Abstract
The process of chromosome duplication faces many obstacles. One way to circumvent blocks is to hop over them by placing a new clamp on a downstream primer. This resembles lagging strand synthesis, where the tight grip of polymerase to the clamp and DNA must be overcome upon completing each Okazaki fragment so it can transfer to new primed sites. This review focuses on recent single-molecule studies showing that E. coli Pol III can hop from one clamp to another without leaving the replication fork. This capability provides a means to circumvent obstacles like transcription or DNA lesions without fork collapse.
Keywords: single-molecule, replisome, replication barriers, trombone model, lagging strand, polymerase, helicase
Introduction
Chromosomal replicases utilize circular sliding clamps for high-processivity during replication (reviewed in [1]). Sliding clamps are ring-shaped homo-oligomers in all cell types (Fig 1A). The rings are opened and closed around DNA by a multi-protein clamp loader that couples the energy of ATP hydrolysis to this task. Crystal structures of the multi-protein clamp loaders from E. coli and yeast show that they are circular heteropentamers comprised of AAA+ subunits (ATPases Associated with a variety of Activities) (Fig 1B) [2, 3]. The subunits of the clamp loader form a central chamber that binds primed DNA in a structure specific manner and positions the duplex through the opened clamp prior to ATP hydrolysis. Hydrolysis ejects the clamp loader allowing the ring to snap shut around DNA (Fig 1C). Sliding clamps slide along duplex DNA and they bind directly to the DNA polymerase, tethering it to the primed site for highly processive synthesis.
Figure 1. Structures of sliding clamps and clamp loaders.
(A) Sliding clamps are ring shaped oligomers in all cell types. E. coli β (PDB ID, 2pol), human PCNA (pdb id: 1AXC), Sulfolobus solfataricus PCNA (pdb id: 2IX2); T4 phage gp45 protein (pdb id: 1CZD). (B) Clamp loaders are circular heteropentamers. E. coli γ3δδ ’ bound to primed DNA (pdb id: 3GLF) (reproduced with permission from Figure 1B in (2). Yeast RFC-PCNA complex (pdb id: 1PLQ). (C) Summary of clamp loading onto a DNA primed site.
Sliding clamps not only bind the chromosomal replicase, but they also function with other proteins including ligase, mismatch repair proteins and several different DNA polymerases that are used for repair and lesion bypass [4–8]. The homo-oligomeric structure of sliding clamps enables them to bind to more than one protein at the same time suggesting they may act as a molecular tool-belt [9, 10]. A functional demonstration of a clamp as a tool-belt is provided by studies in the T4 phage replication system performed by the Benkovic group [11]. Using wild type and mutant T4 gp43 polymerases, they demonstrated that the polymerase trade places with one another through an intermediate complex of two DNA polymerases bound to one sliding clamp. Similar studies in the E. coli system have taken advantage of the different DNA polymerases in the cell that utilize the E. coli β sliding clamp and directly demonstrate formation of the intermediate complex of the replicase, DNA polymerase (Pol) III, and the translesion polymerase IV (TLS Pol IV) bound to one β dimer [10]. Further studies showed that different E. coli DNA polymerases (Pols II, III and IV) rapidly exchange the DNA between them during replication fork movement [12].
This review focuses on the use of the sliding clamp in crossing barriers during replication. We provide a brief overview of how sliding clamps enable polymerase hopping over certain DNA blocks. Then we apply single-molecule analysis to one particular polymerase hopping event, asking if the polymerase stays associated with DNA during this process. The example we use is lagging strand replication which is extended in the direction opposite fork movement; this acts as a barrier to a processive polymerase which must undergo rapid dissociation/reassociation events with each Okazaki fragment. Lagging strand replication is proposed to occur without the escape of the lagging strand polymerase, implying that DNA loops, one for each Okazaki fragment, are formed as a result of the opposite direction of lagging strand synthesis relative to replication fork progression. The repetitive formation of DNA loops on the lagging strand, suggested by the constant association of the lagging strand polymerase with the replication apparatus, is referred to as the “trombone model” of replication. The trombone model, hypothesized 40 years ago by the Bruce Albert’s lab working in the T4 replication system [13] was further defined by a mechanism of polymerase hopping among sliding clamps [14, 15].
Although the trombone model is widely accepted, it is difficult to prove and several laboratories continue to probe the mechanism of this fundamental process.
Examples of polymerase hopping among slide clamps to overcome barriers to replication
Chromosomes contain very long DNA molecules that are riddled with a variety of barriers to replication fork progression. Replication over these barriers is likely to be solved in a variety of ways. One mechanism that is applied to a subset of barriers is the use of two sliding clamps, one in front and one in back of the barrier, followed by polymerase hopping over the block by dissociating from the clamp behind the block and reassociating with the other clamp ahead of the block (Fig 2).
Figure 2. Polymerase hops to new clamps to circumvent replication barriers.
A) Polymerase hopping to new clamps on RNA primers circumvents the opposite directionality of lagging strand synthesis. (B) Polymerase hopping to a clamp on a RNA primer synthesized on the leading strand can circumvent a leading strand lesion. (C) Polymerase hopping to a clamp on an mRNA-DNA hybrid can circumvent a transcribing RNA polymerase.
Polymerase hopping among sliding clamps was first observed in the E. coli system using two different primed DNA substrates [14]. In this instance, the “barrier” was the tight grip of the processive Pol III replicase to DNA. Polymerase release from DNA was measured by assembling Pol III on one DNA substrate and observing its transfer to a second DNA substrate. Further study demonstrated that the Pol III replicase overcame its barrier by dissociating from the circular β clamp, specifically upon completing replication of the first DNA substrate; the clamp was left behind on the completed DNA while Pol III rapidly associated with another β clamp on the second DNA substrate [15]. At the time, this observation was taken as an explanation for how a chromosomal replicase can be highly processive, yet can rapidly recycle from one Okazaki fragment to the start of the next every 1–2 kb on the lagging strand (Fig. 2A).
The ability of Pol III to hop from one clamp to another can be generalized to other processes in which the polymerase must overcome and bypass certain replication barriers (reviewed in [16]. Consider the case of polymerase encountering a DNA lesion on a leading strand (Fig 2B). The Marian’s lab has shown that primase (DnaG protein) can synthesize an RNA primer ahead of a block on the leading strand [17]. This enables the replication fork to move past the block by extending the new RNA primer. The mechanism of this process probably requires assembly of a new β clamp at the primed site to which the blocked Pol III can transfer. Another block that requires a polymerase hopping event is the encounter of a replication fork with an RNA polymerase transcribing the leading strand (Fig 2C). In this case the RNA polymerase is displaced, a β clamp is assembled on the mRNA transcript, and Pol III hops to the new clamp for continued leading strand replication [18].
The trombone model of replication
Pol III hopping over barriers appear to generalize to eukaryotes, since the yeast DNA polymerase δ is able to jump among sliding clamps, by losing its tight grip to DNA by releasing from the eukaryotic PCNA clamp and reassociating with a new PCNA clamp assembled onto a second primed substrate [19]. The underlying mechanism by which highly processive polymerases like E. coli Pol III and yeast Pol δ lose their tight grip to the clamp is still far from understood. But regardless of the mechanism of this process, the polymerase is the component that comes free of the substrate (i.e. the clamp – DNA complex). Reassociation of the polymerase with its clamp is exceedingly rapid and is probably not a rate-limiting step in the polymerase hopping process. The rate-limiting step in this process is most likely to be the formation of the RNA primed site, or the assembly of a clamp on new RNA primers. These considerations bring into question whether the polymerase remains attached to the replisome during bypass of replication blocks by the polymerase hopping process.
The trombone model of replication is so named for the repeated formation of new DNA loops on the lagging strand, one for each Okazaki fragment [13]. As an Okazaki fragment is extended, the loop grows. When the Okazaki fragment is finished the loop disassembles and starts over again at a new RNA primer. The growth and loss of DNA loops resembles the action of a trombone slide as the instrument is played. At the root of this trombone model, and the repeated formation of DNA loops, is a requirement that the lagging strand polymerase remains associated with the replisome during production of multiple lagging strand Okazaki fragments. If the lagging strand polymerase were to dissociate from the replisome, the loop would not form, and another DNA polymerase molecule would associate with a clamp on the new RNA primer for extension of the next Okazaki fragment.
Initial evidence for a processive lagging strand polymerase, and thus DNA looping, was the persistence of Okazaki fragment synthesis after dilution of assembled replisomes in the T4 and E. coli systems [13, 20]. But if the kinetics of polymerase association with clamps is rapid, and not rate-limiting at the dilution used, one cannot distinguish whether the same polymerase or different DNA polymerases are used to extend multiple Okazaki fragments. Electron microscopy studies in the T4 and T7 systems provide strong evidence for DNA looping [21–23]. However, these studies provide only snapshots of a dynamic process and one could argue that transient association of the polymerase to the helicase, or other components at the fork, become trapped by cross-linking reagents used in the analysis. In this case, different polymerases could be utilized for each new lagging strand fragment, yet loops would be observed due to cross-linking of transient intermediates. Single-molecule studies in the T4 and E. coli replication systems by the Van Oijen lab have made conclusions about lagging strand loops by following the movement of a bead attached to the lagging strand [24–26]. The bead movements are consistent with formation of DNA loops, although new data suggests another explanation for bead movements besides Okazaki fragment extension loops [27].
The remainder of this review explains single-molecule experiments that are performed using immobilized rolling circle substrates in the E. coli replication system to measure the number of Okazaki fragments that are synthesized by a single lagging strand DNA polymerase molecule [28].
Architecture of the E. coli Replisome
The organization of proteins at the E. coli replication fork is illustrated in Fig. 3 (reviewed in [1]). The hexameric DnaB helicase encircles the lagging strand and uses ATP to translocate along single-strand DNA and separate the parental duplex. The clamp loader pentamer contains at least two τ subunits which have C-terminal extensions that protrude from the top of the clamp loader; each C-terminal extension binds to a DnaB helicase subunit and a molecule of Pol III. Pol III contains three subunits referred to as Pol III core (α, DNA polymerase;ε, 3′–5′ exonuclease; θ, unknown function). The assembly containing a clamp loader and two molecules of Pol III core is referred to as Pol III*. Each molecule of Pol III core within Pol III* binds a β clamp to form the Pol III holoenzyme. The β clamps confer processive synthesis onto both the leading and lagging strand Pol III cores. One or more molecules of primase interact with DnaB for synthesis of an RNA primer, and then dissociate [29]. Thus primase must be replenished from solution for each Okazaki fragment.
Figure 3. Replisome architecture and function in E. coli.
(A) The E. coli replisome consists of the DnaB homohexameric helicase that encircles the lagging strand, a multiprotein clamp loader that has τ subunit extensions that bind DnaB and two Pol III cores. The Pol III cores are held to the leading and lagging strands by β clamps for high processivity. Primase binds DnaB for synthesis of an RNA primer. The single-strand DNA is bound by SSB. The insets illustrate the action of the lagging Pol III core in overcoming the opposite directionality of the replisome by hopping to a new clamp and leaving the old clamp behind on an extended Okazaki fragment. (B) Rolling circle replication. The illustration shows the production of leading and lagging strands by the replisome on a minicircle. Reactions were performed similar to single molecule experiments (as described) in the legend to Figure 6) except the replication initiation buffer contained either 32P-dATP or 32P-dTTP. The data shows leading and lagging strand product analysis in an alkaline gel.
The illustration in Figure 3A depicts the trombone model, with a DNA loop during Okazaki fragment extension due to the association of the lagging strand Pol III-β complex with the replication fork. The subsequent steps in the figure depict the release of the DNA loop when the polymerase leaves behind the old β clamp and binds to a newly assembled clamp on the RNA primer. The trombone model and DNA loop will not prevail if the lagging strand Pol III does not remain attached to the replisome during production of multiple Okazaki fragments. In this case, the Pol III-β complex detaches from the replisome while it extends an Okazaki fragment and a different Pol III core is recruited from solution for the new β clamp on the next RNA primer.
Rolling circle replication
A common method to study replisomes in vitro is to assemble the replisome on a replication fork substrate. Replisome studies often utilize a “rolling circle” substrate which is a circular duplex DNA with an break in one strand that has a 5′ single-strand (ss) DNA tail (Figure 3B) [30]. The inner circle is the leading strand template, and since a circle has no end, the replisome can progress around the circle indefinitely, thus the name “rolling circle”. The leading strand product is continuously displaced from the circle to produce a long ssDNA tail that serves as template for primase and the lagging strand DNA polymerase.
To assemble the E. coli replisome onto the minicircle replication fork, DnaB is allowed to slide over the 5′ ssDNA tail, followed by the Pol III* and the βclamp that load onto the 3′ terminus of the circle. This assembly step is performed in the presence of only 2 dNTPs to prevent replisome movement. Upon adding dNTPs, rNTPs, primase and SSB the replisome extends the 3′ terminus multiple times around the circle, resulting in a long double-strand (ds) DNA tail; one strand is the product of the leading polymerase while the other strand is comprised of lagging strand Okazaki fragments. In the study we review here, we used a synthetic “minicircle” with a 40 dT 5’ ssDNA tail, and a 100mer inner circle that contains no dT residues [12]. This strategy, pioneered by the Richardson group [21], enables one to label either the leading or lagging strand depending on whether α-32P-dATP or α-32P-dTTP is added to the reaction. An example of the product analysis of a minicircle replication reaction in an alkaline agarose gel is shown in Fig. 3B. The leading strand products are much longer than the DNA standards and migrate slowly in the gel, while the lagging strand Okazaki fragments average around 650 nucleotides in length.
Single-Molecule microscopy
The DNA looping trombone model of replication requires that the lagging strand DNA polymerase remain attached to the replication fork during multiple cycles of Okazaki fragment synthesis. The rationale to access the processivity of the lagging strand polymerase by single-molecule techniques is illustrated in Figure 4A. Single-molecule studies of DNA replication are performed using an immobilized rolling circle substrate in a flow cell. Therefore, when the lagging strand polymerase dissociates from the replication fork, it will be carried away in the buffer flow (which does not contain any Pol III), preventing further lagging strand synthesis. Continued leading strand synthesis will yield only single-strand DNA. A fluorescent DNA intercalator will only detect double-strand DNA, not single-strand DNA. Therefore the length of the duplex DNA tail produced by the replisome on a rolling circle substrate in the presence of a constant buffer flow directly reflects the amount of DNA synthesized by a single lagging strand DNA polymerase that binds and dissociate successively for each Okazaki fragment (Figure 4B). With this experimental strategy in mind we developed the necessary tools to perform rolling circle replication in a flow cell and to detect the double strand DNA product.
Figure 4. Experimental rationale to determine whether Pol III hops to new clamps without leaving DNA.
(A) Rolling circle replication is immobilized at the 5’ tail, and is performed by the E. coli replisome in a flow cell using a buffer that contains nucleotides needed for replication, but does not contain Pol III. Due to the opposite directionality of Okazaki fragment synthesis, the lagging strand Pol III must dissociate from each extended Okazaki fragment, yet remain attached to the replisome in order to extend RNA primers as they are synthesized by primase. (B) If the lagging strand Pol III dissociates from the minicircle, it is carried away by the flow. The leading strand Pol III may continue with DnaB helicase to form ssDNA, but the ssDNA cannot be converted to dsDNA because the lagging strand Pol III has been carried away in the buffer flow. Only the dsDNA, made by the lagging strand Pol III, is detected by a fluorescent intercalator.
To construct a flow cell for these studies we made a negative lithography mold on a glass slide 2.5 mm wide, 25 mm long and 50 microns high. A photoclear elastomer was poured over the mold and allowed to harden into a semisolid. Then a rectangular block was cut around the channel produced by the mold. Holes were punched from the top of the elastomer block at either end of the channel into which tubing was inserted for a buffer flow. The elastomer block was then welded to a cover slip using a plasma oven. A picture of the flow cell is shown in Figure 5A.
Figure 5. Single-molecule flow cell construction and determination of force versus flow rate.
(A) Top: Flow cells are made using a photoclear elastomer poured over a negative lithography mould to form a flow channel. Ports are punched into the elastomer at either end of the channel. A glass coverslip is scored with a diamond scribe to form barriers to lipid diffusion. The elastomer block is welded to the coverslip in a plasma oven to form a flow cell. Middle: Picture of a flow cell with a red solution to visualize the flow chamber. Bottom: A flow cell platform containing valves to form a lipid bilayer inside the flow cell attached to the platform. The platform is screwed to the microscope stage to begin the experiment. (B) Curtains of lambda DNA attached to the lipid bilayer using a biotinylated DNA-neutravidin-biotinylated lipid sandwich. The hydrodynamic flow results in DNA migrating to a diffusion barrier to produce the DNA curtain. As the flow rate is increased, the DNA is stretched to greater lengths. The plot below the curtains shows force versus length of lambda DNA. The observed length at 100 µl/min equates to a force of 1.45 pN.
To visualize individual DNA molecules inside the flow cell, we used an Olympus IX70 microscope and a 488nm solid state laser to excite a fluorescent intercalator bound to double-strand DNA. Fluorescence emission was collected back through the objective and filtered for residual laser light before being captured by a 512 × 512 thin backed Hamamatsu EM CCD camera. A motorized shutter permitted a 100 ms exposure every 1s. The signal to noise ratio was improved by aligning the excitation laser at the proper angle through the objective to obtain total internal reflectance fluorescence (TIRF). TIRF limits fluorescence emission to molecules that are within 100–200 nm of the reflecting interface. To immobilize DNA to the surface of the flow cell, we followed protocols developed in Eric Greene’s laboratory (Columbia University) to form a lipid bilayer [31]. Formation of lipid bilayers inside a flow cell from freshly sonicated liposomes requires several buffer exchanges of a few hours, and is usually performed with the flow cell attached to the microscope. To circumvent this bottleneck we designed “flow cell platforms” to which the flow cell can be tightly fastened during lipid bilayer formation (Fig. 5A). This enabled production of lipid bilayers in several flow cells in parallel.
The lipid bilayer contains a small percent (0.5%) of lipids containing a biotin modified head group, and this provides an attachment point to the 5′ biotinylated DNA rolling circle through a neutravidin sandwich. The DNA-lipid conjugate diffuses in the lipid bilayer, and the hydrodynamic force of the buffer flow on the DNA causes the DNA-lipid conjugate to move in the direction of the buffer flow. To trap the DNA-lipid conjugate, the glass cover slip is etched with a diamond scribe before welding the elastomer flow cell together. These scratches form diffusion barriers across which lipids do not migrate. Thus, the DNA-lipid conjugates form a curtain at a diffusion barrier. Figure 5B shows a curtain of biotinylated lambda DNA-lipid conjugates in presence of the yo-pro1 intercalator at a diffusion barrier under different flow rates. The contour length of lambda DNA is measured at different flow rates, and this quantitate the force at the surface of the flow cell for a given flow rate. In the experiments described here, a force of 1.45 pN was provided by a flow of 100 µl/min which stretches lambda DNA to 88% of its full length.
Visualization of Replisome Action in Real-Time
To visualize replisome action during DNA synthesis, the minicircle replication fork substrate was constructed with a 5’ biotin for attachment to the lipid bilayer (29). To monitor replication in real-time we chose the fluorescent DNA intercalator Yo-Pro1 instead of the more commonly used Yo-Yo stain. Yo-Pro1 is a monomeric version of Yo-Yo, and this lowers its affinity for DNA considerably. This requires use of more Yo-Pro1 to visualize DNA, compared to Yo-Yo, but Yo-Yo binds DNA tightly and significantly inhibits minicircle replication in bulk phase assays, while Yo-Pro1 does not inhibit replication even at high concentrations (29). Furthermore, Yo-Pro1 binds to DNA rapidly and also dissociates rapidly, unlike Yo-Yo which is slow to bind and release DNA. Thus, photobleaching is less of a concern with Yo-Pro1 since bleached Yo-Pro1 molecules rapidly exchange with new molecules of Yo-Pro1, thereby providing a continuous signal during irradiation with a laser.
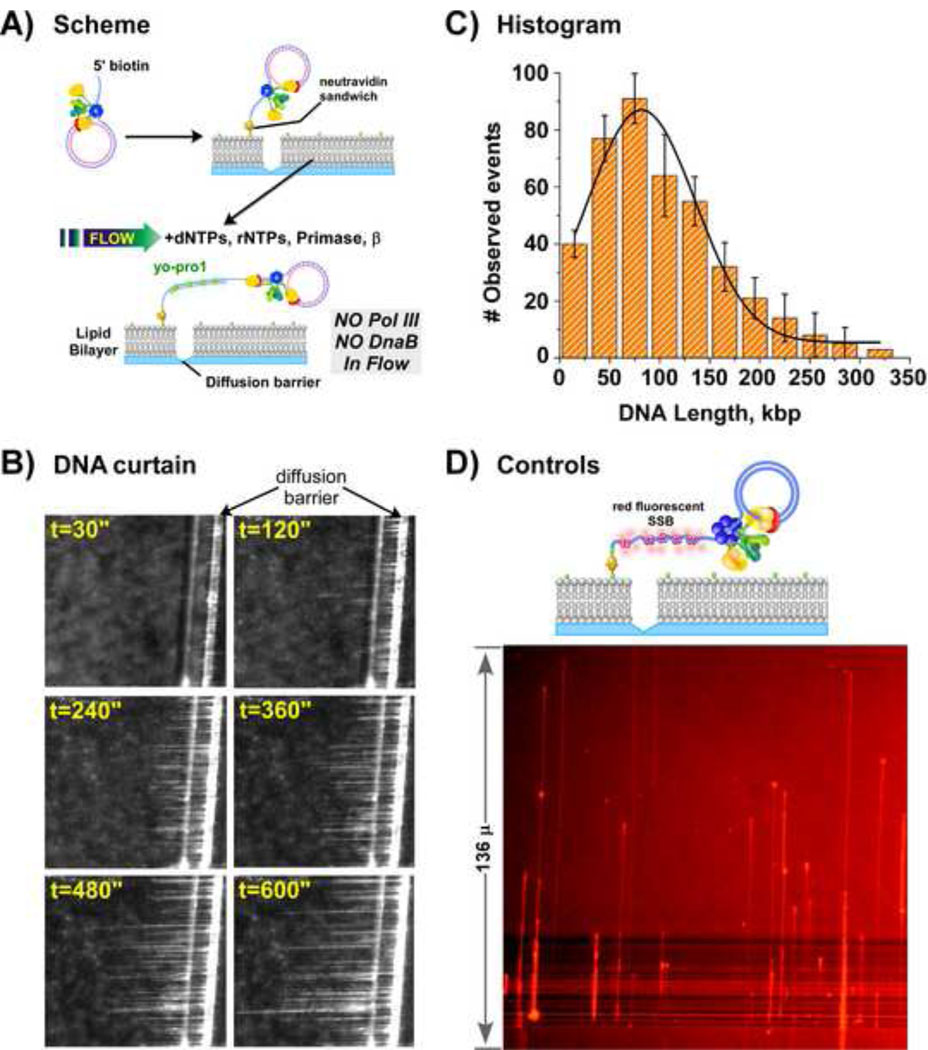
The experimental design of a single-molecule experiment is illustrated in Figure 6A. First, replisomes were assembled onto the minicircle DNA, and then the reaction mix was slowly passed through the flow cell (10 μl/min) for immobilization of the replisome-DNA complex to the lipid bilayer. After immobilization, unbound proteins were washed away using a buffer flow lacking proteins (100 µl/min). The force of the hydrodynamic flow causes the replisome-DNA-lipid conjugate to migrate in the bilayer and stop at a diffusion barrier. Replication is then initiated using a buffer flow that contains the 4 dNTPs, 4 rNTPs, primase, SSB, β and Yo-Pro1, but lacks Pol III* and DnaB.
Figure 6. Single-molecule rolling circle replication on a lipid bilayer produces DNA curtains at a diffusion barrier.
(A) Scheme of single-molecule rolling circle replication on a lipid bilayer. During leading strand synthesis (blue), DNA is displaced from the 100-mer circle as the replisome ‘rolls’ around the template. The newly synthesized 5' ssDNA "tail" is converted to dsDNA by lagging-strand synthesis (red). (B) Images at the indicated times of a growing rolling circle DNA curtain visualized in real-time by TIRF. Replisomes were assembled by mixing DnaB helicase (18.2 pmol, 365 nM) with minicircle DNA (655 fmol, 13.1 nM) in 50 µl Buffer A (20 mM Tris-HCl, pH 7.5, 5 mM DTT, 40 µg/ml BSA, 4% glycerol) containing 8 mM MgOAc2 and 1.25 mM ATP, followed by incubation for 30s at 37°C. Then 25 µl of buffer A containing Pol III* (675 fmol, 27 nM), β2 (1.85 pmol, 74 nM as dimer), 60 µM each dCTP and dGTP and 8 mM Mg(OAc)2 was added. After 6 min at 37°C, 1 μl of the reaction was diluted into 1 ml of Buffer B (Buffer A containing 8 mM MgOAc2, 60 µM each of dCTP and dGTP, and 50 nm Yo-Pro1). The reaction was passed through the flow cell at 10 µl/min for 30s and then replication was initiated upon flowing (100 µl/min) Buffer A containing 60 µM of each dNTP, 250 µM each of CTP, TTP, and UTP, 1 mM ATP, 462 nM SSB4, 300 nM primase, 50 nMβ2, 50 nM Yo-Pro1, 0.8% glucose, 0.01% β-mercaptoethanol, 0.57 U glucose oxidase, and 2.1 U catalase [28]. (C) Histogram of the observed length of 400 DNA molecules and a one-Gaussian fit analysis (black line). (D) A control reaction using a replisome containing DnaB and only one Pol III core-β for leading strand synthesis in the absence of lagging strand synthesis. The ssDNA product is visualized by epifluorescence using SSB labeled with a red fluorophore.
Replication results in the growth of a “curtain” of DNA strands that are visualized in real-time (Fig. 6B). Each DNA thread is the product of an individual replisome and the minicircle is located at the growing point, or tip of the thread. The field of view in Fig. 6B is about 390 kb. The length of individual DNA strands vary considerably; some are less than 30 kb while others are over 300 kb. The length distribution of 400 individual molecules, fit to a single Gaussian function, yields an average length of 86.5 ± 5.3 kb (Fig. 6C).
The production of long strands of dsDNA in the presence of a constant buffer flow has important implications for replisome mechanism and supports the trombone model of replication. A dsDNA molecule that averages 86 kb contains about 132 Okazaki fragments given the size of Okazaki fragments in bulk phase assays under the same conditions. The production of over one hundred Okazaki fragments by one replisome while in a constant buffer flow containing no Pol III, indicates that a single lagging strand Pol III extends all the Okazaki fragments and stays attached to the replication fork the entire time. Therefore each Okazaki fragment must have involved the formation of a DNA loop considering that lagging strand synthesis occurs in the opposite direction of leading strand synthesis.
To confirm that the fluorescent dsDNA product visualized in the experiment depends on a lagging strand Pol III that remains constantly attached to the replisome, and that there was no hidden source of endogenous polymerase, several control experiments were performed. In one control we reconstituted a Pol III* assembly that contained only one Pol III core. During rolling circle replication with DnaB helicase, the “monopolymerase replisome” should produce only the long ssDNA product of leading strand synthesis, even though RNA primers will still be formed on the ssDNA because primase, SSB and β are present in the buffer flow. If there is no source of endogenous Pol III in the system, the RNA primers will not be extended and the leading strand ssDNA product will not become converted to dsDNA. The result shows no fluorescence, confirming that no Pol III is present to extend Okazaki fragments for Yo-Pro1 intercalation. To prove that a long leading strand ssDNA is indeed produced by the monopolymerase replisome, we used an SSB labeled with Texas red that should decorate the ssDNA for visualization by fluorescence. To excite the Texas red-SSB we used a mercury lamp instead of the laser (i.e. epifluorescence). As expected, long SSB coated filaments of ssDNA are detected (Fig 6D). In another control, omission of primase from the buffer flow results in only leading strand products even when leading and lagging strand polymerases are present in the replisome (not shown). In another control performed using the complete replisome with leading and lagging strand polymerases, β was omitted from the buffer flow, (primase and SSB were present). Again, no dsDNA product is detected by Yo-Pro1 (data not shown). This control shows that new β clamps must be assembled on RNA primers during lagging strand synthesis. The single-molecule experiments are performed in the absence of additional clamp loader in the buffer flow, and therefore the clamp loader in the replisome is capable of loading β clamps on RNA primers as they are produced by primase.
DnaB helicase stays in the replisome longer than Pol III
In the single-molecule experiments, replisome function can be terminated by dissociation of either Pol III, the clamp loader or DnaB helicase, since none of these components are present in the buffer flow. In Figure 7, we compare experiments performed in the presence or absence of Pol III* in the buffer flow. If replication fork progression is terminated by DnaB helicase dissociation, the DNA product length should not be affected by the presence of Pol III* in the flow. But if Pol III* dissociates before DnaB, then Pol III* in the buffer flow may bind DnaB for continued fork progression and longer threads of dsDNA will be produced. Indeed, longer DNA products were observed. Analysis of 400 molecules, fit to a single Gaussian function, yields an average length of 140.4 ± 6.8 kb (Figure 7C). This length of DNA is 163% the length observed when Pol III* is omitted from the flow (compare red and green histograms in Fig. 7C). Hence, we conclude that Pol III* often dissociates before DnaB helicase. The reverse experiment, in which DnaB is included in the buffer flow, carries little value since helicase loading factors are required to reassemble DnaB helicase onto DNA in the presence of SSB [32]. Even if the helicase loading factors were included, the kinetics of the helicase loading process is currently unknown and would likely result in a long pause before a functional replisome reassembled. Replisome reassembly after fork collapse is, of course, a fascinating topic and single-molecule analysis using the system reviewed here provides an elegant method to observe the process of replication restart in real-time.
Figure 7. DNA length is enhanced by excess Pol III* in the buffer flow.
DNA curtains produced when (A) Reactions were as described in the legend to Figure 6, where Pol III* is absent from the flow during replication. (B) Pol III* (12 nM) is present in the flow during replication. (C) Histograms of product length when Pol III* is present in the flow (green), or absent from the buffer flow (red) [28]
Summary
It is of paramount importance that a dividing cell duplicates its entire genome. Given the enormous size and complexity of genomic DNA, the duplication process likely encounters a considerable number of obstacles during the replication process. Given the central importance of finishing replication once it has started, natural selection has arrived at solutions that resolve conflicts between the replication apparatus and various blocks to replication fork progression. One method the replisome uses to circumvent blocks is to hop over them by placing a new clamp onto a primed site on the other side of the block. This may, in fact, be the reason clamps and clamp loaders evolved in the first place, rather than simply for processive DNA synthesis. The fact that clamps and clamp loaders are not fundamentally required for processive synthesis is provided by the demonstration of highly processive viral polymerases that do not utilize a sliding clamp and clamp loader [32].
This review has focused on recent single-molecule studies that examine the ability of E. coli Pol III to hop from one clamp to another without leaving the replication fork [12]. To assess this, polymerase hopping to sliding clamps during lagging strand replication was studied. The “barrier” that must be traversed during lagging strand synthesis is the tight grip of Pol III to the β clamp and DNA, which must be overcome upon extending each Okazaki fragment so Pol III can transfer to new primed sites. The experiments reviewed here demonstrate that the polymerase is highly proficient at hopping from one clamp to another without dissociating from the replication fork. Maintaining this association with the rest of the replication fork during polymerase hopping from one clamp to another suggests that movement past blocks like RNA polymerase or DNA lesions, could happen without fork collapse.
Acknowlegement
We are grateful to Eric Green (Columbia University), Steven Kowalczykowski (U. C. Davis), Albert Libchaber (Rockefeller University), Dr. Axel Buguin (Institut Curie), Kathy Lindsay and Martin Eber (Olympus Inc.), and the Rockefeller University machine shop for help in several phases of this project. We also appreciate HHMI and NIH GM38839 for supporting this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 2.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 3.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 5.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 2000;1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci U S A. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci U S A. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha NK, Morris CF, Alberts BM. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J Biol Chem. 1980;255:4290–4293. [PubMed] [Google Scholar]
- 14.O'Donnell ME. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1987;262:16558–16565. [PubMed] [Google Scholar]
- 15.Stukenberg PT, Turner J, O'Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. doi: S0092-8674(94)90662-9 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Yao NY, O'Donnell M. Replisome dynamics and use of DNA trombone loops to bypass replication blocks. Mol Biosyst. 2008;4:1075–1084. doi: 10.1039/b811097b. doi: 10.1039/b811097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. doi: nature07527 [pii] 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langston LD, O'Donnell M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J Biol Chem. 1992;267:4045–4053. [PubMed] [Google Scholar]
- 21.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 22.Nossal NG, Makhov AM, Chastain PD, 2nd, Jones CE, Griffith JD. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J Biol Chem. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- 23.Park K, Debyser Z, Tabor S, Richardson CC, Griffith JD. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J Biol Chem. 1998;273:5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 24.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 25.Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998a. [DOI] [PubMed] [Google Scholar]
- 26.Tanner NA, Loparo JJ, Hamdan SM, Jergic S, Dixon NE, van Oijen AM. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 2009;37:e27. doi: 10.1093/nar/gkp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc Natl Acad Sci U S A. 2009;106:13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CA, Zechner EL, Reems JA, McHenry CS, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. V. Primase action regulates the cycle of Okazaki fragment synthesis. J Biol Chem. 1992;267:4074–4083. [PubMed] [Google Scholar]
- 30.Mok M, Marians KJ. Formation of rolling-circle molecules during phi X174 complementary strand DNA replication. J Biol Chem. 1987;262:2304–2309. [PubMed] [Google Scholar]
- 31.Graneli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 32.Kornberg A, Baker TA. DNA Replication. 2nd edn. New York: W.H. Freeman; 1992. [Google Scholar]