Abstract
Viral diversity and lifecycles are poorly understood in the human gut and other body habitats. Therefore, we sequenced the viromes (metagenomes) of virus-like particles isolated from fecal samples collected from adult female monozygotic twins and their mothers at three time points over a one-year period. These datasets were compared to datasets of sequenced bacterial 16S rRNA genes and total fecal community DNA. Co-twins and their mothers share a significantly greater degree of similarity in their fecal bacterial communities than do unrelated individuals. In contrast, viromes are unique to individuals regardless of their degree of genetic relatedness. Despite remarkable interpersonal variations in viromes and their encoded functions, intrapersonal diversity is very low, with >95% of virotypes retained over the period surveyed, and with viromes dominated by a few temperate phage that exhibit remarkable genetic stability. These results indicate that a predatory viral-microbial dynamic, manifest in a number of other characterized environmental ecosystems, is notably absent in the very distal intestine.
The diversity of viruses in the gut, and their role in the assembly, maintenance and adaptations of the microbiota and its pool of genes (microbiome) remain unclear. In many environments, the dominant ecological relationship between viruses and their microbial hosts is predatory and follows Lotka-Volterra (LV)/ Kill-the-Winner dynamics. LV is characterized by top-down control of microbial communities (i.e., microbial biomass is significantly below the carrying capacity of the habitat), rapid microbial and viral population shifts, and evidence of Red Queen co-evolution (i.e. escape strategies in prey population are countered by predator adaption). One manifestation of this arms race is positive selection on loci like bacterial O-antigens and CRISPR elements1,2, and viral tail fibers3. In contrast to this predator-prey dynamic, there is another viral life cycle where temperate rather than lytic viruses are longer term contributors to microbial host phenotypes through provision of adaptive genes. This latter dynamic can change the metabolic capacities of free living bacteria, obligate intracellular mutualists4, as well as the lifestyles of pathogens5. In fact, many of the differences between closely related microbial strains arise from prophage insertions6,7.
Sampling the DNA virome
Recent studies of the human gut virome have focused on pathogen discovery8, or have analyzed a few individuals without defining the microbial composition of their gastrointestinal tracts9–11. In this report, we characterize the fecal viromes of four pairs of adult female monozygotic (MZ) twins and their mothers. All were healthy and none had received antibiotics in the 6-month period prior to sampling. Fecal samples were obtained at the beginning of the study, two months later (50±9 days) and 1 year later (364±10 days). VLPs were purified from 32 frozen fecal specimens. Since the yield of VLP DNA from 2–5 g of feces averaged 500 ng, we performed random amplification of the purified viromes to obtain sufficient material for shotgun 454 FLX pyrosequencing. After quality filtering, our final dataset contained 280,625,127 nt (Table S1). We verified the reproducibility of the protocol by sequencing replicates from 5 samples (Table S1; Methods). One additional sample was subjected to deeper sequencing (70,157,333 nt). Bacterial taxa represented in fecal samples were characterized by pyrosequencing of their 16S rRNA genes (Table S2). We had previously performed12 shotgun sequencing of total fecal DNA isolated from the 12 frozen samples obtained at the first time point that were now used to prepare purified VLPs (Table S3).
Prominence of prophage and phage
We generated a custom non-redundant viral database (NR_Viral_DB) to facilitate analysis of VLP-derived metagenomic datasets (see Methods). The NR_Viral_DB contained 4,193 genomic sequences (96.2 Mb): 73.3% were eukaryotic viral genomes while 25.8% were phages and prophages; 76.9% of the latter were dsDNA phage, mostly members of the order Caudovirales. The bacterial hosts of these known bacteriophages are principally members of the Proteobacteria (54%), Firmicutes (32%) and Actinobacteria (7%).
A relaxed search against the NR_viral_DB (tblastx; e-value < 1e-3) showed that 81±6% (mean±S.D.) of reads generated in this study did not match to any known viruses (Fig. S1). However, most of the identifiable viruses in the 32 VLP-derived viromes were prophage or phage generally classified as temperate (Fig. 1). The Podoviridae illustrate this point: this family consists of both lytic and temperate members; in the fecal viral community its dominant representatives were temperate (e.g., coliphage P22-like). The predicted hosts of the identifiable phage and prophage were members of the Firmicutes and Bacteroidetes (Fig. 1); these phyla, and in particular the families Ruminococaceae, Lachnospiraceae and Bacteroidaceae, comprised the most abundant bacterial taxa in the sampled fecal microbial communities, as defined by 16S rRNA gene analysis (Fig. S2).
Fig. 1. Classification of viruses present in VLP preparations generated from fecal samples collected from four families of MZ twins and their mothers.
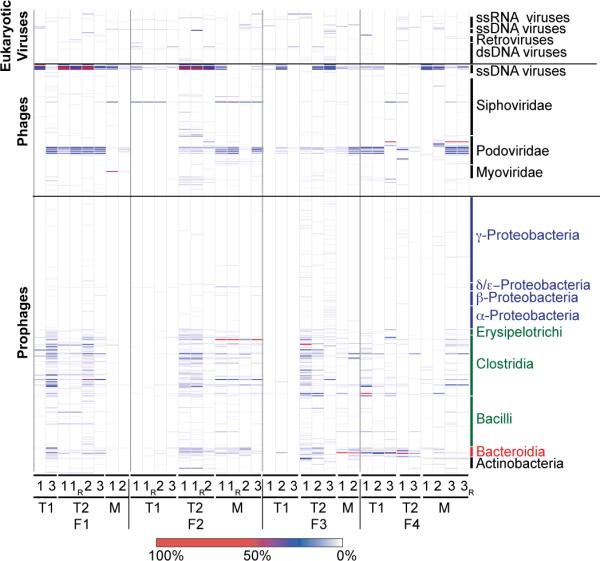
Prophages are classified based on their bacterial host taxonomy. Prominent bacterial phyla are represented by different colors (Proteobacteria, blue; Firmicutes green; Bacteroidetes, red; Actinobacteria, black). Class-level taxa within these phyla are indicated. Phage and eukaryotic viruses are sorted according to taxonomy. Nomenclature used for VLP preparations from fecal biospecimens: F, family; T1, co-twin 1; T2, co-twin 2; M, mother of co-twins. Time points (1–3), and technical replicates (R) produced from a given sample are noted. The color bar at the bottom of the figure provides a reference key for the percent coverage of a viral genome in the NR_Viral_DB by reads from given VLP virome dataset (data normalized using 14,000 randomly selected reads/dataset).
Intra- and interpersonal variation in viromes
Two different approaches were used to analyze `within VLP sample' diversity (i.e. alpha diversity estimates). First, CD-Hit-est13 was used to cluster reads with ≥90% sequence identity over 85% of their length. This allowed us to calculate a cluster-level Shannon index for each VLP sample (Table S4) and to define the expected number of virotypes per VLP preparation using procedures described in Methods (median of 346; range 52–2773). Second, PHACCS (Phage Communities from Contig Spectra) analysis (see Methods) indicated that each sample contained a median of 35 (range 10–984) predicted virotypes (Table S5; average Shannon Index of 3.32±0.71). Analysis of 16S rRNA datasets, where noise due to PCR and pyrosequencing artifacts and chimeras had been removed, indicated that there were ~800 species-level bacterial phylotypes in the first time point fecal communities14. Recent results obtained from deep shotgun sequencing of other fecal microbiomes suggest that the number may be <200 species15. Thus, the ratio of virotype to bacterial phylotype in the fecal microbiota of these healthy adults appears to approach 1.
Two complementary methods were employed to define the percentage of shared viral sequences between fecal samples obtained at different time points from the same individual and from different individuals (beta-diversity estimates). In the first method, we used CD-Hit-est to cluster pooled reads from all of the VLP viromes. Hellinger distances were subsequently calculated based on a matrix of the number of CD-Hit clusters vs VLP samples to determine relationships between samples. The second approach was based on contig assemblies generated from the pooled VLP viromes (Maxiphi16; see Methods). Both beta-diversity estimates showed that the same individual harbors very similar fecal viral communities over at least a one-year period: i.e., within an individual >95% of the viral genotypes were present and their relative abundances showed minimal variation (<8% permutation of the rank abundance). This is different than other comparably characterized ecosystems where almost daily changes in beta diversity occur17.
While intrapersonal variation in viromes was minimal, interpersonal variation was very high (Fig. S3). To analyze the clustering patterns generated by the distance matrices, 100 random sub-samplings of equal number of sequences per sample were used to generate the distance matrices, and a consensus UPGMA tree. These trees showed that the main branching pattern clusters samples from the same individual, while there was no significant clustering of samples from the same family (Fig. S4 and S5).
To further establish that temperate phages are dominant members of fecal VLP preparations, we searched for sequence identity between VLP viromes and 121 sequenced human gut microbial genomes. We identified 13 different bacterial genomes containing prophages present at high abundance in at least one VLP sample (Table S6). For example, Fig. S6 shows a region of the Ruminococcus torques ATCC 27756 genome that contains a predicted ~60Kb prophage. Sequence identity plots demonstrated that this prophage was prominently represented in a VLP sample from the mother of family 2 [F2M.1 and F2M.1 (R)] where it comprised 18–20% of reads at the first time point; at the two later time points (2 months and 1 year) the phage was still present albeit at lower abundance (0.8 and 1.4%, respectively). This phage was not detectable in her twin daughters or other individuals in our study.
Given the low diversity of individual fecal viromes and the apparent representation of a few high abundance phage, we attempted to assemble partial or complete phage genomes using high stringency conditions (see Methods). This effort yielded 5,004 contigs ≥500 bp, 88 of which were 10Kb-71.4Kb (Fig. S7). All VLP-derived pyrosequencing reads from all 32 datasets were then aligned against these large contigs. The results revealed that virotypes represented by the large contigs were present mainly in only one individual where their abundance varied over time (Table S7). The nucleotide sequence conservation of these contigs (expressed as percent identity between reads from a given VLP sample and the contig) during the 1-year period was astonishingly high within an individual (99.74±0.26%). Only 8 of the 88 contigs were present in more than one individual from different families, with an average percent identity of 88.23±1.4% between subjects (Table S8).
Functional variation among viromes
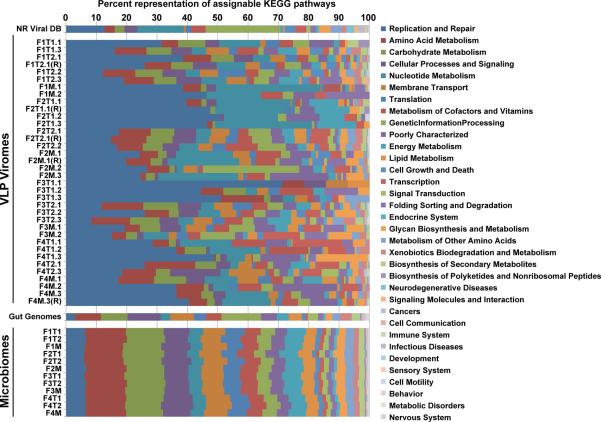
We next defined functions encoded by the 32 fecal VLP-derived viromes by querying the KEGG and COG databases. The same procedure for functional assignments was used for the 12 shotgun fecal microbiome datasets generated from the first time point sample from each individual in the 4 families. The percentage of fecal VLP-derived reads with significant hits to the COG and KEGG databases (BLAST cutoff, E<10−5) was only 3.2±2.8% (mean±S.D.) and 1.7±1.9% (mean±S.D.), respectively, compared to 36.0±6.9% (mean±S.D.) and 23.3±3.1% (mean±S.D.) for reads obtained from the 12 total fecal community DNA samples (Fig. S8). Comparison of VLP-derived viromes and the 12 microbiomes revealed significant functional differences (Fig. S9) in agreement with previous studies of aquatic ecosystems where viruses were significantly enriched for genes related to DNA, RNA synthesis and replication while the corresponding microbial communities were enriched for nitrogen and carbohydrate metabolism, and membrane transport18. Fig. 2 and S10 provide a sample-by-sample view of the proportional representation of KEGG and COG categories in sequenced purified VLP-derived viromes and in fecal microbiomes. There is only modest interpersonal variation in the distribution of KEGG and COG pathways in the microbiomes (R2=0.993±0.005 for KEGG, 0.984±0.013 for COG). Moreover, the distribution of these functions is similar to their distribution in 121 sequenced human gut genomes (R2=0.82 for KEGG, 0.95 for COG; Table S9). In contrast, there is marked variation in these functional categories in the sequenced VLP-associated viromes (Fig. 2 and S10), although further and deeper analysis of these differences was limited by the low percentage of viral reads that were classifiable.
Fig. 2. A sample-by-sample view of the proportional representation of KEGG second level pathways in sequenced VLP-associated viromes and gut microbiomes.
Known or predicted proteins encoded by viruses in the NR_Viral_DB, fecal VLP-derived viromes, 121 sequenced reference human gut-associated microbial genomes, and fecal microbiomes are shown. See Fig. 1 for sample nomenclature.
The 88 VLP-derived large contigs encode 2,440 predicted proteins, 830 of which have significant similarity to viral or bacterial proteins present in the NR_Viral_DB and/or in the 121 gut genomes (blastx e-value < 1e-5). Metastats analysis identified significant differences in the representation of KEGG and COG functions associated with the large contigs compared to the NR_Viral_DB, including pathways related `Glycan metabolism', `Cell Wall Biosynthesis', and `Transcription' (Fig. S11). To identify genes that may confer new and potentially advantageous functions to viruses present in the distal gut microbiota and/or to their microbial hosts, we searched this list of 2,440 proteins, eliminating those with homologs in the NR_Viral_DB, as well as all others whose putative functions suggested a viral origin (e.g., polymerases, capsid proteins, holins, etc). We were left with 23 proteins belonging to 16 protein families (Table 1). These proteins, seven of which use iron or sulfur in their reactions, are involved in a number of processes associated with the anaerobic gut microbiota. Homologs of these proteins present in 121 gut genomes were subsequently retrieved, aligned19 and approximate maximum likelihood trees generated using FastTree20. The results (Fig. S12) indicate that some of the VLP virome-associated proteins are evolving in ways that are distinguishable from homologs present in known sequenced bacterial genomes, as is the case in the few environmental communities that have been subjected to comparable metagenomic studies21.
Table 1. Proteins encoded by 88 large viral contigs assembled from fecal VLP viromes that have no homologs in the NR_Viral_DB and whose functions are involved in processes associated with the anaerobic gut microbiota.
This list of proteins includes (i) two transcriptional regulators (a homolog of ExsB involved in regulation of succinoglycan levels28; and an anaerobic nitric oxide reductase regulator belonging to the sigma 54 family); (ii) an anaerobic ribonucleoside triphosphate reductase activating protein that uses S-adenosylmethionine (SAM), an iron-sulfur cluster, and a reductant for the de-novo anaerobic synthesis of nucleotides29–31; (iii) other SAM-related proteins (Fe-S oxidoreductase, and a SAM-decarboxylase that uses SAM for synthesis of spermidine and spermine, which in turn stimulate RNA polymerases and stabilize the DNA helix respectively32), (iv) three oxidative stress-related proteins (an iron/manganese superoxide dismutase, thioredoxin, and a ferritin Dps family protein); (v) a methylglyoxal synthase homolog involved in pyruvate metabolism, (vi) a thymidylate synthase and a 6-pyruvoyl tetrahydropterin synthase involved in folate metabolism; (vii) a member of the phosphoadenosine phosphosulfate reductase family that participates in the cysteine biosynthesis and uses thioredoxin as electron donor; (viii) cysteine desulfurase (nifS), which plays an important role in Fe-S cluster biosynthesis by catalyzing removal of sulfur from cysteine to produce alanine; and (ix) a group of proteins involved in peptidoglycan synthesis [a member of CAZy Glycosyltransferase family 2 (GT2), a GT25 member, and five N-acetylmuramoyl-L-alanine amidases; acquisition of this last group of enzymes is intriguing in light of evidence that some phages can subvert normal bacterial pathways for surface glycan biosynthesis5].
| No of ORFs | Name |
|---|---|
| 5 | N-acetylmuramoyl-L-alanine amidase [EC 3.5.1.28] |
| 3 | Thymidylate synthase [EC 2.1.1.148] |
| 2 | 6-pyruvoyl tetrahydropterin synthase [EC 4.2.3.12] |
| 1 | Anaerobic nitric oxide reductase transcription regulator (NifA) |
| 1 | Fe-S Oxidoreductase |
| 1 | Anaerobic ribonucleoside-triphosphate reductase activating protein |
| 1 | ExsB |
| 1 | Phosphoadenosine phosphosulfate reductase family member [EC 1.8.99.4] |
| 1 | Ferritin Dps family protein |
| 1 | Glycosyltransferase family 25 |
| 1 | Glycosyltransferases family 2 |
| 1 | Methylglyoxal synthase [EC 4.2.3.3] |
| 1 | Iron/manganese superoxide dismutases, C-terminal domain [EC 1.15.1.1] |
| 1 | Thioredoxin [EC 1.8.4.8] |
| 1 | S-adenosylmethionine decarboxylase [EC 4.1.1.50] |
| 1 | Cysteine desulfurase [EC 2.8.1.7] |
Evidence of a temperate lifecycle
Integrases are markers of temperate phage. We identified 10 ORFs with homology to integrases in the 88 contigs, and 8,955 reads with significant similarity (blastp e-value <1e-4) to 785 different integrases among the 1,386,331 reads comprising the entire VLP dataset. The number of hits to different integrases per VLP sample was then used to construct a distance matrix that showed that (i) the diversity among identified integrases was significantly lower within VLP viromes purified from the same individual over time than between individuals, and (ii) there were no significant differences between individuals regardless of family relationships (Fig. S13).
As noted above, in most ecosystems where phage-host interactions have been studied in detail, lytic lifecycles and Red Queen dynamics appear to dominate22. Metagenomic studies of salterns and sludge ecosystems indicate that many of the most apparent genetic changes over time are at loci that prevent phage attachment (e.g., outer-membrane proteins and polysaccharides). Probably the clearest example of Red Queen dynamics between phage and their hosts are the CRISPR elements in sludge1 and acid mine drainage systems2. Similarly, phage genomes often show evidence of changes in their tail fibers over time3. Given this paradigm, it is striking that we found essentially no evidence of this type of behavior in fecal phages (see Supplementary Discussion for analysis of CRISPRs). In contrast, we found high abundances of dominant phage, present in the same individual for extended periods of time, with no significant divergence or mutations in their genomes. The presence of integrases in the assembled VLP contigs is also consistent with the notion that they represent prominent temperate phage in the fecal microbiota.
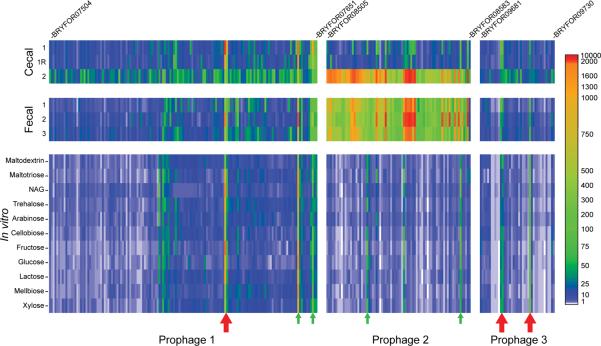
One potential scenario is that phage production occurs via induction of prophages caused by energy limitation in the feces; at this point, fecal microbial hosts are effectively at a dead end for their associated phage, and the viruses may gain an advantage for transmission by `going it alone'. Experimental evidence for this scenario is provided by gnotobiotic mice co-colonized for two weeks with Marvinbryantia formatexigens, a human gut acetogen that contains three predicted prophages, and Bacteroides thetaiotaomicron, a saccharolytic bacterium that harbors two predicted prophages. Normalized RNA-Seq counts, generated from cecal contents and fecal samples harvested at the time of sacrifice (n=3 mice)23, revealed that one of the three prophages in M. formatexigens was completely activated (all ORFs transcribed) in all fecal samples and in a subset of cecal samples (Fig. 3). In the case of the remaining two M. formatexigens prophages, only a few genes were expressed, including a pair of adjacent ORFs encoding a HicA family toxin (BRYFOR7601) and a HicB family anti-toxin (BRYFOR7602) in one of the prophages, and a pair of genes that specify a RelE family toxin (BFYFOR9696) and a PHD family anti-toxin (BRYFOR9697) in the other prophage; these two gene pairs were constitutively expressed in all fecal samples, in all cecal samples, and during in vitro growth in defined medium containing a variety of carbon sources (Fig. 3). Co-expression of toxin-antitoxin genes is known to maintain stable integration of phage DNA in bacterial chromosomes24,25. Importantly, the one prophage that was fully activated (prophage 2 in Fig. 3) does not have a detectable toxin/anti-toxin gene pair. Only two small (2–3 gene) clusters were expressed in the two B. thetaiotaomicron prophages in vivo; all of these clusters encode predicted toxin or anti-toxin genes (Fig. S14). These findings illustrate how a prophage may be liberated from its host cell when that cell is present in a fecal community.
Fig. 3. Gnotobiotic mice reveal in vivo activation of the transcriptome of a Marvinbryantia formatexigens prophage.
Shown are the three predicted prophages present in M. formatexigens and the levels of expression of their ORFs in cecal and fecal micobial communities harvested from gnotobiotic mice co-colonized with Bacteroides thetaiotaomicron. Expression levels for genes in each prophage genome are from normalized RNA-Seq read count data (see color key; normalization based on sequencing effort and length of each predicted ORF). Active expression is defined as a normalized read count >100. R, technical replicate shows the reproducibility of the method for performing RNA-Seq analysis. RNA-Seq data are also presented for each prophage genome in M. formatexigens during mid-log phase growth in defined medium containing different carbon sources (NAG, N-acetylglucosamine). Red arrows indicate the position of toxin/anti-toxin gene pairs. Green arrows denote genes with hypothetical functions that are expressed in more than 50% of the conditions tested. ORF designations for the first and last genes in the predicted genomes of each prophage are provided.
Incorporating virome analyses into HMPs
Human microbiome projects (HMPs) have been initiated throughout the world to define the interrelationships between physiologic status, and/or disease states and microbial community structure and function. Our results suggest that these metagenomic studies should also include VLPs recovered from various body habitat microbiota (Fig. S15 and Supplementary Discussion). Functions embedded in both dominant and sub-dominant phages may provide informative molecular signatures (biomarkers) of a microbiota and its human host, of microbial community responses to impending or fully manifest disease states, and of the extent to which community health or pathology endures after apparent recovery of the human host from a disease or therapeutic intervention. In addition, gnotobiotic mice harboring defined collections of human gut symbionts inoculated with VLP-derived viromes should provide informative models for further dissection of various aspects of the interactions of phage and their microbial hosts in different regions of the gut, including an assessment of whether LV dynamics operate in more proximal regions of the intestine where energy generated from dietary components may be more available.
Methods Summary
Sample collection
The Missouri Adolescent Female Twin Study (MOAFTS26) is composed of female twin pairs born in the state of Missouri between 1975–1986, and their mothers. Procedures for obtaining informed consent and sample collection were approved by the Washington University Human Studies Committee.
DNA extraction and 454 Pyrosequencing
Aliquots of frozen fecal samples (2–5 g) were processed for isolation of VLPs by serial filtration, followed by cesium chloride gradient ultracentrifugation27. VLPs were lysed in a solution containing Proteinase K and 10% SDS. DNA was extracted with 10% cetyltrimethylammonium bromide/0.7M NaCl and amplified using the illustra™ GenomiPhi™ V2 kit (GE Healthcare). The resulting DNA was used for multiplex shotgun 454 FLX pyrosequencing. For further details about VLP purification, extraction of VLP DNA, assembly of pyrosequencer reads, and data analysis see Methods.
Methods
Purification of VLPs
Viral purification was performed with minor modifications of the procedure described in an earlier publication27. In brief, all samples were frozen at −20°C within 30 min after donation, placed at −80°C within 36h, and maintained at −80°C until use. For VLP purification a 2–5 g aliquot of each fecal sample was re-suspended in 25 mL SM buffer [100 mM NaCl, 8 mM MgSO4, 50 mM Tris (pH 7.5) and 0.002% gelatin (wt/vol)]. Following centrifugation (2,500 × g for 10 min at room temperature), the resulting supernatant was removed and passed sequentially through 0.45μm and 0.22μm Whatman filters to remove residual cells. The filtrate was then adjusted with CsCl to a density of 1.12g/mL and deposited on top of a 3 mL step gradient prepared using 1mL CsCl solutions with densities of 1.7 g/mL SM buffer, 1.5 g/mL, and 1.35 g/mL. Samples were centrifuged for 2h at 60,000 × g (4°C) in a SW41 swinging bucket rotor (Beckman). The 1.5 g/mL layer was recovered since material in this density range is known to be enriched for bacteriophages27. At each step of the purification procedure, an aliquot of the sample was viewed under an epifluorescence microscope after viral particles had been stained with SYBR-gold; this allowed us to document the presence of VLPs and note whether a decrease in the representation of bacterial and eukaryotic cellular elements had occurred.
Extraction of viral DNA
After the 1.5 g/mL layer was collected from the step gradient, chloroform was added (0.2 volumes) and the solution was centrifuged for 5 min at 2,500 × g. The aqueous phase was treated with DNAse (Sigma Aldrich; final concentration 2.5U/mL) to remove residual host and bacterial DNA. To extract the virions, 0.1 volume of 2M Tris HCl/ 0.2 M EDTA, 1 volume of formamide, and 100μL of a 0.5M EDTA solution were added per 10mL of sample, and the resulting mixture was incubated at room temperature for 30 min. The sample was subsequently washed with 2 volumes of ethanol and pelleted by centrifugation for 20 min at 8,000 × g at 4°C. The pellet was washed twice with 70% ethanol and re-suspended in 567μL of TE buffer, followed by 30μL of 10% SDS and 3μL of a 20 mg/mL solution of Proteinase K (Fisher Scientific; cat no. AC61182-0500). The mixture was incubated for 1h at 55°C, and 100 μL of 5M NaCl and 80μL of a solution of 10% cetyltrimethylammonium bromide/0.7M NaCl were subsequently introduced. After a 10 min incubation at 65°C, an equal volume of chloroform was added and the mixture was centrifuged (5 min at 8,000 × g; room temperature). The resulting supernatant was transferred to a new tube and an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) was added, followed by centrifugation (5 min at 8,000 × g; room temperature). The supernatant was recovered and an equal volume of chloroform was introduced. Following centrifugation, the supernatant was collected and 0.7 volumes of isopropanol used to precipitate the DNA. After another centrifugation step (15 min at 13,000 × g at 4°C), the material was washed (500μL of cold 70% ethanol), air dried, and resuspended in 50μL of TE. An aliquot of the purified DNA was used as a template in polymerase chain reactions that contained universal primers directed at bacterial 16S rRNA and eukaryotic 18S rRNA genes; this assay was used to confirm the absence of detectable contaminating non-viral DNA.
Amplification of VLP-associated DNA
Shotgun 454 pyrosequencing requires 3–5μg of DNA for library preparation. The typical yield from our fecal VLP DNA isolation procedure was 500ng/sample. Therefore, WGA (Whole Genome Amplification) was performed using reagents and protocols in the illustra™ GenomiPhi™ V2 kit (GE Healthcare) to generate sufficient material for library construction. Ten to 50ng of purified VLP DNA were mixed with 9μL of `Sample Buffer' from the kit and heat denatured at 95°C for 3 min. Nine microliters of `Reaction Buffer' and 1μL of `Enzyme Mix' were then added and the solution incubated for 90 min at 30°C. Three separate WGA reactions were performed for each viral DNA preparation to minimize potential bias in amplification. The amplified products from each sample were subsequently pooled and purified (QIAGEN DNeasy kit).
To test for bias in the amplification and sequencing of VLP-DNA preparations that had been subjected to WGA, we analyzed the VLP sample from individual F3T1.1 where the yield of DNA was sufficient to perform shotgun pyrosequencing with un-amplified as well as with amplified subsamples. We pooled 16,567 reads derived from WGA DNA, and 18,845 reads from the unamplified aliquot and clustered them using the procedures used for alpha diversity calculations (see CD-Hit Clustering below). We found that 98.4% of the sequences from the un-amplified DNA were also present in the WGA reads while 91.96% of the WGA sequences were represented in the un-amplified sample dataset. This difference could be due to sequencing of amplified low abundance viral DNAs that were not sequenced in the unamplified sample. WGA is also known to preferentially amplify small ssDNA viruses17.
Multiplex shotgun pyrosequencing of VLP viromes
DNAs, purified from each of 12 VLP preparations, were labeled with a different MID (Multiplex Identifiers; Roche). Equivalent amounts of the barcoded samples were then pooled prior to a run of 454 FLX pyrosequencing. Shotgun reads were filtered by removing (i) all duplicates (defined as sequences whose initial 20 nucleotides are identical and that share an overall identity of >97% throughout the length of the shortest read; duplicates are a known pyrosequencing artifact33), (ii) reads with degenerate bases (`N's), and (iii) sequences with significant similarity to human reference genomes (BLASTN with e-value < 1E-5) in order to ensure the de-identification of samples.
Bacterial 16S rRNA gene amplification and sequencing
An aliquot (500mg) of each frozen pulverized fecal sample was re-suspended in a solution containing 500μL of extraction buffer [200mM Tris (pH 8.0), 200mM NaCl, 20mM EDTA], 210μL of 20% SDS, 500μL of phenol:chloroform:isoamyl alcohol (25:24:1) and 500μL of a slurry of 0.1-mm diameter zirconia/silica beads (BioSpec Products). Cells were mechanically disrupted using a bead beater (BioSpec Products) set on high for 2 min at room temperature, followed by extraction with phenol:chloroform:isoamyl alcohol and precipitation with isopropanol. DNA obtained from three separate aliquots of each fecal sample were pooled and used for amplification of bacterial 16S rRNA genes.
Approximately 330bp amplicons, spanning variable region 2 (V2) of bacterial 16 rRNA genes were generated by using PCR and (i) modified primer 8F (5′-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3') which consisted of 454 primer B (underlined) and the universal bacterial primer 8F (italics) and (ii) modified primer 338R (5' GCCTCCCTCGCGCCATCAGNNNNNNNNNNNNCATGCTGCCTCCCGTAGGAGT 3') which contained 454 primer A (underlined), a sample specific, error correcting 12-mer barcode34 (N's), and the bacterial primer 338R (italics).
Four replicate polymerase chain reactions were performed for each pooled fecal DNA sample. Each 20μL reaction contained 100ng of gel purified DNA (Qiaquick, Qiagen), 8μL 2.5× HotMaster PCR Mix (Eppendorf), and 0.3 μM of each primer. The PCR program consisted of initial denaturation at 95°C for 2 min followed by 30 cycles of denaturation (95°C for 20 sec), annealing (52°C for 20 sec) and amplification (65°C for 1 min). Replicate PCRs were subsequently pooled and purified using Ampure magnetic purification beads (Agencourt). DNA was quantified using Picogreen (Invitrogen) and an equimolar amount of each sample was used for multiplex 454 FLX amplicon pyrosequencing.
Bacterial 16S rRNA data processing and analysis
16S rRNA reads were analyzed using QIIME35: fasta, quality files and a mapping file indicating the barcode sequence corresponding to each sample were used as inputs. The QIIME pipeline takes this input information and split reads by samples according to the barcode, and classifies reads into OTUs based on sequence similarity. It also performs taxonomical classification using the RDP-classifier36, builds a de-novo taxonomic tree of the sequences based on sequence similarity, and creates a sample x OTUs table that can be used, together with the tree, for calculating alpha and beta diversity.
Custom Non Redundant viral database (NR_Viral_DB)
All complete viral and bacteriophage genomes available in the European Bioinformatics Institute (EBI) database were downloaded, as were all complete prophage genomes present in the SEED database and all entries for the taxid 10239 (Viruses) in RefSeq between 1Kb – 500Kb. The database was complemented with prophage sequences identified from a survey of 396 sequenced microbial genomes (Table S10) using the software tool PhageFinder37. To make the database non-redundant, all sequences were compared against each other and only those with <95% identity throughout their length were retained.
CD-Hit Clustering
CD-Hit-est is a software tool designed for clustering nucleotide sequences by similarity13. We used CD-Hit-est to cluster the pooled reads obtained from all viral samples. Hierarchical clustering was performed based on continuous reduction of the required percentage overlap between reads (from 99% to 85%) while maintaining a sequence identity of ≥90%. A sample x CD-Hit cluster table was subsequently generated, analogous to an OTU table. The table was analyzed using QIIME to generate alpha diversity estimates (as Shannon indices) as well as beta diversity matrices based on Hellinger distances.
Viral alpha and beta diversity
These estimates were based on a pipeline composed of several software programs: GAAS38, Circonspect16, PHACCS39, and MaxiPhi16. Circonspect (http://sourceforge.net/projects/circonspect/) was used to form cross-contig spectra of 3× coverage. To ensure stringent assembly, contigs in Circonspect were determined by Minimo (available in the AMOS package at http://sourceforge.net/projects/amos/) instead of TIGR Assembler. Contig assembly parameters were 98% similar sequences overlapping by at least 35 bp. Viral average genome length was then estimated using GAAS (http://sourceforge.net/projects/gaas/) (tblastx against complete NCBI RefSeq viral genomes, minimum 30% similarity, minimum 70% relative length).
For each pair of samples to compare, the input for MaxiPhi was their cross-contig spectrum and average genome length. Building on PHACCS (http://sourceforge.net/projects/phaccs/), MaxiPhi ran a Monte-Carlo simulation to determine how many virotypes samples had in common (percent shared), and how many of the most abundant ones changed their abundance rank (percent permuted). Using VLP reads from each individual sample assembled against themselves as internal controls, the best average genome length for each beta-diversity computation was found as the length within 20% of the input value that produced the percent shared and percent permuted closest to 100 and 0% respectively for both controls.
The entire viral diversity analysis was done at several levels: between time points for each individual, between twins for each family, between twin and mother for each family, and between all families. Input viral metagenomes were pooled as necessary, e.g., for the co-twin comparison, sequences from all 3 time points from each twin were merged.
PHACCS calculates expected number of virotypes by estimating Shannon indexes from contig spectra. To compare the results with an independent method, Shannon indexes derived from CD-Hit clustering were determined: the expected number of clusters per sample was divided by the expected number of clusters per virotype, as determined by the average genome size given by GAAS and the mapping of clusters per Kb of viral contigs.
Assembly and analysis of viral genomes
The 454 Newbler assembler Software Release 2.0.01.14 was used for assembly of viral genomes. Default parameters were employed except for minimum identity between the sequences (98%) and minimum overlap (100bp). These stringent conditions diminish the risk of false assembly between reads from different viruses40.
We created an online tool for visualizing assembled viral contigs (http://gordonlab.wustl.edu/phage_omics/). Each contig with a length >10Kb generated from the assembly was blasted against a non-redundant set of proteins encoded by viruses present in our NR_Viral_DB (contains entries deposited in public databases as of May, 2009), as well as translated ORFs present in 121 sequenced microbial genomes representing cultured representatives of the human gut microbiota. ORF prediction was performed using glimmer341. ORFs were subsequently annotated based on blastx searches (e-value < 1E-5) of the KEGG (v51), COG/String (v842), PFAM (v2343), and TIGRFAM (v744) databases. All features and annotations for each contig were included in a MySQL database and displayed using lightweight genome viewer45.
All processed pyrosequencing reads from each VLP sample were blasted (blastn e-value <1e-5) against each of the contigs. Significant hits were recorded and the positions used for plotting cumulative coverage. The length of the alignment was used to calculate a normalized coverage value46. Percent similarity of each read to the contig was also calculated and averaged for total percent identity calculations.
Functional assignment of reads and statistical analyses
All available pyrosequencing reads from fecal microbiomes and VLP-derived viromes were used to query (blastx e-value <1e-5) the KEGG (v51) and COG/String (v842) databases. The same databases were queried (blastp, e-value < 1e-5) with known and predicted proteins encoded by the 121 reference sequenced human gut microbial genomes, and by all viral genomes (excluding prophages) in our NR_Viral_DB. After best blast hits were assigned to COG categories or KEGG second level pathways, Metastats18 was used to identify significant functional differences (p<0.05) between fecal virome and microbiome datasets.
Prophage Coverage Plots
Prophage present in the 121 gut microbial genomes were identified using PhageFinder. Each identified prophage was then extracted in silico together with 50Kb of flanking bacterial genomic sequences. Nucmer47 was subsequently used to map all VLP pyrosequencer reads (defaults settings) onto this set of extracted sequences. Mummerplot, which like Nucmer is part of the Mummer package47, was employed to generate sequence identity plots (threshold, > 80% similarity). The prophage genome coordinates of the matches and the sequence identity with VLP reads were used to generate tables of `percent coverage' and `fold-coverage'.
Search for integrase genes
All integrase protein sequences were extracted from the NR_Viral_DB and from the 121 sequenced human gut-associated microbial genomes. VLP pyrosequencing reads were blasted against this database of extracted sequences (blastp, e-value <1e-4) and the best blast hit stored for every read. The number of reads from a given sample that were similar to a given integrase in the database was recorded and used to generate a Hellinger-based distance matrix between samples (QIIME).
CRISPR spacers represented in viral metagenomes
Seventy-four available human gut microbial genomes, representing members of the most predominant bacterial families present in the human fecal microbiota, were used to search for CRISPR elements with CRISPR-Finder48. CRISPRs were identified in 48 of these genomes: they contained a total of 95 different repeat sequences and 2,196 spacers. The direct repeats were subsequently compiled into a database, which in turn was used to search each of our fecal microbiome datasets (Program cross_match49; parameters: -minmatch 7 -maxmatch 12 -gap1_only -screen -minscore 10). Spacers were then extracted from all microbiome pyrosequencer reads where at least 2 matches for the same CRISPR repeat were identified and the intervening spacer was ≥10 nucleotides. All of these spacers from the microbiomes and 121 sequenced reference genomes were pooled together, and used to screen all VLP-derived pyrosequencing reads using cross_match (parameters: -minmatch 14 -maxmatch 14 -gap1_only -screen -minscore 10). Virome reads with hits to microbiome or reference genome CRISPR spacers over >90% of the length of their spacers were recorded.
Gnotobiotic mouse experiments
All studies with mice used protocols approved by the Washington University Animal Studies Committee. Methods for co-colonization of adult germ-free adult male C57Bl/6J mice with Marvinbryantella formatexigens and Bacteroides thetaiotaomicron, harvesting their cecal contents, preparation of rRNA-depleted RNA from cecal contents and fecal samples for subsequent cDNA synthesis, Illumina GA-IIx sequencing of cDNA, plus mapping and normalization of the resulting reads are described in another publication23. RNA-Seq datasets used for our analysis of prophage gene expression can be found under GEO accession numbers GSM544893, GSM544900, GSM544858, GSM544866, GSM544940, and GSM544944 (in vivo data) as well as GSM544856, GSM544873, GSM544835, GSM544947, GSM544872, GSM544917, GSM544931, GSM544871, GSM544883, GSM544863, GSM544865 (in vitro data).
Statistical tests
Statistical tests were performed and heatmaps were produced using the R package50.
Supplementary Material
Acknowledgements
We thank Sabrina Wagoner and Jill Manchester for superb technical assistance, Jeremiah Faith for help developing Phage_omics and together with Federico Rey for microbial RNA-Seq datasets, Peter Turnbaugh for assistance with fecal metagenomic studies and Beltran Rodriguez-Mueller and Dana Willner for valuable discussions. This work was supported in part by grants from the NIH (American Recovery and Reinvestment Act supplemental funding of DK78669), the Crohn's and Colitis Foundation of America, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. A.R is the recipient of an International Fulbright Science and Technology Program award.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare that they have no competing interests.
References
- 1.Kunin V, et al. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 3.Angly F, et al. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol. 2009;11:2863–2873. doi: 10.1111/j.1462-2920.2009.02021.x. [DOI] [PubMed] [Google Scholar]
- 4.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuch R, Fischetti VA. The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS One. 2009;4:e6532. doi: 10.1371/journal.pone.0006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 7.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkbeiner SR, et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitbart M, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Brito B, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. doi: 10.1038/ismej.2010.1. advanced online publication, 11 Feb 2010 (DOI:10.1038/ismej.2010.1) [DOI] [PubMed] [Google Scholar]
- 18.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharon I, et al. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 23.Rey FE, et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. doi: 10.1074/jbc.M110.117713. Published May 5, 2010 as doi:10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeShazer D. Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J Bacteriol. 2004;186:3938–3950. doi: 10.1128/JB.186.12.3938-3950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heath AC, et al. Ascertainment of a mid-western US female adolescent twin cohort for alcohol studies: assessment of sample representativeness using birth record data. Twin Res. 2002;5:107–112. doi: 10.1375/1369052022974. [DOI] [PubMed] [Google Scholar]
- 27.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 28.Becker A, Kuster H, Niehaus K, Puhler A. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol Gen Genet. 1995;249:487–497. doi: 10.1007/BF00290574. [DOI] [PubMed] [Google Scholar]
- 29.Gon S, Faulkner MJ, Beckwith J. In vivo requirement for glutaredoxins and thioredoxins in the reduction of the ribonucleotide reductases of Escherichia coli. Antioxid Redox Signal. 2006;8:735–742. doi: 10.1089/ars.2006.8.735. [DOI] [PubMed] [Google Scholar]
- 30.Padovani D, Thomas F, Trautwein AX, Mulliez E, Fontecave M. Activation of class III ribonucleotide reductase from E. coli. The electron transfer from the iron-sulfur center to S-adenosylmethionine. Biochemistry. 2001;40:6713–6719. doi: 10.1021/bi002936q. [DOI] [PubMed] [Google Scholar]
- 31.Garriga X, et al. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem Biophys Res Commun. 1996;229:189–192. doi: 10.1006/bbrc.1996.1778. [DOI] [PubMed] [Google Scholar]
- 32.Tabor CW, Tabor H. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu Rev Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- 33.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 34.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. doi: 10.1038/nmeth.f.303. advanced online publication 11 april 2010 (DOI:10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouts DE. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angly FE, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angly F, et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breitbart M, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci U S A. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen LJ, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faith JJ, Olson AJ, Gardner TS, Sachidanandam R. Lightweight genome viewer: portable software for browsing genomics data in its chromosomal context. BMC Bioinformatics. 2007;8:344. doi: 10.1186/1471-2105-8-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohwer F, Seguritan V, Choi DH, Segall AM, Azam F. Production of shotgun libraries using random amplification. Biotechniques. 2001;31:108–112. 114–106, 118. doi: 10.2144/01311rr02. [DOI] [PubMed] [Google Scholar]
- 47.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.