Abstract
The vertebrate retina is very well conserved in evolution. Its structure and functional features are very similar in phyla as different as primates and teleost fish. Here we describe the spatio-temporal characteristics of neurogenesis in the retina of a teleost, medaka, and compare them to other species, primarily the zebrafish. Several intriguing differences are observed between medaka and zebrafish. For example, photoreceptor differentiation in the medaka retina starts independently in two different areas, and at more advanced stages of differentiation, medaka and zebrafish retinae display obviously different patterns of the photoreceptor cell mosaic. Medaka and zebrafish evolutionary lineages are thought to have separated from each other 110 million years ago, and so the differences between these species are not unexpected, and may be exploited to gain insight into the architecture of developmental pathways. Importantly, this work highlights the benefits of using multiple teleost models in parallel to understand a developmental process.
Keywords: Teleost, Medaka, Retina, Mosaic, Photoreceptor, Opsin, Neurogenesis
INTRODUCTION
The vertebrate retina consists of six classes of neurons and one type of glia, arranged in three major layers: the photoreceptor cell layer (PRCL), the inner nuclear layer (INL), and the ganglion cell layer (GCL). Each of these contains multiple cell classes and cell types, many or which are arranged in finer sublaminae (Masland, 2001). Similar to other parts of the central nervous system, despite its complexity the retina develops from a single sheet of neuroepithelial cells (Sauer, 1935; Hinds and Hinds, 1974; Das et al., 2003). During neurogenesis, these cells gradually exit the cell cycle and differentiate. Retinal ganglion cells are the first to become postmitotic in all vertebrate species investigated so far, while other cells exit the cell cycle in a variable sequence (Young, 1985; Holt et al., 1988; Hu and Easter, 1999). Following the specification of retinal cell classes and types, neurons differentiate synaptic connections as well as other features that allow them to collect visual information, process it, and relay it to the brain. Thus photoreceptors differentiate photosensitive structures, known as outer segments, while ganglion cells extend long processes into several areas of the midbrain (Attardi and Sperry, 1963; Burrill and Easter, 1994; Lemke and Reber, 2005; Chuang et al., 2007).
The development of the retina has been studied using numerous experimental strategies, including genetic, genomic, proteomic, biochemical, and tissue culture methods (Watanabe and Raff, 1990; Blackshaw et al., 2001; Gustincich et al., 2004). Although this diversity of approaches is very valuable, the most reliable conclusions are drawn from studies performed in intact organisms. Hence the importance of animal models in the studies of the nervous system. The exceptional structural and functional conservation of the vertebrate retina makes it possible to combine findings from diverse and otherwise very different species to arrive at developmental principles that apply to eyes of all vertebrates. Retinae of amphibians (Xenopus levis), birds (chicken), and mammals (mainly mouse and rat, but also dog) have been widely studied with the help of developmental and genetic approaches. Primate retinae have been mostly used in neuroanatomical and physiological studies as genetic manipulation in this group of species is cumbersome at best. More recently, the zebrafish and medaka have been developed as model organisms to study the retina (reviewed in Malicki, 2000; Wittbrodt et al., 2002; Avanesov and Malicki, 2004). Small size, short generation time, rapid external development, and optical transparency of their embryos during early stages of embryogenesis, along with the availability of molecular and genetic techniques have helped these two teleost species to gain a considerable importance as model systems. One of the best indicators of how successful the zebrafish model has become is the number of retinal mutants reported in the last decade (Malicki et al., 1996; Fadool et al., 1997; Neuhauss et al., 1999; Golling et al., 2002; Muto et al., 2005). Mutations in these lines of animals affect numerous aspects of eye biology, including both developmental and physiological processes, such as the morphogenesis of the optic primordium, cell fate decisions, differentiation of photoreceptor outer segments, retino-tectal pathfinding, and phototransduction. In numerous cases, mutant genes have been cloned, opening a way for the molecular analysis of developmental pathways (Fricke et al., 2001; Kay et al., 2001; Wei and Malicki, 2002; Tsujikawa and Malicki, 2004; Tsujikawa et al., 2007). Genetic screens in medaka, combined with the completion of its genome project are likely to result in a similar wave of informative cloning experiments.
Studies of the medaka eye have largely focused on early stages of its development, the formation of the optic primordium in particular. Thus cell fate analysis traced the position of cells that contribute to the optic primordium from the early gastrula period through optic lobe formation at stages 17-19 (Hirose et al., 2004), and a 3-D analysis of cell movements in mosaic animals revealed that optic vesicle evagination is driven by active migration of individual cells (Rembold et al., 2006). In addition, a number of genetic manipulations showed that Six3, Geminin, as well as Rx3 are all important regulators of optic lobe morphogenesis (Loosli et al., 2001; Carl et al., 2002; Del Bene et al., 2004). Apart from the analysis of the optic primordium, the studies of medaka retinal neurogenesis have been largely limited to ganglion cells and the formation of the retinotectal projection (Yoda et al., 2004). These studies revealed, for example, a number of potential downstream target genes of a well-studied regulator of ganglion cell specification, ath5 (Del Bene et al., 2007).
To lay the foundation for further studies of retinal neurogenesis in medaka, we chose to determine its main spatio-temporal features. We describe the time course of retinal layer formation, the spatial coordinates of cell differentiation in the plane of the photoreceptor cell layer, and the arrangement of cells in the photoreceptor mosaic. Although closely similar in their overall organization, medaka and zebrafish retinae differ in several important aspects: they differentiate following different spatiotemporal parameters, and their photoreceptor cell mosaics are patterned differently. A description of these differences in two relatively closely related species, combined with the analysis of the underlying genetic circuitry may become a source of valuable insights into the logic of developmental pathways and their evolution.
RESULTS
Formation of neuronal lamination
As the timing of retinal pattern formation has not been described in medaka so far, we decided to characterize this process by preparing histological sections through retinae at several developmental stages. Based on previous analyses of ganglion cell differentiation, neurogenesis in the medaka retina begins by stage 26 (2 days and 6 hrs, 22 somites) (Yoda et al., 2004; Del Bene et al., 2007). At stages 25, 27 (not shown) and 28 (Fig. 1A, Fig S1A), however, we do not see lamination in the inner retina on histological sections. The first histologically distinguishable indication of ganglion cell layer formation is the appearance of the optic nerve by stage 29 (3 days, 2 hours, Fig. 1B and Fig. S1C). Despite the presence of optic nerve fibers, retinal lamination is still not visible at this time (Fig. 1B, Fig S1B). Only 8 hours later, however, by stage 30, both inner and outer plexiform layers are noticeable in the central retina, dorsal to the optic nerve (Fig. 1C and Fig. S1D, Fig S1E). By stage 34 (5 days 1 hrs), all 3 nuclear layers as well as 2 plexiform layers that separate them are well differentiated and extend up to the retinal margin (Fig. 1D). This pattern persists largely unchanged throughout later stages of retinal development. Among changes that occur later is the thickening of the photoreceptor cell layer, which takes place as the result of outer segment differentiation. This is already quite evident by stage 39 (Fig. 1E) and becomes even more pronounced in the adult eye (Fig. 1F). Although larval and adult retinae contain the same cell layers, their contributions to the overall thickness of the retina are very different: at stage 34 the inner nuclear layer is nearly 4 times thicker than the photoreceptor cell layer. The opposite is true in the adult: the photoreceptor cell layer, including the outer segments, is ca. 5 times thicker than the INL. In addition to the development of robust outer segments, other processes, such as cell rearrangement or cell death may contribute to this dramatic change of proportions.
Fig. 1. Differentiation of laminar architecture in the medaka retina.
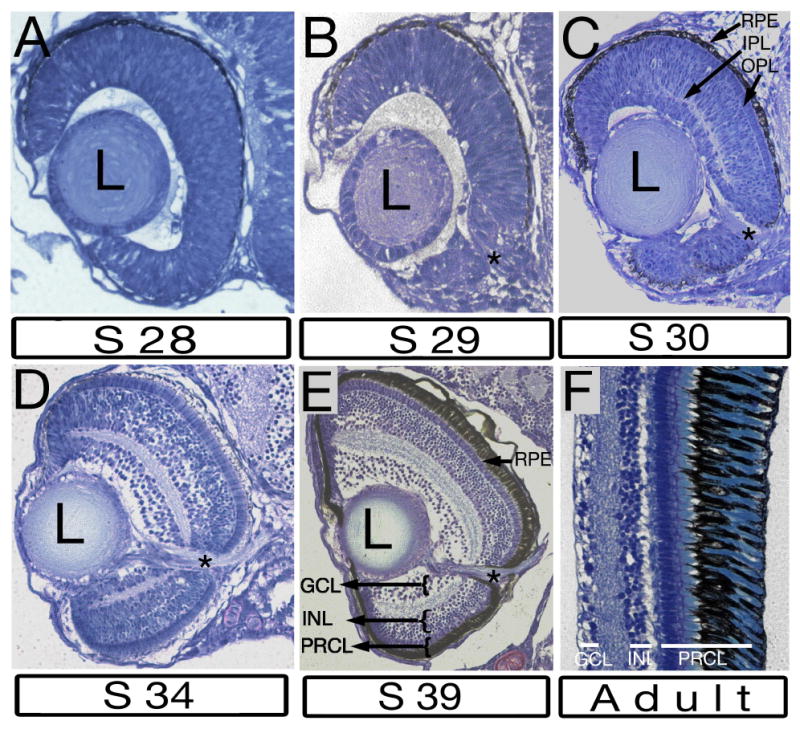
Tansverse plastic sections stained with methylene blue. A: Retinal lamination is absent at stage 28 (2 days 16 hours, 19 somite stage). B: At stage 29 (3 days 2 h, 30 somite stage) the optic nerve is clearly visible, however the retina is not subdivided into layers. C: Both plexiform layers are seen in the central retina above the optic nerve at stage 30 (3 days 10 h, 35 somite stage). An array of elongated photoreceptor cells is visible between the OPL and the RPE in the central retina, but not in the retinal periphery at this stage. D: Both plexiform layers extend into the retinal periphery by stage 34 (5 days 1hrs). E: All retinal laminae are clearly visible at stage 39 (6 days). The retinal pigment epithelium is thickened to accommodate the differentiation of photoreceptor outer segments. F: The retina of adult medaka features the same array of 3 nuclear and 2 plexiform layers as the larval retina. The retinal pigment epithelium and the photoreceptor cell layer are much more prominent relative to other laminae at this stage. In all panels, dorsal is up, lateral is left. The optic nerve is indicated by asterisks. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; L, lens; OPL, outer plexiform layer; PRCL photoreceptor cell layer; RPE, retinal pigment epithelium.
These histological observations were confirmed using antibodies to ganglion and photoreceptor cells. In agreement with previous reports (Yoda et al., 2004; Del Bene et al., 2007), staining with an antibody against acetylated α-tubulin revealed retinal ganglion cells at stage 26 (Fig. 2A) but not at stage 25 (not shown). By stage 28, the optic nerve is clearly seen to exit the retina (Fig. 2B, arrows). Consistent with gradual increase in the number of differentiated ganglion cells, the optic nerve thickens considerably by stage 30 (Fig. 2C. arrows). In addition to ganglion cells, we analyzed photoreceptors. Zpr-1 is an antibody commonly used to stain double cone photoreceptors in the zebrafish retina (Larison and Bremiller, 1990; Schmitt and Dowling, 1996; Doerre and Malicki, 2002; Tsujikawa et al., 2007). In the medaka eye, Zpr1-positive double cones are seen throughout the entire photoreceptor cell layer by stage 40 (Fig. 2J). This staining pattern persists in the adult retina (Fig. 2K). Similar to zebrafish, Zpr1 stains the entire surface of the medaka photoreceptor cell, including outer segments and synaptic termini (Fig. 2L).
Fig. 2. Patterns of retinal neurons in the larval medaka.
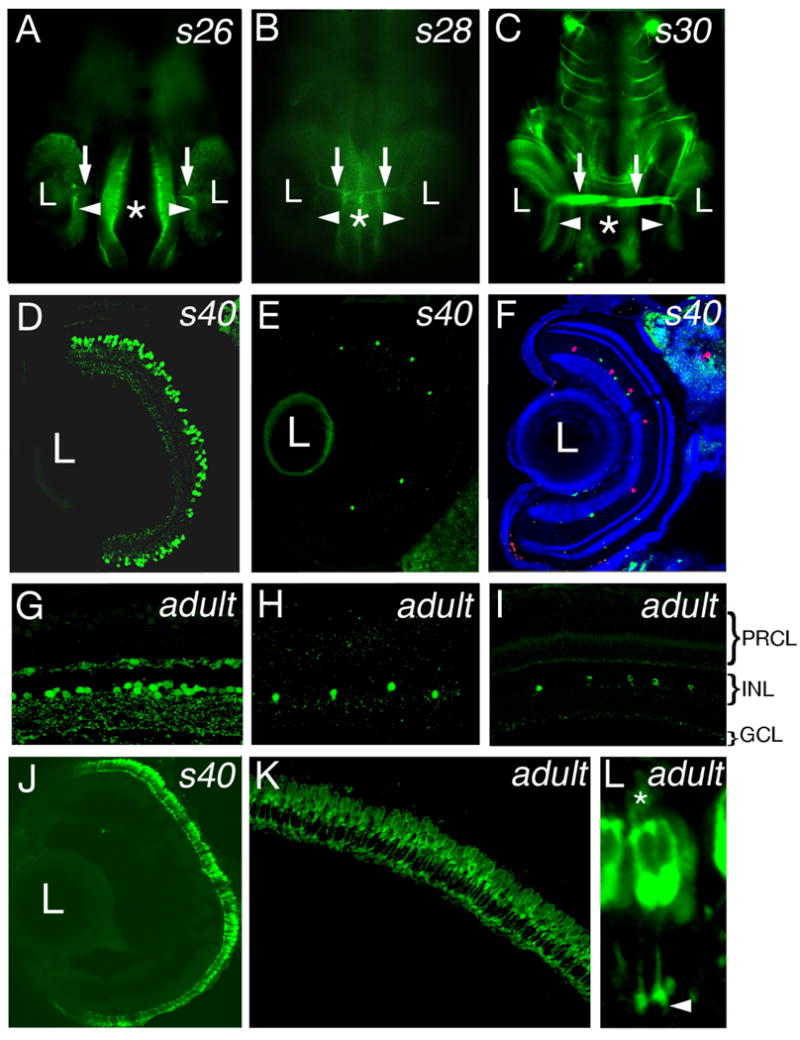
A-C: Ventral view of medaka embryos stained with anti-acetylated tubulin antibody. At stage 26 (A), staining is detectable in a small group of ganglion cell bodies (arrowhead) and their axons (arrows). By stage 28 (B), ganglion cell bodies and the optic nerve (arrows) are clearly visible. By stage 30 (C), the ganglion cell layer (arrowheads) as well as the optic nerve (arrows) display prominent staining. D-I: Immunostaining of plastic sections (D and G) and cryosections (E, F, H, I) through the stage 40 (D - F) and the adult (G - I) medaka retina with antibodies against GABA (D, G), serotonin (E, H, green in F), and tyrosine hydroxylase (I, red in F). J-L: Immunostaining of transverse cryosections through the retina with the Zpr1 antibody at stage 40 (J) and in the adult (K) reveals regular arrangement of photoreceptors cell layer. Cell bodies, outer segments (asterisk), and synaptic termini (arrowhead) of photoreceptor cells are immunoreactive (L). In (A-C), asterisks indicate the midline. In (D-F, and J) dorsal is up, lateral is left. In (G-I and K-L) the ventricular surface of the retina is up. L, lens.
The six major classes of neurons in the vertebrate retina can be subdivided into many cell types (MacNeil and Masland, 1998; MacNeil et al., 1999; Rockhill et al., 2002). To investigate how some of these less numerous cell populations distribute in the medaka retina, we used antibodies to subclasses of interneurons characterized by different neurotransmitter content. Three different groups of amacrine cells were detected in the medaka retina with antibodies against neurotransmitters GABA, serotonin, and tyrosine hydroxylase, an enzyme involved in dopamine biosynthetic pathway. All 3-cell types analyzed are fairly evenly distributed in the plane of the inner nuclear layer at stage 40 and in the adult (Fig. 2D-I, and Fig. S2). Similar to what has been observed in zebrafish, GABA-positive amacrine cells are much more frequent, compared to Serotonin- and TH-positive neurons. We have observed ca. 120 GABA-positive cells per section through the central retina of medaka at stage 40 (n = 8). The numbers of Serotonin-, and TH-positive cell are roughly the same in zebrafish at 6 dpf and in medaka at 10 dpf: 5 cells per transverse section through the central retina (n=8 for medaka) (compare to Avanesov et al., 2005). Although GABA immunostaining is detected predominantly in the inner nuclear layer on plastic sections both at S40 and in the adult (Fig. 2D, G), staining of cryosections reveals the presence of GABA-positive cells also in the ganglion and horizontal cell layers (Fig. S2). This, again, is similar to the zebrafish retina, which contains GABA-immunoreactive cells in both the ganglion cell and horizontal cell layers at 6 dpf and in the adult (Sandell et al., 1994; Avanesov et al., 2005).
Orientation of cell divisions in retinal neuroepithelium
Prior to the onset of neuronal differentiation, the retina consists of proliferating progenitor cells organized into a single epithelial sheet (Das et al., 2003). These epithelial cells divide at the apical surface of the neuroepithelium (Fig. 3), and during neurogenesis gradually exit the proliferative cycle and assume a variety of neuronal fates. The relationship between mitotic spindle orientation in a dividing neural progenitor cell and the fate of its descendants has been a subject of intensive and in some cases controversial studies (reviewed in Malicki, 2004). In some mammalian retinae, cell divisions that are characterized by vertically oriented mitotic spindles are more frequent during neurogenesis (Cayouette et al., 2001). It has been suggested that these divisions give rise to asymmetric outcomes, characterized by two different cell fates. In contrast to that, horizontal spindle orientation was suggested to precede symmetric cell divisions that produce cells of the same fate (Cayouette and Raff, 2003; Zigman et al., 2005). Thus early proliferative cell divisions in the retina would have a symmetric result: two mitotically active cells, whereas neurogenic cell divisions would frequently produce an asymmetric outcome: one mitotically active cell and one postmitotic neuron. In contrast to mammals, where such a scenario may appear probable, divisions characterized by vertically oriented mitotic spindles are very rare in chick and zebrafish retinae and thus are unlikely to account for the appearance of the vast majority of postmitotic neurons (Silva et al., 2002; Das et al., 2003).
Fig. 3. Mitotic spindle orientation in the neuroepithelium of the medaka retina.
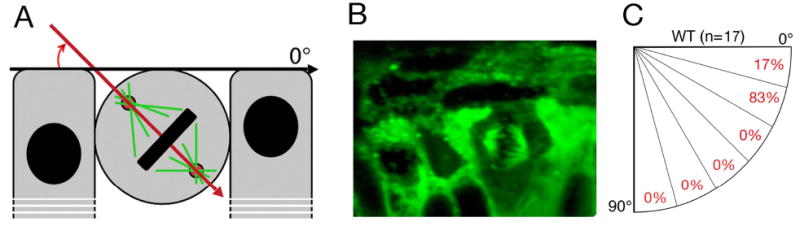
A: A diagram showing how the angle of mitotic spindle orientation was evaluated. B: Immunostaining with anti-α tubulin antibody reveals a mitotic spindle near the apical surface of the retinal neuroepithelium in the medaka eye at stage 29. C: Quantitative evaluation of mitotic spindle orientations in retinae from 10 embryos at stage 29 (n = 17). In (B), apical is up.
To test whether vertically oriented mitotic spindles are present in the medaka retina during neurogenesis, we used anti-α tubulin antibody to stain transverse sections at stage 29 (Fig. 3). At this developmental stage, neurogenesis is already in progress as evidenced by the presence of ganglion cells (Fig. 1B). Anti-α-tubulin staining identifies mitotic spindles and allows one for a rough determination of their orientation relative to the apical surface of the retinal neuroepithelium (illustrated in Fig. 3A). During the analysis of this staining pattern, the angle value of 0° was recorded when the mitotic spindle axis was parallel to the apical surface. On the opposite extreme, an angle of 90° would be recorded for a cell featuring mitotic spindle axis perpendicular to the apical surface (Fig. 3A). Analysis of cell divisions in the central retina dorsal to the optic nerve revealed that the majority (83%) feature angle values between 15° and 30°. Fewer cells (17%) were seen dividing at an angle below 15°. We did not observe any dividing cells characterized by mitotic spindle orientation exceeding 30° (Fig. 3C, n = 17). These results are similar to data obtained in zebrafish, where the angle of division did not exceed 40° (Das et al., 2003), and argue that a change in the orientation of the mitotic spindle cannot account for the switch from proliferative to neurogenic cell divisions in the medaka eye.
Formation of the photoreceptor cell layer
The sequence of photoreceptor differentiation across the ventricular surface of the retina varies in different vertebrate species (reviewed in Malicki, 2004). In zebrafish, retinal neurogenesis progresses in a wave- like fashion: it starts in a small group of ventrally located cells, the so-called ventral patch, spreads into the ventro-nasal region, and then dorsally and temporally to cover the entire retina (Raymond et al., 1995; Schmitt and Dowling, 1996; Masai et al., 2000). To characterize the spatial parameters of photoreceptor differentiation in the medaka eye, we used wholemount in situ hybridization with a rod opsin probe, and immunostaining with the Zpr-1 antibody. Photoreceptor differentiation in the medaka retina starts in two areas. First, rod opsin expression (Fig. 4A, B) and Zpr-1 staining (Fig. 4I, J) are seen in the central retina dorsal to the optic nerve. Zpr-1 expression is first seen at stage 33 (Fig. 4I), preceding the appearance of the opsin transcript at stage 35 (Fig. 4A) by ca. 24 hours. By stage 36, rod opsin in situ signal expands mostly dorsally, and remains confined to cells dorsal to the optic nerve (Fig. 4C). Interestingly, by stage 37 the second area of rod opsin expression becomes detectable at a considerable distance from the optic nerve in a small group of ventrally located cells (Fig. 4D. arrowhead). This domain is reminiscent of the so-called ventral patch, seen in the zebrafish (Larison and Bremiller, 1990). It also expands, and by stage 38 it margin reaches the optic nerve area (Fig. 4E-F). By stage 39, rod opsin transcript is present throughout the entire retina, although differences in the intensity of the in situ signal persist in different sectors of the retina at this stage and later (Fig. 4G). The adult retina shows clear expression of rod opsin in the photoreceptor layer (Fig. 4H).
Fig. 4. The progression (wave) of photoreceptor differentiation visualized via in situ hybridization with the rod opsin probe and immunostaining with the Zpr-1 antibody.
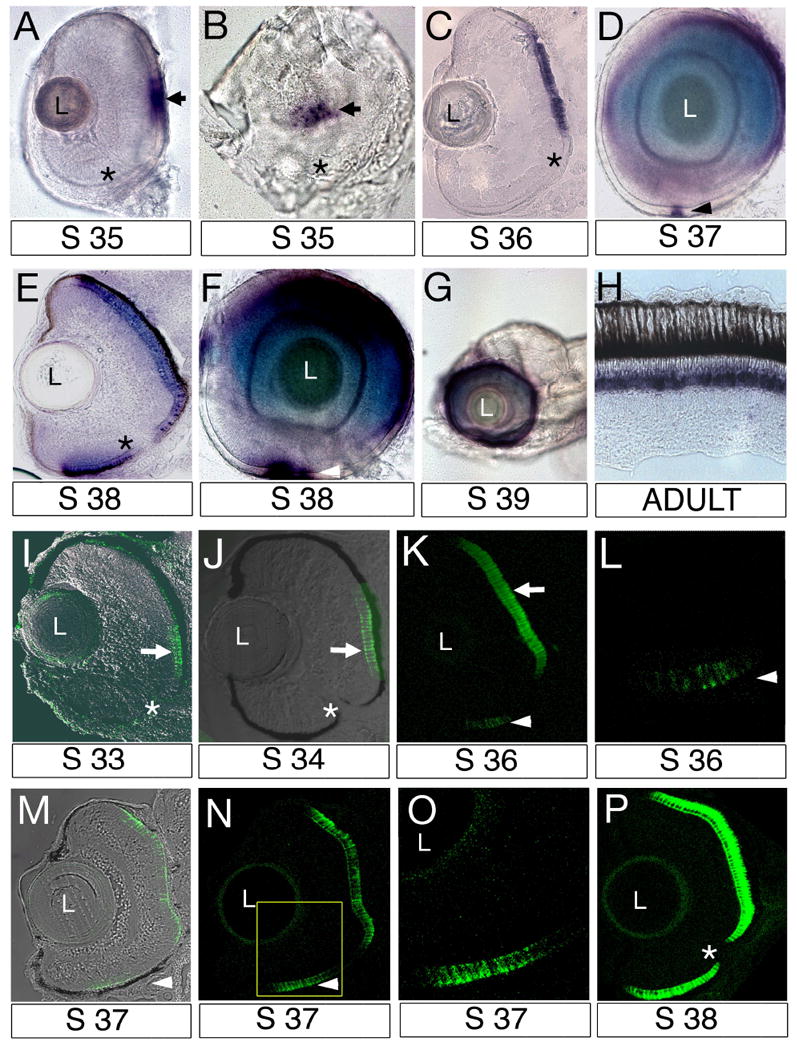
A, B: Anterior (A) and medial (B) view of the eye following whole mount in situ hybridization with the rod opsin probe at stage 35. Expression is detected in the central retina (arrows) dorsal to the optic nerve (asterisk). C: Cryosection through the retina following in situ hybridization with the rod opsin probe at stage 36. Although detectable in a fairly broad domain, rod opsin signal is present dorsal to the optic nerve only. D: Lateral view of the eye following whole mount in situ hybridization with the rod opsin probe at stage 37. The second domain of rod opsin expression appears in the ventral retina (arrowhead). E: Transverse cryosection through the retina following in situ hybridization with the rod opsin probe at stage 38. F: Lateral view of the eye following whole mount in situ hybridization with the rod opsin probe at stage 38. G: By stage 39, the expression of rod opsin is seen around the entire circumference of the eye. H: In situ hybridization with a rod opsin probe on cryosections of the adult retina. As expected, expression is seen in the nuclear region of the photoreceptor cells layer (blue signal). I-P: Confocal images of transverse cryosections through the retina stained with the Zpr-1 antibody. I: Immunostaining is detected in the central retina at stage 33. J: Zpr-1 expression domain remains restricted dorsal to the optic nerve. K: The second domain of Zpr-1 staining is detected in the ventral retina (arrowhead) by stage 36. L: Magnification of the ventral retina at stage 36. M, N: Zpr-1 expression domain in the ventral retina expands by stage 37. Transmitted light image is superimposed on immunofluorescence signal (green). O: Magnification of the ventral retina enclosed in a box in panel (N). P: Zpr-1 is detected throughout the entire retina by stage 38. In all panels dorsal is up. Asterisks indicate the optic nerve. L, lens.
Immunostaining with the Zpr-1 antibody reveals a pattern of photoreceptor differentiation similar to that detected via in situ hybridization with a rod opsin probe. Zpr-1 signal is first detected in the central retina dorsal to the optic nerve (Fig. 4I). By stage 36, the second domain of Zpr-1 expressing cells is seen in the ventral eye (Fig. 4K, L), and remains clearly separated from the dorsal one at least until stage 37 (Fig. 4M-O). By stage 38, Zpr-1 staining signal is detected in the entire photoreceptor cell layer (Fig. 4P). The spatio-temporal progression of photoreceptor differentiation as revealed by both in situ hybridization with a rod opsin probe and Zpr-1 antibody staining clearly differs from that seen in the zebrafish eye. The most obvious difference is that medaka photoreceptor differentiation initiates in the central and not ventral retina as in zebrafish. The appearance of a small ventral group of opsin- and Zpr-1-positive cells is similar to zebrafish, but takes place surprisingly late in development.
Photoreceptor mosaic
In the retinae of many teleosts, cone photoreceptors form a regular pattern, referred to as the photoreceptor mosaic (Marc and Sperling, 1976; Bowmaker and Kunz, 1987; Larison and Bremiller, 1990; Cameron and Easter, 1993; Hisatomi et al., 1997). In most cases, four double cone pairs are arranged around one single cone forming a cruciform configuration. This kind of organization has been previously reported in many species, including the medaka adult, and is referred to as the square mosaic. It has not been established, however, whether this arrangement is present in the larval medaka. We took advantage of our finding that the Zpr-1 antibody recognizes double cones in the medaka retina, and determined that these cells form a regular array in the central retina of medaka larvae already by stage 40 (Fig. 5A). Both members of the double cone pair are of the same size, and on tangential sections each double cone pair appears nearly square in shape. Each corner of a double cone pair appears to touch four other double cone pairs (Fig. 5A). In contrast to the central retina, we did not see a square pattern of cones in the retinal periphery at S40. In some cases, however, photoreceptor cells appeared to be organized in rows (Fig. 5B).
Fig. 5. Photoreceptor mosaic in the larval medaka.
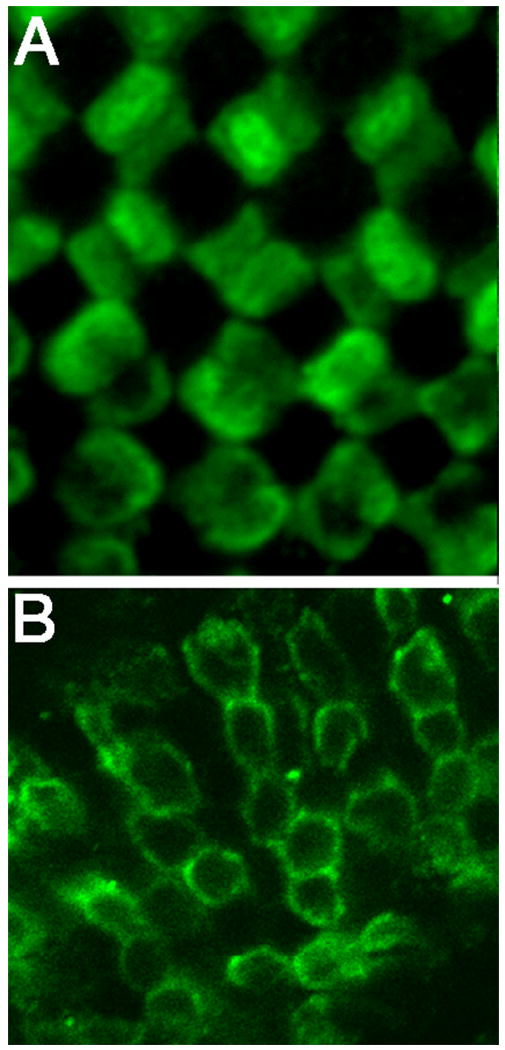
Confocal images of tangential sections through the retina stained with the Zpr-1 antibody at stage 40. A: Tangential section through the central retina, region above the optic nerve. Double cones form a regular pattern. B: Tangential section through the rostral retina. In contrast to the central retina, double cones are organized in rows.
To further characterize the photoreceptor mosaic, we used electron microscopy. This analysis confirmed Zpr-1 staining results, showing that in the adult retina four double cones assemble around one centrally located cone cell, forming a cruciform arrangement (Fig. 6C-E). Again, the two members of a double cone pair are roughly the same size, closely adhere to each other, and their interface follows a straight line (Fig. 6 D, indicated with arrows in E, H). Four additional single cones, sometimes referred to as corner cones, are positioned around the central single cone, each between a different pair of double cones. The corner cones are roughly equidistant from each other, and appear to occupy corners of a square. Curiously, cell-cell interface of the double cone pair contains several cell membranes closely sandwiched together (Fig. 6G, H). Such an arrangement of membranes has been previously reported in double cone pairs of the guppy retina (Berger, 1967). In that species, subsurface membranes are thought to lack continuity with the plasma membrane and are presumed to form two closed pouches, one in each of the two neighboring cells (Berger, 1967). Supernumerary membranes are absent in the area of contact between different cone types (Fig. 6I). In agreement with the Zpr-1 staining data, cross section through a double cone pair is roughly oval at this stage (Fig 6, compare D and E). Transverse sections through the retina of the medaka adult reveal features typical of most vertebrates: the outer segments of photoreceptor cells are arranged in multiple strata (Fig. 6A), the inner segments contain numerous mitochondria (Fig. 6A, B), and their surface is connected to the apical processes of Muller glia by belts of cell junctions (arrowheads in Fig. 6B, F).
Fig. 6. Photoreceptor mosaic in the adult medaka.
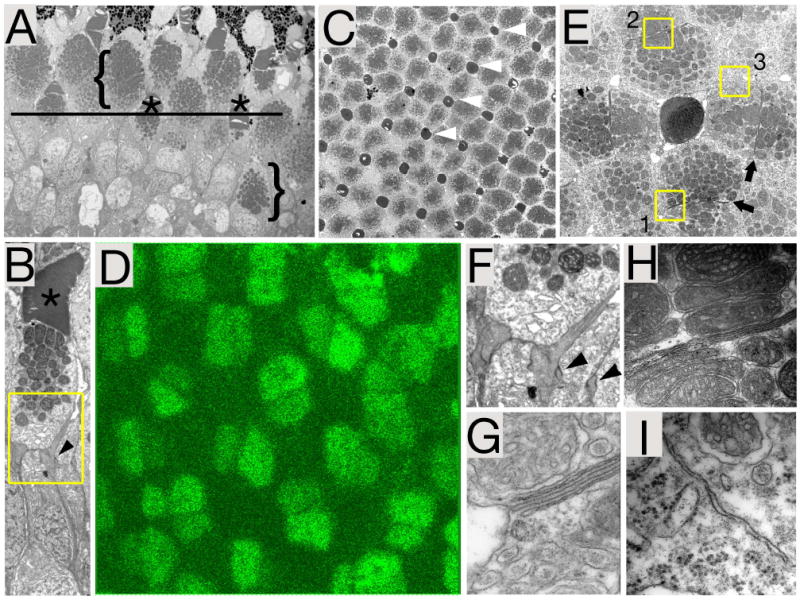
A, B: The ultrastructure of a transverse section through the photoreceptor cell layer. Typical features are present: outer segments (asterisks), mitochondria-rich inner segments (brackets), and junctions of the inner limiting membrane (arrowhead in B). C: The ultrastructure of a tangential section through the photoreceptor cell layer in the central retina. Note the regular distribution of short single cone outer segments (arrowheads). Black line in (A) indicates the approximate position of the tangential section shown in (C). D: Tangential section through the central retina stained with the Zpr-1 antibody. The regular arrangement of double cones is visible. E: An enlargement of the photoreceptor pattern shown in (C). Arrows point to the interface between the two members of double cone pairs. Boxes indicate regions of the photoreceptor mosaic enlarged in panels (H-I). F: Detail of image shown in panel (B). Cell junctions of the outer limiting membrane are indicated with arrowheads. G, H: Details of the cell-cell interface between two members of the double cone pair. Note the presence of 6 parallel membranes, four of which most likely belong to subsurface cisternae. Areas shown correspond to box 1 and 2 in panel (E). I, Cell-cell interface between a double cone and a single cone. Area shown corresponds to box 3 in panel (E).
DISCUSSION
As medaka and zebrafish are the only fish species broadly used in genetic and embryological studies, it is valuable to compare retinal development in these two organisms. One obvious difference between medaka and zebrafish is the length of embryogenesis, which in zebrafish takes considerably less time (summarized in Fig. 7A). For example, the zebrafish optic primordium forms by 12 hpf, whereas the analogous process in medaka occurs after 26 hours of embryogenesis (Kimmel et al., 1995; Iwamatsu, 2004). Similarly, based on DiI labeling experiments, the differentiation of the retino-tectal projection in zebrafish takes less than 2 days, from ca. 30 to 72 hpf. (Burrill and Easter, 1995). In the medaka, on the other hand, the same phase of development has been documented to take about four days, from 54 hpf to 6 dpf (Yoda et al., 2004; Del Bene et al., 2007). Thus the medaka retina requires almost twice as much time to complete this process, compared to zebrafish. We also observed a lengthening of developmental events while studying photoreceptor cell differentiation. In the zebrafish retina, two photoreceptor markers, Zpr-1 and rod opsin, appear in the photoreceptor cell layer almost simultaneously (Larison and Bremiller, 1990; Raymond et al., 1995). In contrast to that, in the medaka eye their onsets of expression are ca. 24 hours apart. The extended length of medaka embryogenesis is both an asset and a liability. An advantage of a slower development is that it allows one to better differentiate the onset of expression or the onset of mutant phenotypes for different genes. This is valuable when studying developmental pathways as it allows one to form hypotheses about regulatory interactions. In other types of experiments, slow development is a disadvantage as it becomes more difficult to manipulate gene function through the injections of mRNA or antisense compounds into early embryos. One has to note, however, that not all events are obviously slower in medaka development, compared to zebrafish. When studied by histological criteria, the time period between the optic nerve formation and the appearance of the inner plexiform layer takes approximately 8 hours in medaka, not much longer than the analogous process in the zebrafish retina.
Fig. 7. Summary of selected events occurring during retinal neurogenesis in medaka.
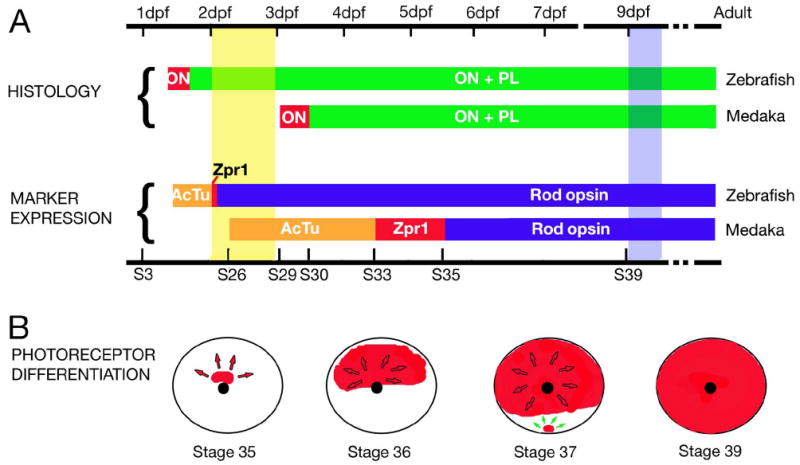
A: A schematic representation of the time course of events during retinal neurogenesis in medaka, compared to that in the zebrafish. Time is provided in days post fertilization (dpf, top scale bar). Developmental stages (S3 – S39) are also provided for medaka (bottom scale bar). The two top horizontal bars represent the time course of histological differentiation in zebrafish and medaka, color-coded to indicate when the optic nerve (ON, red) and the plexiform layers (PL, green) differentiate. The two bottom bars illustrate the onset of expression for markers of ganglion cell and photoreceptor differentiation: acetylated tubulin (AcTu, orange), Zpr-1 (Zpr1, red), and rod opsin (opsin, blue). The two vertical bars indicate the hatching period in zebrafish (yellow) and medaka (blue). (B) A schematic representation of the photoreceptor differentiation in the retina of medaka. Circle represents lateral view of the eye, and the black dot indicates the optic nerve. The distribution of the rod opsin transcript at 4 stages is shown in red. Arrows indicate the directions of wave expansion.
Photoreceptor differentiation in the medaka retina displays complex spatio-temporal characteristics as evidenced by in situ hybridization with the opsin probe, and immunostaining with the Zpr-1 antibody (summarized in Fig. 7B). This is not unexpected as spatio-temporal parameters of opsin expression are complex in many vertebrates (Bruhn and Cepko, 1996). One intriguing aspect of this process in the medaka retina is the onset of photoreceptor differentiation in two spatially separate domains. The first one forms in the center of the retina, dorsal to the optic nerve. The second, much smaller, appears close to the ventral margin of the eye. This pattern is very different from what was reported in zebrafish and goldfish. In these two related species, opsin expression is first present in a small ventral group of cells and only later spreads dorsally (Raymond et al., 1995; Stenkamp et al., 1996). For some markers of differentiation, such as atonal or cone opsin, the expression expands from the ventral retina first to the nasal, then to dorsal, and finally to the temporal region of the zebrafish eye (Masai et al., 2000). In all layers of the zebrafish retina, however, the first differentiating neurons are present in a small group of ventrally located cells (Burrill and Easter, 1995; Schmitt and Dowling, 1996; Passini et al., 1997; Hu and Easter, 1999). Interestingly, the location of these cells is very reminiscent of the second differentiation domain in medaka. One is thus tempted to speculate that this is the ancestral origin of photoreceptor differentiation, and that the central domain of expression in medaka is a more derived feature. To place such hypothesis on a firmer ground, however, one needs to consider spatio-temporal patterns of photoreceptor differentiation in the retinae of other fish species. In salmonid fishes, opsin expression originates in a single area, which is, however, located in the centro-temporal, and not ventral retina (Cheng et al., 2007). Unfortunately, to our knowledge no systematic descriptions of photoreceptor differentiation are available for other groups of teleosts. The progression of neurogenesis from a small ventral group of cells to other regions of the zebrafish retina has been shown to involve the function of the atonal and sonic hedgehog genes (Masai et al., 2000; Neumann and Nuesslein-Volhard, 2000; Stenkamp et al., 2000). It will be interesting to see whether they act in a similar manner in either of the two photoreceptor differentiation domains of the medaka eye.
Photoreceptor cells in the retinae of many teleosts as well as in some higher vertebrates are arranged in regular patterns, referred to as a mosaic. The regularity of fish photoreceptor mosaics is most obvious in the arrangement of cone cells. Although many exceptions are known, most teleost cone mosaics consist of 3 elements: the double cone, and two types of the single cone. This is true for the zebrafish (Larison and Bremiller, 1990), trout (Bowmaker and Kunz, 1987), perch (Loew and Wahl, 1991), salmon (Cheng et al., 2006), guppy (Kunz et al., 1983), and many species of the series antherimorpha (Reckel and Melzer, 2003), which includes medaka. These three elements can be organized in many ways, including row arrangement, twisted row arrangement, square pattern, and less frequently pentagonal and hexagonal patterns. The variety of arrangements is further confounded by the presence of less typical cone morphologies, such as miniature cones, triple, and even quadruple cones (Reckel and Melzer, 2003). The organization of cones is frequently inconsistent across the retina, and in many species square mosaic may exist in the center of the retina while row mosaic is present in its periphery (Cameron and Easter, 1993; Reckel and Melzer, 2003). We have found this also to be true at stage 40 in medaka larvae. Yet another complication stems from the fact that in some groups of animals cones switch fates in the course of life. In salmonid fishes, for example, single cones switch from UV to blue opsin expression during development (Cheng et al., 2006). Our analysis reveals that the medaka photoreceptor mosaic features a square arrangement of double cones. This is consistent with previous reports, which based on different criteria reached the same conclusion (Nishiwaki et al., 1997). In this type of mosaic, four double cones are arranged in a cloverleaf pattern around a single cone, and are surrounded by four other evenly spaced cones, which appear to occupy corners of a square, and are sometimes referred to as corner cones. A very similar arrangement is detected via Zpr-1 immunostaining of the central retina both in the adult and at hatching, which indicates that the medaka photoreceptor mosaic is established at an early stage, and its basic pattern does not change during development. This pattern of photoreceptor cells is also seen in the adult medaka on electron micrographs. Consistent with studies in other species, the two members of the medaka double cone pair are separated from each other by an array of several membranes. Supernumerary membranes most likely belong to subsurface cisternae, which have been previously reported to exist on both sides of the interface between double and triple cones (Berger, 1967; Reckel and Melzer, 2003). Smaller structures of this type were also reported in other neurons of the retina and elsewhere in the nervous system (Fisher and Goldman, 1975). Although they were suggested to provide insulation between neighboring cells, the function of subsurface cisternae is far from being fully understood.
The use of two teleost model organisms, the zebrafish and medaka, in parallel is likely to be productive for several reasons. In each of these models, numerous mutations affecting eye development have been isolated and characterized (Malicki et al., 1996; Neuhauss et al., 1999; Loosli et al., 2004; Muto et al., 2005). While some of them are likely to affect the same genes in both species, others will be unique to one organism. This will be the case even in the unlikely event that saturation mutagenesis is achieved, because homologous developmental pathways are likely to have evolved differently in each species and will, for example, display redundancy of different features. Similarly, functional differences are likely to exist between paralogous loci in each species given that they evolved independently for 110 million years in zebrafish and medaka lineages (Nelson, 1994). This is because paralogous genes are likely to take over different aspects of ancesteral gene’s activity, and this subdivision of functions between different paralogs is most probable to have occured differently in medaka and zebrafish evolutionary lineages (Postlethwait, 2007). Comparisons of expression patterns between two species may also be informative as they will reveal which domains of expression are conserved in both organisms and thus likely to be of functional importance. In this work, we have focused on the medaka retina, which so far received less attention, compared to zebrafish. Data presented in this paper will aid further genetic analysis of retinal development using the two genetic teleost model organisms, medaka and zebrafish, in parallel.
EXPERIMENTAL PROCEDURES
Laboratory animals
Wild-type CAB strain of Medaka was kindly provided by Dr. Wittbrodt’s lab. Animals were maintained under 14/10 hour light/dark cycle. Wild-type embryos were obtained by natural mating and staged according to Iwamatsu (1994). For effective PTU (N-Phenylthiourea) treatment, the chorion of embryos was pierced in several locations with a sharp borosilicate glass microinjection needle, and PTU solution was added to the final concentration of 0.03%.
Histology
Medaka embryos were staged according to Iwamatsu (1994) and fixed overnight in 4% paraformaldehyde (w/v) in PBST (Phosphate buffered saline, 0.1% Tween) at 4°C. Embryos were dehydrated in 50%, 75%, 85%, and 95% aqueous solution of ethanol, 15 min. each, and embedded in JB4 resin (Polysciences, Inc) as described previously (Doerre and Malicki, 2002). Sections, 5 μm thick, were prepared using a microtome (Ultracut E, Reichert Jung) and photographed with a digital camera (Axiocam, Zeiss), mounted on a microscope (Axioscope, Zeiss). Images were processed using Photoshop software.
Antibody staining
Embryos were fixed overnight in 4% paraformaldehyde (or a combination of 2% paraformaldehyde and 2% gluteraldehyde for anti-GABA staining) in PBST at 4°C, washed 2 times, 5 min. each, with PBST and infiltrated in 30% sucrose (w/v) in PBST overnight at 4°C. Subsequently, embryos were embedded in frozen section medium (Neg-50, Richard-Allan Scientific) and 5 μm sections were collected on glass slides (superfrost, Fisher). Antibody staining was performed as described previously (Avanesov et al., 2005) using the following primary antibodies: Zpr-1 (1:150, Oregon Monoclonal Bank), anti-GABA (1:500, Sigma), anti-tyrosine hydroxylase (1:100, Chemicon), anti-serotonin (1:250, Sigma), and anti-α-tubulin (1:500, Sigma).
For whole mount immunostaining experiments, embryos were fixed in 80% methanol and 20% dimethylsulfoxide (Sigma) for 5h, bleached in 10% hydrogen peroxide in methanol/DMSO solution for 2h, and stored in methanol at -20°C. Embryos were then washed 2 times, 12 min. each, with PBST, blocked with 10% goat serum (Gibco) for 30 min., incubated with mouse anti-acetylated α tubulin primary antibody (1:1000, Sigma) for 5h, washed 3 times, 30 min. each in PBST, and incubated with a fluorophore-conjugated secondary antibody for 5h. Subsequently, embryos were washed with PBST, 2 times, 30 min. each, and photographed with a digital camera as above. Photoshop software (Adobe) was used to process digital images.
In situ hybridization
PTU-treated embryos were used for in situ hybridization experiments. Rod opsin cDNA was obtained from Dr. Kawamura (Matsumoto et al., 2006). Digoxigenin labeled RNA probes were generated using Dig RNA labeling mix (Roche). Whole-mount in situ hybridization was performed according to a previously published protocol (Thisse et al., 2004). Photographs were taken with a digital camera as described above. For in situ hybridization on sections, a zebrafish whole-mount in situ protocol (Thisse et al., 2004) was used with several modifications as specified below. Medaka embryos were staged and fixed overnight at 4°C in 4% PFA in PBST. Following fixation, embryos were washed twice with PBST, transferred into 30% sucrose, and incubated at 4°C overnight for cryoprotection. Embryos were then transferred into cryoprotecting medium (Neg-50, Richard-Allan Scientific) and 5μm frozen sections were collected on superfrost slides (Fisher Scientific). Sections were dried at room temperature for 1 hour, washed twice with PBST, and digested with proteinase K (Sigma, 10 μg/ml) for 1 minute. Following proteinase digestion, sections were quickly washed twice in PBST and then incubated in PBST for 5 minutes. After washes, sections were fixed in 4% PFA in PBST for 10 minutes and then washed in PBST 3 times, 5 minutes each. 500 μl of prehybridization mix (Thisse et al., 2004) was deposited on each slide and incubated at 60 °C for 30 min. Prehybridization mix was then replaced with 150 μl of the hybridization mix and incubated overnight at 60°C. Incubations were performed in a humidified environment of a water bath chamber to prevent concentration changes of the hybridization solution. Following hybridization, slides were washed once in 2x SSC for 20 minutes at 60°C, twice in 0.2x SSC, 5 min. each, and once for 10 min. at room temperature in PBST. Sections were then incubated for 30 min. with 200 μl of blocking solution (2% sheep serum and 2mg/ml BSA in PBST), and following that overnight with 100μl of anti-DIG antibody in PBST in a humidified environment at 4 °C. Slides were then washed 3 times, 10 minutes each, with PBST, and incubated in the staining buffer (100mM Tris-HCl pH 9.5, 50mM MgCl2, 100 mM NaCl, 0.1% Tween 20) for 5 min. Staining was performed in 100 μl of BM purple substrates (Roche) according to manufacturer’s instructions. Following staining, sections were washed in PBST, coverslipped and immediately photographed as described above.
Electron Microscopy
Adult Medaka fish were anesthetized with 0.1% Tricane, placed on ice, and decapitated with a razor blade. Eyes were extracted and fixed overnight in 2.5% glutaraldehyde and 2% formaldehyde in 0.1M cacodylate buffer and 0.08M calcium chloride at 4°C. Fixed tissue was washed 3 times, 5 min each, in the cacodylate buffer and postfixed in 2% osmium tetraoxide in cacodylate buffer for 1.5 h. Following postfixation, eyes were washed for 5 min. in distilled water followed by 30 min. staining in 2% uranyl acetate. Tissue was then dehydrated in an ethanol series (25% ethanol for 5 min., 50% for 7 min., 70% for 10 min., 95% for 5 min., 95% for 10 min., 100% for 5 min., 100% for 10 min. and finally 100% for 15 min.) and embedded in Epon (Marivac Ltd., Que., Canada). Ultrathin transverse and tangential sections of the eye photoreceptor layer were obtained using standard protocols. Digital electron micrographs were produced using Philips CM-102 electron microscope and processed with Photoshop software.
Supplementary Material
Higher magnification images of laminar architecture in the medaka retina. Transverse plastic sections stained with methylene blue. A: The central retina at stage 28. Lamination is absent at this stage. B: The dorsal retina at stage 29. Plexiform layers are absent at this stage. C: The optic nerve at stage 29. D: Both the inner and the outer plexiform layer are clearly differentiated in the central retina at stage 29. E: The optic nerve at stage 30. In all panels, dorsal is up and lens is to the left. Asterisks indicate the optic nerve. Black arrows point to the inner plexiform layer. The white arrow indicates the outer plexiform layer. L, lens.
Retinal interneurons in the larval medaka. Cryosections through medaka retinae at stage 40. A: GABA signal is present in the inner portion of the inner nuclear layer, in the ganglion cell layer, as well as in horizontal cells. B: Serotonin-positive cells (green) are sparsely distributed at the inner rim of the INL. C: Similarly, tyrosine hydroxylase-positive cells (red) are also found in the inner portion of the INL. Sections in (B, C) are counterstained with phalloidin to visualize plexifom layers (blue). In all panels, dorsal is up and the midline is right. L, lens.
Acknowledgments
We thank Dr. Joachim Wittbrodt for kindly providing us with wild-type medaka adults, and Dr. Shoji Kawamura for supplying opsin cDNA. Drs. Deborah Stenkamp, Arindam Majumdar, and Novales Flamarique commented on previous versions of this manuscript. Drs. Jan Åke Gustafsson, Agneta Mode, Lotta Hambraeus, Sodertorns Hogskola, and the Department of Biosciences and Medical Nutrition at Karolinska Institutet provided support and funding for Satish Kitambi. Andreas Warkentin helped us to perform several experiments. This project was funded in part by a National Eye Institute award RO1 EY016859 (to JM).
References
- Attardi D, Sperry R. Preferential Selection of Central Pathways by Regenerating Optic Fibers. Experimental Neurology. 1963;7:46–64. doi: 10.1016/0014-4886(63)90093-1. [DOI] [PubMed] [Google Scholar]
- Avanesov A, Dahm R, Sewell WF, Malicki JJ. Mutations that affect the survival of selected amacrine cell subpopulations define a new class of genetic defects in the vertebrate retina. Dev Biol. 2005;285:138–155. doi: 10.1016/j.ydbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Avanesov A, Malicki J. Approaches to study neurogenesis in the zebrafish retina. Methods Cell Biol. 2004;76:333–384. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- Berger ER. Subsurface membranes in paired cone photoreceptor inner segments of adult and neonatal Lebistes retinae. J Ultrastruct Res. 1967;17:220–232. doi: 10.1016/s0022-5320(67)80044-3. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Bowmaker JK, Kunz YW. Ultraviolet receptors, tetrachromatic colour vision and retinal mosaics in the brown trout (Salmo trutta): age-dependent changes. Vision Res. 1987;27:2101–2108. doi: 10.1016/0042-6989(87)90124-6. [DOI] [PubMed] [Google Scholar]
- Bruhn SL, Cepko CL. Development of the pattern of photoreceptors in the chick retina. J Neurosci. 1996;16:1430–1439. doi: 10.1523/JNEUROSCI.16-04-01430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill J, Easter S. The first retinal axons and their microenvironment in zebrafish cryptic pioneers and the pretract. J Neurosci. 1995;15:2935–2947. doi: 10.1523/JNEUROSCI.15-04-02935.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill JD, Easter SS., Jr Development of the retinofugal projections in the embryonic and larval zebrafish (Brachydanio rerio) J Comp Neurol. 1994;346:583–600. doi: 10.1002/cne.903460410. [DOI] [PubMed] [Google Scholar]
- Cameron DA, Easter SS., Jr The cone photoreceptor mosaic of the green sunfish, Lepomis cyanellus. Vis Neurosci. 1993;10:375–384. doi: 10.1017/s095252380000376x. [DOI] [PubMed] [Google Scholar]
- Carl M, Loosli F, Wittbrodt J. Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development. 2002;129:4057–4063. doi: 10.1242/dev.129.17.4057. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Raff M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development. 2003;130:2329–2339. doi: 10.1242/dev.00446. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Whitmore AV, Jeffery G, Raff M. Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J Neurosci. 2001;21:5643–5651. doi: 10.1523/JNEUROSCI.21-15-05643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CL, Flamarique IN, Harosi FI, Rickers-Haunerland J, Haunerland NH. Photoreceptor layer of salmonid fishes: transformation and loss of single cones in juvenile fish. J Comp Neurol. 2006;495:213–235. doi: 10.1002/cne.20879. [DOI] [PubMed] [Google Scholar]
- Cheng CL, Gan KJ, Flamarique IN. The ultraviolet opsin is the first opsin expressed during retinal development of salmonid fishes. Invest Ophthalmol Vis Sci. 2007;48:866–873. doi: 10.1167/iovs.06-0442. [DOI] [PubMed] [Google Scholar]
- Chuang JZ, Zhao Y, Sung CH. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell. 2007;130:535–547. doi: 10.1016/j.cell.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/s0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Ettwiller L, Skowronska-Krawczyk D, Baier H, Matter JM, Birney E, Wittbrodt J. In vivo validation of a computationally predicted conserved Ath5 target gene set. PLoS Genet. 2007;3:1661–1671. doi: 10.1371/journal.pgen.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- Doerre G, Malicki J. Genetic analysis of photoreceptor cell development in the zebrafish retina. Mechanisms of Development. 2002;110:125–138. doi: 10.1016/s0925-4773(01)00571-8. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Brockerhoff SE, Hyatt GA, Dowling JE. Mutations affecting eye morphology in the developing zebrafish (Danio rerio) Dev Genet. 1997;20:288–295. doi: 10.1002/(SICI)1520-6408(1997)20:3<288::AID-DVG11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fisher SK, Goldman K. Subsurface cisterns in the vertebrate retina. Cell Tissue Res. 1975;164:473–480. doi: 10.1007/BF00219939. [DOI] [PubMed] [Google Scholar]
- Fricke C, Lee JS, Geiger-Rudolph S, Bonhoeffer F, Chien CB. astray, a zebrafish roundabout homolog required for retinal axon guidance. Science. 2001;292:507–510. doi: 10.1126/science.1059496. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Contini M, Gariboldi M, Puopolo M, Kadota K, Bono H, LeMieux J, Walsh P, Carninci P, Hayashizaki Y, Okazaki Y, Raviola E. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc Natl Acad Sci U S A. 2004;101:5069–5074. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds J, Hinds P. Early ganglion cell differentiation in the mouse retina: an electron microscopic analysis utilizing serial sections. Dev Biol. 1974;37:381–416. doi: 10.1016/0012-1606(74)90156-0. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Varga ZM, Kondoh H, Furutani-Seiki M. Single cell lineage and regionalization of cell populations during Medaka neurulation. Development. 2004;131:2553–2563. doi: 10.1242/dev.01140. [DOI] [PubMed] [Google Scholar]
- Hisatomi O, Satoh T, Tokunaga F. The primary structure and distribution of killifish visual pigments. Vision Res. 1997;37:3089–3096. doi: 10.1016/s0042-6989(97)00115-6. [DOI] [PubMed] [Google Scholar]
- Holt C, Bertsch T, Ellis H, Harris W. Cellular Determination in the Xenopus Retina is Independent of Lineage and Birth Date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev Biol. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kunz YW, Ennis S, Wise C. Ontogeny of the photoreceptors in the embryonic retina of the viviparous guppy, Poecilia reticulata P. (Teleostei). An electron-microscopical study. Cell Tissue Res. 1983;230:469–486. doi: 10.1007/BF00216193. [DOI] [PubMed] [Google Scholar]
- Larison K, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109:567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- Lemke G, Reber M. Retinotectal mapping: new insights from molecular genetics. Annu Rev Cell Dev Biol. 2005;21:551–580. doi: 10.1146/annurev.cellbio.20.022403.093702. [DOI] [PubMed] [Google Scholar]
- Loew ER, Wahl CM. A short-wavelength sensitive cone mechanism in juvenile yellow perch, Perca flavescens. Vision Res. 1991;31:353–360. doi: 10.1016/0042-6989(91)90088-m. [DOI] [PubMed] [Google Scholar]
- Loosli F, Del Bene F, Quiring R, Rembold M, Martinez-Morales JR, Carl M, Grabher C, Iquel C, Krone A, Wittbrodt B, Winkler S, Sasado T, Morinaga C, Suwa H, Niwa K, Henrich T, Deguchi T, Hirose Y, Iwanami N, Kunimatsu S, Osakada M, Watanabe T, Yasuoka A, Yoda H, Winkler C, Elmasri H, Kondoh H, Furutani-Seiki M, Wittbrodt J. Mutations affecting retina development in Medaka. Mech Dev. 2004;121:703–714. doi: 10.1016/j.mod.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Loosli F, Winkler S, Burgtorf C, Wurmbach E, Ansorge W, Henrich T, Grabher C, Arendt D, Carl M, Krone A, Grzebisz E, Wittbrodt J. Medaka eyeless is the key factor linking retinal determination and eye growth. Development. 2001;128:4035–4044. doi: 10.1242/dev.128.20.4035. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH. The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J Comp Neurol. 1999;413:305–326. [PubMed] [Google Scholar]
- MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998;20:971–982. doi: 10.1016/s0896-6273(00)80478-x. [DOI] [PubMed] [Google Scholar]
- Malicki J. Harnessing the power of forward genetics - analysis of neuronal diversity and patterning in the zebrafish retina. Trends in Neurosciences. 2000;23:531–541. doi: 10.1016/s0166-2236(00)01655-6. [DOI] [PubMed] [Google Scholar]
- Malicki J. Cell fate decisions and patterning in the vertebrate retina: the importance of timing, asymmetry, polarity and waves. Curr Opin Neurobiol. 2004;14:15–21. doi: 10.1016/j.conb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Marc RE, Sperling HG. The chromatic organization of the goldfish cone mosaic. Vision Res. 1976;16:1211–1224. doi: 10.1016/0042-6989(76)90044-4. [DOI] [PubMed] [Google Scholar]
- Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–263. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes) Gene. 2006;371:268–278. doi: 10.1016/j.gene.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Muto A, Orger MB, Wehman AM, Smear MC, Kay JN, Page-McCaw PS, Gahtan E, Xiao T, Nevin LM, Gosse NJ, Staub W, Finger-Baier K, Baier H. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. Fishes of the World. New York: John Wiley & Sons, Inc; 1994. [Google Scholar]
- Neuhauss SC, Biehlmaier O, Seeliger MW, Das T, Kohler K, Harris WA, Baier H. Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J Neurosci. 1999;19:8603–8615. doi: 10.1523/JNEUROSCI.19-19-08603.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- Nishiwaki Y, Oishi T, Tokunaga F, Morita T. Three-Dimenshional reconstitution of cone arrangement on the spherical surface of the retina in the medaka eyes. Zool Sci. 1997;14:795–801. [Google Scholar]
- Passini MA, Levine EM, Canger AK, Raymond PA, Schechter N. Vsx-1 and Vsx-2: differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J Comp Neurol. 1997;388:495–505. doi: 10.1002/(sici)1096-9861(19971124)388:3<495::aid-cne11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zoolog B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Raymond P, Barthel L, Curran G. Developmental patterning of rod and cone photoreceptors in embryonic zebrafish. J Comp Neurol. 1995;359:537–550. doi: 10.1002/cne.903590403. [DOI] [PubMed] [Google Scholar]
- Reckel F, Melzer RR. Regional variations in the outer retina of atherinomorpha (Beloniformes, Atheriniformes, Cyprinodontiformes: Teleostei): photoreceptors, cone patterns, and cone densities. J Morphol. 2003;257:270–288. doi: 10.1002/jmor.10122. [DOI] [PubMed] [Google Scholar]
- Rembold M, Loosli F, Adams RJ, Wittbrodt J. Individual cell migration serves as the driving force for optic vesicle evagination. Science. 2006;313:1130–1134. doi: 10.1126/science.1127144. [DOI] [PubMed] [Google Scholar]
- Rockhill RL, Daly FJ, MacNeil MA, Brown SP, Masland RH. The diversity of ganglion cells in a mammalian retina. J Neurosci. 2002;22:3831–3843. doi: 10.1523/JNEUROSCI.22-09-03831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell J, Martin S, Heinrich G. The development of GABA immunoreactivity in the retina of the zebrafish. J Comp Neurol. 1994;345:596–601. doi: 10.1002/cne.903450409. [DOI] [PubMed] [Google Scholar]
- Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- Schmitt EA, Dowling JE. Comparison of topographical patterns of ganglion and photoreceptor cell differentiation in the retina of the zebrafish, Danio rerio. J Comp Neurol. 1996;371:222–234. doi: 10.1002/(SICI)1096-9861(19960722)371:2<222::AID-CNE3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Silva AO, Ercole CE, McLoon SC. Plane of cell cleavage and numb distribution during cell division relative to cell differentiation in the developing retina. J Neurosci. 2002;22:7518–7525. doi: 10.1523/JNEUROSCI.22-17-07518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL, Frey RA, Prabhudesai SN, Raymond PA. Function for Hedgehog genes in zebrafish retinal development. Dev Biol. 2000;220:238–252. doi: 10.1006/dbio.2000.9629. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL, Hisatomi O, Barthel LK, Tokunaga F, Raymond PA. Temporal expression of rod and cone opsins in embryonic goldfish retina predicts the spatial organization of the cone mosaic. Invest Ophthalmol Vis Sci. 1996;37:363–376. [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Malicki J. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron. 2004;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc Natl Acad Sci U S A. 2007;104:14819–14824. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Raff M. Rod Photoreceptor Development in Vitro: Intrinsic Properties of Proliferating neuroepithelial Cells Change as Development Proceeds in the Rat Retina. Neuron. 1990;2:461–467. doi: 10.1016/0896-6273(90)90058-n. [DOI] [PubMed] [Google Scholar]
- Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nature Genetics. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Shima A, Schartl M. Medaka--a model organism from the far East. Nat Rev Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- Yoda H, Hirose Y, Yasuoka A, Sasado T, Morinaga C, Deguchi T, Henrich T, Iwanami N, Watanabe T, Osakada M, Kunimatsu S, Wittbrodt J, Suwa H, Niwa K, Okamoto Y, Yamanaka T, Kondoh H, Furutani-Seiki M. Mutations affecting retinotectal axonal pathfinding in Medaka, Oryzias latipes. Mech Dev. 2004;121:715–728. doi: 10.1016/j.mod.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Zigman M, Cayouette M, Charalambous C, Schleiffer A, Hoeller O, Dunican D, McCudden CR, Firnberg N, Barres BA, Siderovski DP, Knoblich JA. Mammalian inscuteable regulates spindle orientation and cell fate in the developing retina. Neuron. 2005;48:539–545. doi: 10.1016/j.neuron.2005.09.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Higher magnification images of laminar architecture in the medaka retina. Transverse plastic sections stained with methylene blue. A: The central retina at stage 28. Lamination is absent at this stage. B: The dorsal retina at stage 29. Plexiform layers are absent at this stage. C: The optic nerve at stage 29. D: Both the inner and the outer plexiform layer are clearly differentiated in the central retina at stage 29. E: The optic nerve at stage 30. In all panels, dorsal is up and lens is to the left. Asterisks indicate the optic nerve. Black arrows point to the inner plexiform layer. The white arrow indicates the outer plexiform layer. L, lens.
Retinal interneurons in the larval medaka. Cryosections through medaka retinae at stage 40. A: GABA signal is present in the inner portion of the inner nuclear layer, in the ganglion cell layer, as well as in horizontal cells. B: Serotonin-positive cells (green) are sparsely distributed at the inner rim of the INL. C: Similarly, tyrosine hydroxylase-positive cells (red) are also found in the inner portion of the INL. Sections in (B, C) are counterstained with phalloidin to visualize plexifom layers (blue). In all panels, dorsal is up and the midline is right. L, lens.


