and polymerase-μ (a related enzyme). The proposed early V(D)J receptor lymphocyte is based on observations in both jawed and jawless vertebrate lymphocytes, although jawless vertebrates use variable lymphocyte receptors (VLRs), as well as unrelated mechanisms of immune receptor diversification and membrane anchoring. A role has been proposed for activation-induced cytidine deaminase (AID)-related molecules in the generation of VLR diversity in lampreys; however, the relationship to AID-based somatic hypermutation and class-switch recombination in jawed vertebrate V(D)J receptor systems has not been resolved. Some aspects of the diversification of early lymphocytes into B cell- and T cell-like type cells may be a general feature of both jawed and jawless vertebrates or reflect convergent evolution. At this stage in the evolution of jawed vertebrate adaptive immunity, a genetic event that disrupted a V-type immunoglobulin domain formed a site for genetic recombination and subsequent receptor diversification. The characteristics of the jawed vertebrate V(D)J receptor lymphocyte represent the main cellular and structural features that are widespread among various taxonomic groups. Diversification of lymphocyte form and function reflects a complex integration of diverse biological features.
Expansion of the potential of V(D)J recombination
Although segmental rearrangement of an undefined receptor may have greatly increased the recognition potential for antigen, a mechanism to further amplify the repertoire by selectively increasing joining diversity using pre-existing cellular DNA repair pathways would be both parsimonious and advantageous in terms of repertoire expansion. The RAG1–RAG2 recombinase complex introduces DNA breaks that result in DNA hairpins, which are inconsistent with the function of general DNA repair machinery. However, enzymatic opening of the hairpins a few nucleotides away from the tip forms incompatible single strand overhangs that undergo untemplated processing before joining. A limited degree of sequence diversity is introduced through this process.
Extensive studies in higher vertebrates have highlighted the role of terminal deoxynucleotidyl transferase (TdT), a lymphocyte-specific template-independent DNA polymerase, to introduce untemplated nucleotides at the junctions of V, D and J segments. It is probable that a gene regulatory mechanism that enabled controlled expression of a TdT paralogue in an early lymphocyte created a means to mediate extensive and complex genetic variation in complementarity determining region 3 (CDR3) of the immunoglobulin and TCR ancestor. The importance of TdT for conventional adaptive immunity is apparent in the low level of variation in fetal progenitors of B-1 B cells, which do not express this polymerase49. The gene duplication event that gave rise to TdT and polymerase-μ (a related enzyme) in jawed vertebrates probably occurred in a vertebrate ancestor, as only a single polymerase-μ–TdT homologue is found in the genomes of the sea urchin36 and amphioxus35. This gene duplication event was a crucial step in the development of a highly diverse immune repertoire (FIG. 3) and along with other duplicate specialization for the emergence of the jawed vertebrate adaptive immune system2.
Immunoglobulin: surface receptor and soluble effector
There is little information from which to predict the nature of the initial rearranging events of the earliest monomeric or multimeric antigen-binding receptors. Antigen-binding receptors consisting only of immunoglobulin heavy chains (as a homodimer) have been described in species as phylogenetically separated as camelids and sharks, but these are probably derived evolutionary features6. Membrane-bound immunoglobulins are expressed earlier in ontogeny than the secreted forms; both forms are found in jawed vertebrate lymphocytes.
Differences between membrane-bound and secreted forms of immunoglobulin arise from alternative RNA processing and a choice of alternative poly(A) sites50–53. The functionality of the one gene (that is, one antigen specificity)–two isoforms (membrane-bound and secreted) system is that the membrane-bound receptor allows for selection, diversification and affinity maturation, in which continuous feedback occurs between the immunoglobulin receptor and the nucleus of the B cell. In all jawed vertebrates, these two receptor isoforms are associated with the heavy chain isotypes and give a broad physiological advantage. The programmed switch between membrane-bound and secreted immunoglobulin occurs independent of V(D)J recombination and is a shared character of other signalling receptors such as FAS (also known as CD95), granulocyte–macrophage colony-stimulating factor (GM-CSF) receptor and interleukin-4 (IL-4) receptor (reviewed in REF. 54). A more elaborate innovation is seen in the terminal cellular differentiation event associated with the shift from transmembrane expression to secretion in jawed vertebrates, in which the rate of immunoglobulin transcription in plasma cells is several orders of magnitude higher than in resting B cells. Although it remains to be proven, similar changes occur in the VLRB-expressing cells of agnathans (see below).
Further diversification of immunoglobulin
Because clonally expressed antigen-binding receptors arise by chance, several rounds of modification and selection offer an efficient means for achieving high affinity. Furthermore, in the context of an immunological response to disease, the function of a cell type expressing a receptor could be neutralized by antigenic shifts in pathogens. The ability to modify and enhance the function of clonally expressed receptors offers enormous selective advantages, as would effector cell memory. Rearranging immunoglobulin genes undergo affinity maturation, which selects for B cells that encode higher affinity antibody molecules specific for the respective antigen. Both SHM and gene conversion, which are AID-mediated mechanisms of somatic variation, can alter the affinity and specificity of a recombined antibody V region. In avians, as well as in various other mammalian species55, gene conversion driven by AID is the primary mechanism of immunoglobulin diversification56,57. AID and SHM are features of all living jawed vertebrates, including cartilaginous fish, which show affinity maturation58,59. Furthermore, the APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 3) family of cytidine deaminases, which includes AID, has a long evolutionary history and can function as innate defence mechanisms against retroviruses and retrotransposons60. It is possible that all cytidine deaminases have a common ancestor that functioned in some aspect of immunity.
AID also mediates CSR, which involves a change to a clonally committed immunoglobulin receptor that is unrelated to the primary binding functions of its V regions. Specifically, CSR effectively grafts a ‘selected’ primary antigen-binding specificity onto polypeptide sequences that mediate different secondary biological functions (FIG. 1). Relative to mammals, the most divergent forms of CSR have been shown to occur in amphibians61, and it occurs throughout the tetrapods1,6. To a certain degree, CSR is driven by the nature of the antigen and/or pathogen that is encountered. Given that SHM can be traced to cartilaginous46,48 and bony fish62, which lack CSR, and assuming that these main phylogenetic groups did not evolve another system to somatically vary rearranged genes, it is possible that AID functioned in heavy chain CSR in a tetrapod common ancestor, making it a secondary function of AID. The presence of IgD throughout tetrapods, and possibly other jawed vertebrates 11, suggests that AID may have been co-opted into the primordial CSR system to expand the number of constant regions beyond the limitations of alternative splicing.
Notwithstanding the role of MHC class I and II molecules in presenting antigen to αβ TCRs, it is the role of AID in B cells and not in T cells that defines many differences in the immune functions of these cell types. The absence of an effect of AID on the loci encoding the TCR α- and β-chains in mammals could be interpreted to indicate that V(D)J recombination arose first. However, AID-mediated receptor diversification could have been lost in the TCRs as a means to lessen the likelihood of generating receptors that recognize self antigens.
In a departure from our general view of TCR diversification, in cartilaginous fish, the TCR γ-chain can undergo somatic mutation63. This could be a derived feature of sharks but, alternatively, it might reflect an ancestral state that is similar to antigen recognition by immunoglobulins and γδ TCRs. γδ TCRs vary considerably across vertebrates, ranging from highly specialized, virtually innate canonical receptors64 to receptors that are highly diversified in species with high number of circulating γδ T cells65. Studies in one species of cartilaginous fish revealed a high degree of diversity and abundant expression of γδ TCRs13. Further phylogenetic investigations of this poorly studied receptor class may shed new light on ‘primitive’ V(D)J receptor systems.
As suggested earlier, the identification of AID-like deaminases in lamprey lymphocytes, combined with the apparent absence of RAG1 and RAG2 in jawless vertebrates, led to the suggestion that deaminase-mediated diversification of antigen receptors might have preceded transposition-based generation of immune repertoires30. This particular interpretation assumes that the absence of the V(D)J system in lampreys and hagfish is not the result of gene loss. Current available data do not allow firm phylogenetic ordering of whether AID-type diversification already influenced in the genes encoding a primordial receptor displayed on the surface of immune cells or was adapted after the specification of distinct lymphocyte lineages.
Antigen presentation
It is difficult to determine whether antigen presentation preceded or succeeded the emergence of a selectable, rearranging αβ TCR, as we cannot identify clear MHC class I and II homologues outside of the jawed vertebrates, which is also the point in phylogeny at which MHC components involved in antigen processing arose2. One appealing argument is that as immunoglobulin and diversified γδ TCRs recognize native antigen independently of MHC class I and II molecules, these diversified antigen-binding receptors are the primitive mode of function. The cellular recombination and antigen processing mechanisms that are integral to αβ TCR function may have originated through the co-option of phylogenetically ancient processes of phagocytosis and lysosomal processing as a means to direct the enormous functional advantage of V(D)J-mediated recognition to intracellular or endocytosed antigens, although alternative explanations for the origins of antigen presentation have been proposed66. It is particularly notable that both antigen-binding receptors and MHC class I molecules are members of a limited family of molecules that contain C1 immunoglobulin-like domains. The interactions between αβ TCRs and MHC molecules might be a highly derived system that originated in the cross-regulation of lymphocytes mediated by receptor affinity for different epitopes of a common antigen. In such a case, recognition specificity brought cells expressing different clonal receptors into close proximity for cross-regulatory signalling. The system could have originated from one that was similar to the current αβ T cell–APC mechanisms of cross-regulation.
The origin of lymphocytes
Lymphoid organs and lymphocytes have three distinct roles: to provide suitable microenvironments for the development of immune effector cells, mediate quality control (positive and negative selection) and regulate the efficacy of the immune response (memory and suppression)67. In jawed vertebrates, the B and T cell lineages define adaptive immunity, although some exceptional differences in tissue-specific distribution14 and, in the case of B cells, alterative functions, including primary phagocytosis and antimicrobial activities, have occurred68.
In lampreys, lymphoid-like cells22 are present in the pharyngeal margins, supraneural body (known as protothonephros), intestine (known as typhlosole) and kidney (known as opisthonephros). Several transcription factors and other regulatory factors that function in lymphoid cell development in jawed vertebrates have been identified in lamprey lymphocyte-like cells (reviewed in REF. 20). Lamprey lymphocyte-like cells are classified by their rearranging surface receptors VLRA and VLRB, which notably show antigen responses and cytokine expression profiles that are in some ways similar to jawed vertebrate T and B cells, respectively. Cell transfection studies with recombinant VLRs showed that VLRA functions as a membrane-bound molecule, whereas VLRB is secreted69. This notable parallel to jawed vertebrate immunity suggests that diversifying systems of immune receptors either tend towards the emergence of cells expressing membrane-bound or secreted receptors as a necessary mechanism of regulation (albeit one achieved through mechanisms unrelated to those used for immunoglobulins) or that the lamprey lymphocyte-like cells reflect aspects of a close homology to B and T cells, which nonetheless express unrelated forms of somatically diversifying receptors69 (BOX 3).
Box 3. Gene regulatory networks.
Immune cells are ubiquitous among bilaterians but it remains unclear how these cells relate among disparate phyla and how lymphocyte systems emerged from these cells. To understand the origins of lymphocytes, we need unambiguous and specific characteristics by which to compare cell types and functions. Cell morphology is highly dynamic, and immune mechanisms evolve too quickly for this purpose. Shared expression of transcription factors is useful as evolutionary relationships among this class of genes are easy to reconstruct. Nonetheless, virtually all transcription factors have several functions at different developmental stages and in different cell types. Interpretation of the importance of shared patterns of transcription factors is confounded further by gene duplication and paralogue specialization especially across invertebrate and vertebrate phyla. It is often difficult to distinguish co-option of dedicated modular genetic programmes that may be useful to immune functions (for example, cell motility programmes) but independently derived in the animal lineages being compared from a case where shared use truly reflects a common ancestral function. So the meaning of shared transcription factor use among invertebrate immune cells and vertebrate lymphocytes will become definitive only as their interactions are assembled into more specific gene regulatory networks. In this way, basal elements of regulatory circuitry95 can be defined and used for reconstructing what probably is a complex history of immune cell evolution and systems co-option that gave rise to lymphocytes.
Insights from specialized B and T cell subsets
Some classes of B and T cells express antigen-binding receptors with recognition features that are more closely related to those of innate immune receptors than immunoglobulins and αβ TCRs. Limited or fully restricted use of segmental elements and junctional diversity are two mechanisms to generate such receptors. Such receptors are expressed by canonical γδ T cells, invariant natural killer T (iNKT) cells and B-1 B cells; cells that often arise early in development. Although it is tempting to view the genetic simplicity of these antigen-binding receptors and early ontogeny as a carryover from a primitive state, these cells might also be more recently derived developmental shortcuts that can contribute to an early immune response. Joined and partially joined immunoglobulin genes in the germline of sharks and skates reflect a well-defined case of reversion to innate immunity1. These germline-joined genes could be the ultimate form of restricted diversity among antigen-binding receptors, but they are generally thought to be a derived feature of gene organization that is possible in the presence of multiple gene loci. Innate immunity has its definitive strengths and the prejoined immunoglobulin genes, B-1 B cells and canonical γδ T cells might reflect a systematic capacity for revision and specialization.
Earliest conventional adaptive immune receptor
Despite the remarkable insights into the origins of adaptive immunity that have been gained from in-depth studies of somatic rearrangement, the absence of key intermediate species and the rapid evolution of rearranging immune receptor elements confound our ability to infer the nature of the evolutionary predecessors of immunoglobulins and TCRs and the relative order of events in their divergence. Nevertheless, assuming protection from infection as the primary driver of receptor selection, viral or bacterial receptor molecules encoding either a V or closely related intermediate (I)-type domain would be attractive candidates for the earliest receptors70. Although molecules such as viral receptors can be viewed as pathogen adaptation to a host cell, they could function in protection if they were adapted to a secretory function that could block viral attachment to the cell70. Any number of other related molecular types could also have been an effective antigen-binding receptor precursor into which a rearrangement mechanism was introduced. Some attention has been directed to ‘simpler’ non-rearranging V-type receptors such as CD8β, VpreB (also known as Igι), THY1 and monomorphic V-encoding NK cell receptors as primordial immunoglobulin-type receptors. Although these molecules have limited diversity in their current forms and may reflect derived rather than primitive systems, further investigation of their functions is warranted.
Similarly, the non-rearranging V gene-containing molecules such as novel immune-type receptors (NITRs)71, which are highly diversified and show allogeneic-binding specificity72, are more likely to have diverged from an immunoglobulin or TCR gene cluster than be a modern manifestation of an ancient receptor family. Where in the evolutionary tree the diversified V regions are first seen is potentially relevant to species that diverged before the emergence of the vertebrates and may retain some aspect of ‘primitive’ immune receptor structure and function.
The variable region-containing chitin-binding proteins (VCBPs) found in cephalochordates73 (such as amphioxus) and urochordates37 (such as the sea squirt Ciona intestinalis) (FIG. 2) are a diverse multigene family of proteins with a V domain and a chitin-binding domain, both of which can potentially recognize pathogens. The V domains have a high level of regionalized polymorphisms74 localized to a portion of the V domain that is not used in primary antigen recognition by immunoglobulins and TCRs75. VCBPs are expressed by cells that localize to the gut (in amphioxus and the sea squirt)73, by other immune-type cells (haemocytes in the sea squirt) and in discrete areas of pharyngeal tissues (in amphioxus), a distribution consistent with immune-type surveillance (L. J. Dishaw, R. DeSantis, M. R. Pinto and G.W.L., unpublished observations). In addition, amphioxus VCBPs have pronounced haplotype variability, which shares many characteristics of the variation seen in immune effector molecules such as killer cell immunoglobulin-like receptors (KIRs) in humans76 (L. J. Dishaw and G.W.L., unpublished observations).
The high level of regionalized sequence variation in VCBPs is consistent with immune function. In some ways this effect, described here as a putative innate immune receptor, is reminiscent of the high level of complexity seen in other innate immune receptors in amphioxus and sea urchins. Other diversified multigene systems of non-rearranging V-region genes have also been identified in the sea urchin36 and are of considerable interest (BOX 1).
Once the basic mechanisms of immunoglobulin and TCR diversification arose, the B cell receptor (BCR) and TCR systems seem to have evolved on different trajectories; the TCR system has remained generally similar throughout vertebrates, whereas the immunoglobulin system has adopted specialized characteristics in different species. There is no clear explanation for the marked differences in the form and function of BCRs in different vertebrates other than the possibility that the gene loci, which to varying degrees are genomically organized to facilitate somatic rearrangement in every B cell, may be particularly susceptible to marked interspecies variations; cases as extreme as the recombining of immunoglobulin VH regions with TCR Vδ gene segments have been reported77–79.
Cellular support systems for receptor diversity
Somatic diversification of receptors is one of many molecular mechanisms that evolved to support the clonally dependent vertebrate adaptive immune system. The cell types and cell communication mechanisms that are required to select, implement and control recognition diversity reflect an even richer and more complex set of biological innovations than receptor recombination itself; the origins of which are likely to predate the V(D)J systems. Many questions remain as to the nature of lamprey lymphocyte-like cells, but several lines of evidence suggest that these cells have similar regulatory mechanisms to jawed vertebrate lymphocytes. Furthermore, the vertebrate adaptive immune system can be viewed as a case where V(D)J or VLR recombination mechanisms of receptor diversification (it is not clear which came first) were integrated into a pre-existing mechanism of cellular selection that preceded the modern vertebrate form of the V(D)J system69.
These observations raise the more important question of whether cellular selection or a somatic receptor diversification system arose first, although it seems unlikely that useful receptor diversity could be achieved without some form of cellular selection already in place. It is probable that a system of extensive receptor diversification would be detrimental in the absence of a system that selects useful specificities. To date, there is no compelling evidence that lymphocyte-like cells exist outside of vertebrates; however, elements of the gene regulatory networks that operate in lymphocytes may be part of morphologically distinct immune cells of invertebrate deuterostomes. Some of these regulatory programmes might have been co-opted from more general progenitor cell programmes80. The question arises as to how such an intricate cell system could have arisen in an incremental manner. Diversity in the context of conventional multigene families is a common feature of immune systems, such as lectins and the invertebrate V-region immunoglobulin-domain genes mentioned earlier. Although molecular variation in such cases is orders of magnitudes below that generated by the adaptive immune system, they probably require similar cellular selection mechanisms, a hypothesis that is supported in part by the elucidation of the mechanisms underlying the restricted expression of members of some moderate size gene families encoding NK cell receptors81.
Recent genome sequences suggest that vertebrates arose from a chordate ancestor with innate immune diversity that exceeds that of modern vertebrates35,36,82,83 (BOX 1). Large innate multigene families of this type may have provided the initial pressure to evolve cell-based mechanisms of selection. Diversifying receptors might then have emerged within this pre-adapted framework. It is of note that the genome of amphioxus reflects a capacity to induce changes in innate receptor signal transduction pathways through domain shuffling; such processes could rapidly alter immune networks in evolving immune cells84. Functional characterization of more divergent immune cells in invertebrate deuterostomes will help to clarify the origins of vertebrate adaptive immunity and may extend our concepts of the evolutionary period over which most parts of the cellular machinery of the jawed vertebrate adaptive immune system emerged.
Conclusions
Detailed analyses of the somatic events that give rise to receptor variation in vertebrate species have provided considerable insight into the evolution of adaptive immunity. The dynamic nature of receptor evolution is apparent even among classes of jawed vertebrates, in which structural variants that share a single general form but exhibit entirely different mechanisms of diversification have been characterized. In jawed and jawless vertebrates, extraordinary selective forces have promoted the evolution of adaptive immune receptors that differ in structural class and mechanism of recombination but seem to have converged on fundamentally similar functions. The cellular networks that underlie the functions of these unrelated receptors may be homologous and reflect a system for specificity selection that is far more ancient and abstract than previously realized. Alternatively, the similarities may reflect a case for evolutionary convergence and shed light on limitations of the underlying basis for the evolution of adaptive immune systems.
Genomic investigations of other deuterostome invertebrate groups indicate that immune receptor diversity and the elements that create structural variation in vertebrate adaptive immune responses have far longer evolutionary histories than previously envisioned. In this regard, looking beyond the vertebrates has brought additional insight into the ancient nature of immunity and the variation in the pathways and patterns that create, sustain and select for immune receptor diversity. Despite the intrinsic differences in immunity within groups of species, there is no reason to assume that vertebrates require a complex immune system any more than do complex invertebrates85. Acquisition, integration and regulation of basic physiological processes, which typically are not associated with immunity, coupled with the capacity to effect large generational change within genomes extended the potential for recognition of non-self beyond conventional innate immunity. Further studies of the mechanisms underlying these changes and detailed studies of the mechanisms that regulate the development and function of immune cells in jawless vertebrates and closely related invertebrates have elucidated, and continue to elucidate, the fundamental basis of immunity as an integrated biological process.
Acknowledgments
The work in the laboratory of G.W.L. is supported by US National Institutes of Health grants AI23338 and AI57559. The work in the laboratory of J.P.R. is supported by Canadian Institutes for Health Research grant MOP74667 and National Science and Engineering Research Council of Canada grant NSERC 458115/211598. The work in the laboratory of S.D.F. is supported by the Intramural Research Program of the US National Institutes of Health, National Institute on Aging.
- Somatic hypermutation
(SHM). The process by which point mutations are introduced in the heavy- or light-chain variable region gene segments, resulting in a change in the expressed protein, which may alter affinity or specificity for antigen
- V(D)J recombination
The lineage-specific RAG1- and RAG2-mediated assembly of functional immunoglobulin and T cell receptor genes from individual variable (V), diversity (D) and joining (J) gene segments. This process occurs exclusively in B and T cell progenitors
- Recombination signal sequences
(RSSs). Highly conserved heptamer and nonomer sequences that flank recombining segmental elements in immunoglobulin and T cell receptor genes. These sequences are separated by 12 or 23 base pair spacers that are required to direct the recombination of compatible elements
- Recombination-activating gene 1
(RAG1). A protein that, along with RAG2, forms a DNA recombinase complex that catalyses V(D)J recombination. During the joining phase additional ubiquitous DNA repair factors are recruited. Both genes are exclusively expressed in developing lymphocytes
- Gene conversion
An immunoglobulin gene diversification process closely related to somatic hypermutation. In general, U:G base mismatches are repaired by copying sequence information from upstream pseudo V segments into the rearranged VJ and VDJ exon of the IgL and IgH genes, respectively. In birds and rabbits it occurs before antigen encounter
- Tetrapods
Four-limbed vertebrates that include the amphibians and amniotes (reptiles, birds and mammals). These groups share some common immune features, such as class-switch recombination
- Activation-induced cytidine deaminase
(AID). An enzyme that acts on single-stranded DNA and converts cytidines to uracils. It is typically expressed in B cells after antigen encounter. Its mutagenic activity is restricted to immunoglobulin loci
- Phylogeny
The study of evolutionary relatedness among groups or species of organisms. Similar analyses can be applied to relatedness among genes
- Lampreys and hagfish
Two extant groups of the jawless vertebrate lineage that have anatomical and physiological differences from jawed vertebrates. Lampreys undergo metamorphosis from a larval form, termed an ammocoete, and approximately half of the species are parasitic in adult life. Hagfish are ocean dwelling species that feed on decaying animal matter and small crustaceans
- Leucine-rich repeat (LRR)
A protein structural motif that is comprised of 20–30 amino acid regions that are unusually rich in the hydrophobic amino acid leucine and form a characteristic structural fold
- Copy choice mechanism of recombination
A genetic recombination mechanism in which a new DNA sequence is generated by replication using multiple DNA sequences as templates. The DNA polymerase continues to synthesize a single DNA molecule while jumping from one template strand to another. The choice of templates is largely driven by sequence homologies, allowing the previously synthesized DNA to anneal to similar sequences elsewhere in the genome and thereby redirecting DNA synthesis
- Chordates
An animal phylum that comprises both jawed and jawless vertebrates, and two subphyla of invertebrates: the cephalochordates (such as amphioxus) and the urochordates (such as the sea squirt Ciona intestinalis)
- Deuterostomes
An animal superphylum composed of four phyla: the chordates (which include vertebrates), the echinoderms (consisting of starfish, sea urchins and allied species), the hemichordates (acorn worms) and Xenoturbellida (containing two marine worm-like species). Genomes from each of the three major deuterostome phyla have been sequenced
- Transposons
(Also are known as ‘jumping genes’ and ‘selfish DNA’). DNA sequences that encode transposases, the enzymes required to excise the transposon from its original chromosomal location and to integrate it in a different position with the genome. The ends of transposons consist of DNA repeats that serve as recognition sites for the transposase itself
- Purple sea urchin (Strongylocentrotus purpuratus)
. An echinoderm that is a well established model system for developmental biology. Sequencing of its genome increased the interest of the broader scientific community (including immunologists) in this invertebrate model system
- Complementarity determining region 3 (CDR3)
The region in B and T cell receptors that interacts with the antigen, in which hypervariable sequences are located. CDR3 typically is derived somatically, whereas CDR1 and CDR2 are germline encoded
- B-1 B cells
In mice, this B cell subset is found mainly in the peritoneal and pleural cavities. B-1 B cells are a self-renewing subset with a restricted repertoire of B cell receptors that respond to common bacterial antigens and have a role in autoimmunity
- Derived evolutionary features
Anatomical structures, genes or functional systems are designated as derived when they originate within a sub-lineage. Primitive evolutionary features refer to those that originate from similar features in a common ancestor
- Invariant natural killer T (iNKT) cells
Lymphocytes that express a particular variable gene segment, Vα14 (in mice) and Vα24 (in humans), precisely rearranged to a particular Jα gene segment to yield T cell receptor α-chains with an invariant sequence
- Novel immune-type receptors (NITRs)
Activating or inhibitory receptors that are encoded by large diversified multigene families in bony fish. All NITRs have a variable region and most have a transmembrane region and cytoplasmic tail with signalling motifs
Footnotes
Competing interests statement.
The authors declare no competing financial interests.
DATABASES
UniProtKB: http://www.uniprot.org
AID | CD8β | RAG1 | RAG2 | TdT | THY1 | VpreB
FURTHER INFORMATION
Gary W. Litman’s homepage: http://health.usf.edu/medicine/pediatrics/bios/bio_litman.asp
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nature Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nature Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 5.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 6.Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nature Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- 7.Kokubu F, Litman R, Shamblott MJ, Hinds K, Litman GW. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J. 1988;7:3413–3422. doi: 10.1002/j.1460-2075.1988.tb03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson MK, Shamblott MJ, Litman RT, Litman GW. Generation of immunoglobulin light chain gene diversity in Raja erinacea is not associated with somatic rearrangement, an exception to a central paradigm of B cell immunity. J Exp Med. 1995;182:109–119. doi: 10.1084/jem.182.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci USA. 2003;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, et al. Identification of IgF, a hinge-region-containing Ig class, and IgD in Xenopus tropicalis. Proc Natl Acad Sci USA. 2006;103:12087–12092. doi: 10.1073/pnas.0600291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci USA. 2006;103:10723–10728. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards MH, Nelson JL. The evolution of vertebrate antigen receptors: a phylogenetic approach. Mol Biol Evol. 2000;17:146–155. doi: 10.1093/oxfordjournals.molbev.a026227. [DOI] [PubMed] [Google Scholar]
- 13.Rast JP, et al. α, β, γ, and δ T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 14.Miracle AL, et al. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int Immunol. 2001;13:567–580. doi: 10.1093/intimm/13.4.567. [DOI] [PubMed] [Google Scholar]
- 15.Zapata A, Amemiya CT. Phylogeny of lower vertebrates and their immunological structures. Curr Top Microbiol Immunol. 2000;248:67–107. doi: 10.1007/978-3-642-59674-2_5. [DOI] [PubMed] [Google Scholar]
- 16.Pollara B, Litman GW, Finstad J, Howell J, Good RA. The evolution of the immune response. VII. Antibody to human “O” cells and properties of the immunoglobulin in lamprey. J Immunol. 1970;105:738–745. [PubMed] [Google Scholar]
- 17.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Ann Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 18.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 19.Alder MN, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nature Immunol. 2008;9:319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 20.Saha NR, Smith J, Amemiya CT. Evolution of adaptive immune recognition in jawless vertebrates. Semin Immunol. 2010;22:25–33. doi: 10.1016/j.smim.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uinuk-Ool T, et al. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci USA. 2002;99:14356–14361. doi: 10.1073/pnas.212527699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer WE, et al. Isolation and characterization of lymphocyte-like cells from a lamprey. Proc Natl Acad Sci USA. 2002;99:14350–14355. doi: 10.1073/pnas.212527499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajoghli B, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–197. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. An important paper that reports the existence of a new diversified antigen receptor that is the basis for adaptive immunity in jawless vertebrates. [DOI] [PubMed] [Google Scholar]
- 25.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102:9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velikovsky CA, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nature Struct Mol Biol. 2009;16:725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nature Immunol. 2007;8:206–213. doi: 10.1038/ni1419. The authors provide the first experimental data in support of a copy choice model for the assembly of VLR receptor genes in jawless vertebrates. [DOI] [PubMed] [Google Scholar]
- 30.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID–APOBEC family cytosine deaminase. Nature Immunol. 2007;8:647–656. doi: 10.1038/ni1463. The authors identify two cytidine deaminases with homology to AID and, based on the expression pattern, suggest that these enzymes are involved in the diversification of the VLR repertoire in lampreys. [DOI] [PubMed] [Google Scholar]
- 31.Kishishita N, et al. Regulation of antigen-receptor gene assembly in hagfish. EMBO Rep. 2010;11:126–132. doi: 10.1038/embor.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasumi S, et al. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA. 2009;106:12891–12896. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 34.Marchalonis JJ, Schluter SF, Bernstein RM, Hohman VS. Antibodies of sharks: revolution and evolution. Immunol Rev. 1998;166:103–122. doi: 10.1111/j.1600-065x.1998.tb01256.x. [DOI] [PubMed] [Google Scholar]
- 35.Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hibino T, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 37.Azumi K, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- 38.Eason DD, Litman RT, Luer CA, Kerr W, Litman GW. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. doi: 10.1002/eji.200425224. [DOI] [PubMed] [Google Scholar]
- 39.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. This is the first report of a RAG1- and RAG2-like gene cluster outside the jawed vertebrate lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. This report identifies the striking similarity between RAG1 and transib transposons, a widespread family of mobile DNA elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fugmann SD. The origins of the Rag genes — from transposition to V(D)J recombination. Semin Immunol. 2010;22:10–16. doi: 10.1016/j.smim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson DR, Norton DD, Fugmann SD. The PHD domain of the sea urchin RAG2 homolog, SpRAG2L, recognizes dimethylated lysine 4 in histone H3 tails. Dev Comp Immunol. 2008;32:1221–1230. doi: 10.1016/j.dci.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji Y, et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinds-Frey KR, Nishikata H, Litman RT, Litman GW. Somatic variation precedes extensive diversification of germline sequences and combinatorial joining in the evolution of immunoglobulin heavy chain diversity. J Exp Med. 1993;178:825–834. doi: 10.1084/jem.178.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz M, Greenberg AS, Flajnik MF. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci USA. 1998;95:14343–14348. doi: 10.1073/pnas.95.24.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity. 2002;16:571–582. doi: 10.1016/s1074-7613(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 49.Hayakawa K, Hardy RR. Development and function of B-1 cells. Curr Opin Immunol. 2000;12:346–353. doi: 10.1016/s0952-7915(00)00098-4. [DOI] [PubMed] [Google Scholar]
- 50.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 51.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nature Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sciammas R, Davis MM. Blimp-1; immunoglobulin secretion and the switch to plasma cells. Curr Top Microbiol Immunol. 2005;290:201–224. doi: 10.1007/3-540-26363-2_9. [DOI] [PubMed] [Google Scholar]
- 53.Sciammas R, Davis MM. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J Immunol. 2004;172:5427–5440. doi: 10.4049/jimmunol.172.9.5427. [DOI] [PubMed] [Google Scholar]
- 54.Heaney ML, Golde DW. Soluble receptors in human disease. J Leukoc Biol. 1998;64:135–146. doi: 10.1002/jlb.64.2.135. [DOI] [PubMed] [Google Scholar]
- 55.Butler JE. Immunoglobulin diversity, B-cell and antibody repertoire development in large farm animals. Rev Sci Tech. 1998;17:43–70. doi: 10.20506/rst.17.1.1096. [DOI] [PubMed] [Google Scholar]
- 56.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 58.Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 59.Malecek K, et al. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 2008;6:e157. doi: 10.1371/journal.pbio.0060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 61.Zarrin AA, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nature Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 62.Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. Sequencing the zebrafish immune repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen H, et al. Characterization of arrangement and expression of the T cell receptor γ locus in the sandbar shark. Proc Natl Acad Sci USA. 2009;106:8591–8596. doi: 10.1073/pnas.0811283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Brien RL, Born WK. γδ T cell subsets: A link between TCR and function? Semin Immunol. 2010 May 5; doi: 10.1016/j.smim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Rhijn I, et al. Highly diverse TCR δ chain repertoire in bovine tissues due to the use of up to four D segments per δ chain. Mol Immunol. 2007;44:3155–3161. doi: 10.1016/j.molimm.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Boehm T. Co-evolution of a primordial peptide-presentation system and cellular immunity. Nature Rev Immunol. 2006;6:79–84. doi: 10.1038/nri1749. [DOI] [PubMed] [Google Scholar]
- 67.Boehm T, Bleul CC. The evolutionary history of lymphoid organs. Nature Immunol. 2007;8:131–135. doi: 10.1038/ni1435. [DOI] [PubMed] [Google Scholar]
- 68.Li J, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nature Immunol. 2006;7:1116–1124. doi: 10.1038/ni1389. [DOI] [PubMed] [Google Scholar]
- 69.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. This work reports that VLR-expressing lymphocyte-like cells can be divided into two classes that are reminiscent of the humoral (B cells) and cellular (T cells) arms of the conventional adaptive immune system in jawed vertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dermody TS, Kirchner E, Guglielmi KM, Stehle T. Immunoglobulin superfamily virus receptors and the evolution of adaptive immunity. PLoS Pathog. 2009;5:e1000481. doi: 10.1371/journal.ppat.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoder JA, et al. Resolution of the NITR gene cluster in zebrafish. Proc Natl Acad Sci USA. 2004;101:15706–15711. doi: 10.1073/pnas.0405242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cannon JP, et al. A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity. 2008;29:228–237. doi: 10.1016/j.immuni.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nature Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. This article describes the identification of a highly diverse family of immunoglobulin receptors in amphioxus and raises the possibility that invertebrates may also rely on diversified antigen receptors for immunity, a characteristic previously thought to be exclusive to jawed vertebrates. [DOI] [PubMed] [Google Scholar]
- 74.Cannon JP, Haire RN, Schnitker N, Mueller MG, Litman GW. Individual protochordates possess unique immune-type receptor repertoires. Curr Biol. 2004;14:R465–R466. doi: 10.1016/j.cub.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 75.Hernandez Prada JA, et al. Ancient evolutionary origin of diversified variable regions revealed by crystal structures of an immune-type receptor in amphioxus. Nature Immunol. 2006;7:875–882. doi: 10.1038/ni1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dishaw LJ, et al. Genomic complexity of the variable region-containing chitin-binding proteins in amphioxus. BMC Genomics. 2008;9:78. doi: 10.1186/1471-2156-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci USA. 2006;103:5036–5041. doi: 10.1073/pnas.0507074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parra ZE, et al. A unique T cell receptor discovered in marsupials. Proc Natl Acad Sci USA. 2007;104:9776–9781. doi: 10.1073/pnas.0609106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller RD. Those other mammals: the immunoglobulins and T cell receptors of marsupials and monotremes. Semin Immunol. 2010;22:3–9. doi: 10.1016/j.smim.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rothenberg EV, Pant R. Origins of lymphocyte developmental programs: transcription factor evidence. Semin Immunol. 2004;16:227–238. doi: 10.1016/j.smim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 81.Anderson SK. Transcriptional regulation of NK cell receptors. Curr Top Microbiol Immunol. 2006;298:59–75. doi: 10.1007/3-540-27743-9_3. [DOI] [PubMed] [Google Scholar]
- 82.Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pancer Z, Cooper MD. The evolution of adaptive immunity. Ann Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Q, et al. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9:R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hedrick SM. The acquired immune system: a vantage from beneath. Immunity. 2004;21:607–615. doi: 10.1016/j.immuni.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 86.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 87.Huang S, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davidson CR, Best NM, Francis JW, Cooper EL, Wood TC. Toll-like receptor genes (TLRs) from Capitella capitata and Helobdella robusta (Annelida) Dev Comp Immunol. 2008;32:608–612. doi: 10.1016/j.dci.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 89.Messier-Solek C, Buckley KM, Rast JP. Highly diversified innate receptor systems and new forms of animal immunity. Semin Immunol. 2010;22:39–47. doi: 10.1016/j.smim.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 90.Flajnik MF, Du PL. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weller GR, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 93.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 94.Gozalbo-Lopez B, et al. A role for DNA polymerase μ in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol Cell Biol. 2009;29:1266–1275. doi: 10.1128/MCB.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]