Abstract
Objective
To discover common variants in 6 lipid metabolic genes and construct and validate a genetic risk score (GRS) based on the joint effects of genetic variants in multiple genes from lipid and other pathobiologic pathways.
Background
Explaining the genetic basis of coronary artery disease (CAD) is incomplete. Discovery and aggregation of genetic variants from multiple pathways may advance this objective.
Methods
Premature CAD cases (N=1,918) and CAD-free controls (N=1,032) were selected from our angiographic registry. In a discovery phase, single nucleotide polymorphisms (SNPs) at 56 loci from internal discovery and external reports were tested for associations with biomarkers and CAD: 28 promising SNPs were then tested jointly for CAD associations, and a genetic risk score (GRS) consisting of SNPs contributing independently was constructed and validated in a replication set of familial cases and population-based controls (N=1,320).
Results
Five variants contributed jointly to CAD prediction in a multigenic GRS model: odds ratio (OR) =1.24 (95% CI 1.16–1.33) per risk allele, p=8.2×10−11, adjusted OR=2.03 (1.53–2.70), 4th vs. 1st quartile. GRS5 had minor impact on area under the receiver-operating characteristic curve (p>0.05) but resulted in substantial net reclassification improvement: 0.16 overall, 0.28 in intermediate risk patients (both p<0.0001). GRS5 predicted familial CAD with similar magnitude in the validation set.
Conclusions
CorGen demonstrates the ability of a multigenic, multipathway GRS to improve discrimination of angiographic CAD. Genetic risk scores promise to increase understanding of the genetic basis of CAD and improve identification of individuals at increased CAD risk.
Keywords: Coronary artery disease, genetics, risk, risk score
INTRODUCTION
Much of the genetic basis of coronary heart disease (CHD) remains to be discovered 1. However, steady progress is occurring using both candidate gene and genome-wide association studies (GWAS)2–3. Insights into pathobiologic pathways has guided candidate gene testing2, 4, whereas GWAS makes no a priori assumptions about genetic site and provides broader, genome-wide coverage 3. We hypothesized that combining discoveries from both of these complementary approaches and extending consideration to multiple pathobiologic pathways could further advance this effort.
To date, the risk attributable to any individual variant has been modest. However, discovering and combining multiple loci with modest effects into a global genetic risk score (GRS) could improve the identification of high-risk populations and improve individual risk assessment 5–7. Only a limited number of GRS studies for CHD have been reported, and these have focused mostly on lipid-related genes and especially on low density lipoprotein cholesterol (LDL-C) related variants 5, 7. However, CHD pathogenesis involves multiple stages and other mechanisms and biopathways, including HDL-C, inflammation, thrombosis, and vascular development 1–2, 4.
METHODS
Study Objectives
The primary objectives of the Intermountain Healthcare’s Coronary Genetics (CorGen) study were: 1) to discover all common SNPs among a set of 6 key genes in the reverse cholesterol transport system and test them for associations with angiographic CAD, and 2) to construct and validate a multivariant GRS, based on the joint effects of these and other (literature reported) genes in lipid and 3 other pathobiologic pathways, to discriminate CAD using precise angiographic phenotyping.
Study Participants
Study subjects for the primary association study were selected from Intermountain Healthcare’s ongoing Angiographic Registry and DNA Bank8. This Registry is approved by the hospital’s institutional review board, and study subjects give written consent prior to enrollment. Fasting blood is sampled at the time of angiography; DNA and plasma are extracted and stored; demographic and angiographic information is collected and entered into an electronic database; and patients are followed prospectively8–9.
To optimize genetic susceptibility, we studied younger subjects: men aged ≤60, women ≤70 years. Approximately 3000 subjects (~2000 CAD cases and ~1000 angiographically normal controls, matched 2:1 for sex, age, and date of registry entry) were selected. Clinical diagnoses preceding angiography included stable disease (angina equivalent or other) in 56%, unstable angina in 25%, and acute MI in 19%.
A separate set of cases with highly familial premature CAD (first degree relative with CHD onset <55 in men, <65 in women) from the University of Utah Cardiovascular Genetics Family Tree Registry1 and a separate set of controls (randomly invited from a public records database) were enrolled as a replication set10.
Study Design and Selection of Genetic Variants
The study consisted of 3 discovery phases and a validation phase (Supplementary Figure 1), each consisting of separate, independent datasets. In discovery phase 1 (SNP discovery), 6 genes with key roles in reverse cholesterol transport, (cholesteryl ester transfer protein [CETP], hepatic lipase [LIPC], lipoprotein lipase [LPL], lecithin-cholesterol acyltransferase [LCAT], scavenger receptor class B type I [SR-BI], and apo-lipoprotein F [apo F]) were scanned in 62 volunteers; 81 SNPs were discovered, and all variant genomic segments were sequenced. In discovery phase 2 (haplotype or tagging [t]SNP discovery), a separate set of 339 Euro-American volunteers was tested to establish tSNPs using the Horne and Camp method of principal components analysis (supplemental materials and 11–12). A total of 38 tSNPs were determined to characterize the linkage disequilibrium groups of the 6 genes. Of these, 10 found to be associated univariably with lipoprotein markers or nominally (p<0.2) with CAD in a discovery set of cases (n=915) and controls (n=522) (discovery phase 3) were selected for multivariable GRS modeling.
To complement and expand GRS candidates to other pathways and genome-wide, literature studies were reviewed as of April 2008. Eighteen SNPs showing the most robust and independent associations at genome-wide significance (p<5 × 10−8) with a CAD-related biomarker (i.e., lipids—divided among LDL-C, HDL-C, TG—or CRP or MCP1) and/or CAD per se (i.e., 9p21.3) and diversified among lipid/lipoprotein, thrombosis, inflammation, vascular function/unassigned pathways were added to the 10 internal candidates SNPs for GRS modeling (Supplementary Figure 1, Supplemental Table 1).
The risk allele for SNP candidates was designated a priori based on associations with a recognized risk marker (e.g., lipid levels) or, preferentially, when reported (for literature SNPs) or angiographically determined (for internal SNP candidates), on associations with CAD directly, prior to entry into multivariable genetic risk score modeling.
Multivariable GRS modeling using backward logistic regression then was used in an angiographically-phenotyped set of 1,918 cases and 1,032 controls to reduce the 28 candidate SNPs from the 2 sources to a set of SNPs contributing jointly to CAD prediction.
In validation testing, the reduced set GRS determined in the discovery set was prospectively tested in a completely independent set of familial CAD cases (n=312) and population controls (n=1,008).
Definition of Angiographic Characteristics and Clinical Covariables
The presence of CAD was determined by coronary angiographic analysis masked to genetic information. Patients were categorized as free of CAD (no lesions noted angiographically), mild/moderate CAD (i.e., most severe lesion <70% stenosis), or severe CAD (i.e., at least one lesion ≥70% stenosis). Patients with mild/moderate CAD were excluded as indeterminate.
Standard clinical criteria and specific therapies were used to define the presence of diabetes, hypertension, and hyperlipidemia9. Smoking was defined as current smoking or a >10 pack-year smoking history. Family history was positive if a parent, sibling, or child manifested CAD or myocardial infarction by age 55 in males or 65 in females.
DNA Extraction and Genotyping
DNA was extracted from blood samples, quality and quantity assessed, and genotyping performed using standard techniques as detailed in Supplementary Materials. Call rates for SNPs in the GRS were 95–97% and required to be >90% for all tested SNPs.
Computation of Genetic Risk Scores
Two methods were used to create the multivariable GRS: a simple, unweighted count method (count GRS) and a weighted method (weighted GRS) 6, 13. Both methods assumed each SNP to be independently associated with risk. (The independence of SNP associations with CAD/CAD risk markers was recently tested in this dataset and found to be valid 14). An additive genetic model was assumed: weightings of 0, 1, and 2 were given according to the number of risk alleles present15.
The count method assumed that each SNP contributed equally to CAD risk and was calculated by summing the number of risk alleles across the panel of SNPs tested. This produced a score between 0 and twice the number of SNPs, i.e., representing the total number of risk alleles. The weighted GRS was calculated by multiplying each ϐ-coefficient for the CAD phenotype from the discovery set by the number of corresponding risk alleles (0, 1, or 2) and then summing the products. The GRS was modeled as a continuous variable and as quartiles.
Statistical Analysis
Statistical analyses were performed using SPSS version 15.0 (Chicago, IL). Chi-square tests (Armitage 1 df test-of-trend for additive genetic model) and t tests were used for comparing proportions and means, respectively, between cases and controls. Logistic regression was used to determine the effect of each variant separately and combined on risk for angiographic CAD. Odds ratios (OR) and 95% confidence intervals (CI) are reported for the high- versus low-risk alleles assuming an additive risk model. Multivariable analyses were used to adjust for history of hypertension, hyperlipidemia, smoking, diabetes, family history, ethnicity/race, and body mass index (BMI). Sex and age were matched by design. Power calculations used nQuery Advisor v.4.0 (Statistical Solutions, Saugus, MA). For risk alleles with minor allele frequency ≥10%, the study had 80% power to detect an OR for CAD of ≥1.3 and >90% for OR>=1.35, for 2-sided alpha of <0.05 for 2000 cases and 1000 controls. Associations with CAD for individual SNPs were considered significant at p≤0.05, and in aggregate for GRS models at p≤0.01.
We plotted receiver-operating characteristic (ROC) curves and calculated areas under the curve (AUC) for logistic regression models including conventional risk factors without and with GRS16. We classified Framingham risk scores (FRS) into 4 categories, with intermediate risk categories defined as 10-year risks of 5 to 9% (low-intermediate) and 10% to 19% (high-intermediate)17–18. The potential of GRS to improve individual risk stratification then was measured using the net reclassification improvement (NRI) method, using flow-limiting CAD (>70% stenosis) as the clinical CHD equivalent and excluding patients with diabetes (an a priori high-risk equivalent and not included in FRS scoring) 16, 19–20.
Sources of funding
This study was funded by a grant from the National Heart, Lung, and Blood Institute, R01- HL071878. The authors are solely responsible for the design and conduct of this study, all study analyses, and drafting and editing of the paper.
RESULTS
Patient Characteristics
Characteristics of the angiographic set of cases and controls are summarized in Table 1. Age averaged 53 years, and 1/3 were women. By design, cases were matched to controls for age and sex. Other traditional risk factors were more prevalent in cases. However, (treated) lipid profiles and blood pressures were similar in cases and controls.
Table I.
Characteristics of Discovery Set Angiographic CAD Cases and Controls
| Variable | Angiographic | Angiographic |
|---|---|---|
| CAD Cases | Normal Controls | |
| N | 1,947 | 1,036 |
| Age (y) (mean [SD]) | 53.1±8.0 | 53.2±8.2 |
| Sex (% women) | 35% | 36% |
| Race/ethnicity† (% White) | 94% | 94% |
| BMI (kg/m2) | 30.1±6.2 | 29.9±6.5 |
| H/o Hyperlipidemia (%) | 67%* | 34% |
| H/o Hypertension (%) | 61%* | 44% |
| H/o Diabetes (%) | 28%* | 12% |
| H/o Smoking (%) | 26%* | 14% |
| Family History CHD | 50%* | 29% |
| Systolic BP (mmHg) | 140±25* | 138±22 |
| Diastolic BP (mmHg) | 82±14 | 81±14 |
| Total Cholesterol (mg/dL) | 193±52* | 185±44 |
| Triglycerides (mg/dL) | 209±196* | 181±152 |
| LDL-C (mg/dL) | 111±40 | 108±37 |
| HDL-C (mg/dL) | 38.0±12.7* | 41.6±13.7 |
| Glucose (mg/dL) | 131±67* | 110±45 |
p<0.05 for cases vs. controls.
No blacks included.
SNP Discovery and Association with CAD
A total of 38 tSNPs in the 6 key lipoprotein metabolic genes were identified by exhaustive scanning (Supplementary Figure 1, Supplementary Table 1); of these, 10 were significantly associated with lipid levels (i.e., at p≤0.002) or at least nominally (p<0.2) with CAD. For initial GRS modeling, these 10 were added to 18 leading literature SNPs reported from GWAS to be associated with CAD-related biomarkers or CAD-risk. These 28 SNPs, with allelic frequencies in cases and controls, are shown in Supplementary Table 1. All SNPs were in Hardy-Weinberg equilibrium (i.e., p>0.05 per SNP).
Genetic Risk Score Modeling
The multivariable genetic model could be simplified by operator-interactive stepwise elimination from a 28- to a 5-SNP genetic risk score (GRS5) without loss of discrimination (Table 2). Five other SNPs showed multivariable association trends (0.05<p<0.2) but were eliminated in the final model (Table 2, footnote). The Hosmer-Lemeshow statistics for the 5 SNP model (7.92, p=0.24, 6 df) and for the model using GRS5 as a single variable (3.91, p=0.27, 3 d.f.) both suggested a good fit to the data.
Table II.
Logistic Regression Model for Genetic Risk Score-5 in Angiographic Case/Control Set
| Gene / locus (rs) | Risk Allele | ϐ | Sig | OR (95% CI) |
|---|---|---|---|---|
| CELSR2 / rs599839 | Major | 0.223 | <0.001 | 1.25 (1.11–1.40) |
| 9p21.3 / rs2383206 | Minor | 0.191 | <0.001 | 1.21 (1.09–1.34) |
| CETP / rs289715* | Major | 0.217 | 0.005 | 1.24 (1.07–1.44) |
| ApoF / rs78739461* | Minor | 0.307 | 0.015 | 1.36 (1.06–1.74) |
| F2 (PT) / rs1799963 | Major | 0.474 | 0.046 | 1.61 (1.01–2.56) |
ϐ =beta coefficient; Sig= significance (p-value); OR=odds ratio for angiographic CAD; CI=confidence interval.
Internally-discovered SNP. Candidate SNPs contributing trend associations in the multivariable model (0.05>p<0.2) but eliminated in the final model were: ApoC1/E rs4420638, CETP rs180075, CRP rs2794520, GCKR rs780094, and LIPC rs36041167. Effect estimates are for all variables considered simultaneously in the model.
The simple count method GRS5 yielded an OR=1.24 per risk allele (CI, 1.16–1.33, p=8.2×10−11; model χ2mv=44.8, model χ2GRS-5 =42.8)(Table 2). GRS5 was predictive in diabetic (OR 1.34 [CI 1.13–1.59], N=668) as well as non-diabetic subgroups of cases/controls (OR 1.23 [CI 1.14–1.32], N=2282).
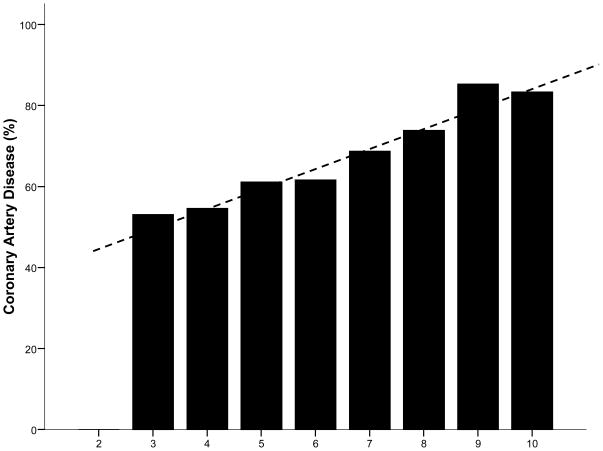
Results for continuous GRS5 and for quartile GRS5 are shown in Table 3. Angiographic CAD prevalence across the spectrum of count-GRS5 scores is presented in Figure 1. Comparisons of CAD prevalence in fourth versus first quartile subjects yielded an unadjusted OR=2.07 (CI 1.59, 2.70) and a risk factor adjusted OR=2.03 (1.53–2.70) for count-GRS5 (Table 3). Weighted GRS5 results were similar: unadjusted OR=2.06 (CI 1.61–2.65), adjusted OR=2.02 (CI 1.55–2.64).
Table III.
Associations between Count Genetic Risk Score-5 and Angiographic CAD
| Count GRS | Continuous GRS | Quartile of Continuous GRS |
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| No. Subjects | 2,983 | 630 | 969 | 930 | 454 |
| Median GRS (Range) | 6 (2–10) | 5 (2–5) | 6 (6) | 7 (7) | 8 (8–10) |
| OR (95% CI)/allele | 1.24 (1.16, 1.33) | 1.0 (-----) | 1.12 (0.91, 1.37) | 1.54 (1.24, 1.90) | 2.07 (1.59, 2.70) |
| Adjusted* | 1.21 (1.13, 1.30) | 1.0 (-----) | 1.20 (0.96, 1.49) | 1.49 (1.18, 1.87) | 2.03 (1.53, 2.70) |
Adjusted for hyperlipidemia, hypertension, BMI, diabetes, smoking, family history. (Cases, controls matched for age, sex by design.)
Figure 1. Prevalence of Angiographic CAD by Count GRS5 Score.
Percent CAD across GRS5 categories was highly significant (p-trend=8.2×10−11). Reference mean count GRS score is 6.3.
GRS Validation Testing
For validation testing, the GRS5 was prospectively tested in a completely separate set of 312 unrelated familial coronary disease cases and 1,008 apparently healthy population controls (Supplementary Table 2). In this independent familial case/control set, the GRS5 remained highly significantly predictive of coronary disease, with a similar magnitude to that observed in the angiographic case/control set, i.e., OR 1.23 (CI 1.11–1.38) per risk allele, p=2.0×10−4. Also, each risk allele except the rare ApoF variant individually predicted risk in these familial cases, with the common CELSR2 and 9p21.3 polymorphisms showing the strongest associations (Supplementary Table 3).
In addition, an internal analysis of reliability within the primary case/control set was performed by dividing the set into equally sized “discovery” and “replication” subsets. This analysis demonstrated a high degree of internal reliability, with GRS5 showing consistent and highly significant predictive value in both subsets (ORs 1.26, 1.23, respectively, both p<10−5).
Incremental Value of Multiple Source SNPs in GRS Modeling
Of the 5 SNPs meeting criteria for GRS5 membership, 2 (i.e., ApoF rs78739461* and CETP rs289715), one novel*, were from the internal lipoprotein gene discovery effort, and 3 were from external candidates from other loci/pathways (i.e., 9p21 rs2383206, CELSR2/PSRC1 rs599839, F2 rs1799963). SNPs from both sources contributed in a complementary fashion to overall risk prediction; both a GRS3 (limited to the 3 external SNPs; χ2GRS-3 =30.6) and a GRS2 (2 internal SNPs; χ2GRS-5 =13.6) predicted CAD but were inferior to the combined GRS5 (χ2GRS-5 =42.8).
Multivariable Predictive Model
A multivariable predictive model for angiographic CAD incorporating genetic and clinical information is presented in Table 4. The contribution of GRS5 is intermediate, similar to family history and greater than hypertension. (The impact of age and sex cannot be determined, given the matching design, and the standard use of statins in CAD patients likely impacts the diagnosis of hyperlipidemia.)
Table IV.
Multivariable Predictive Model for CAD Using Genetic Variables and Standard Risk Factors
| Variable | Wald χ2 | OR (CI) | p Value |
|---|---|---|---|
| Hyperlipidemia | 123.51 | 2.75 (2.30, 3.29) | 1.1×10−28 |
| Diabetes | 46.12 | 2.21 (1.76, 2.77) | 1.1×10−11 |
| Smoking | 39.12 | 1.98 (1.60, 2.46) | 4.0×10−10 |
| Family History | 30.01 | 1.64 (1.37, 1.96) | 4.3×10−8 |
| GRS5 / risk allele | 28.66 | 1.21 (1.13, 1.30) | 8.7×10−8 |
| Hypertension | 2.71 | 1.16 (0.97, 1.39) | 0.10 |
| BMI (per kg/m2) | 0.02 | 1.001 (0.985, 1.018) | 0.88* |
BMI was eliminated from the final model. Effect estimates are for all variables considered simultaneously.
Incremental Value of GRS in Individual Risk Assessment
When count GRS5 was added to Framingham Risk Score in the full angiographic set, net reclassification fraction was found to be 0.233 in CAD cases (p<0.0001) and 0.073 in no-CAD controls (p=0.005), which when combined yielded a net reclassification improvement (NRI) of 0.160 (p<0.0001)(Supplementary Table 3). NRI restricted to intermediate risk subjects yielded an NRI of 0.283 (p<0.0001)(Supplementary Table 4)17–18.
As with others’ experience with novel (and most traditional) risk factors7, 13, 18, the addition of GRS5 had only minor impact on area under the receiver-operating characteristic curve: conventional model C-statistic=0.723 (CI 0.703–0.742), GRS-augmented C-statistic=0.731 (CI 0.712–0.750) (p>0.05).
DISCUSSION
Summary of Key Study Results
CorGen demonstrates the feasibility and potential utility of simultaneously considering the joint effects of common genetic variants from multiple pathobiologic pathways, aggregated as a GRS, to predict the risk of premature CAD. CorGen also demonstrates the complementary effects of combining tSNPs derived from high-definition scanning of candidate genes (i.e., those associated with lipoprotein metabolism) with biomarker-and other risk-related SNPs discovered through high-density GWAS.
Features of CorGen that provide assurance of a valid result include the angiographic characterization of CAD (defining a precise phenotype) and replication in an independent set of cases and controls. On average, each high-risk allele increased risk by 24%, and considered jointly as GRS5, a fourth quartile score increased CAD risk by over 2-fold. GRS also contributed independently to standard risk factors in multivariable modeling. GRS improved the Framingham risk score category in a net of 16% of subjects overall, which compares favorably with the 12% reported NRI for HDL cholesterol16–17 and 10% NRI for systolic blood pressure20. Further, application restricted to subjects in the 2 intermediate risk categories improved classification in a net of 28%. These findings suggest its potential clinical utility in individual risk classification, despite (as with others’ experience) its minor impact on population-level AUC7, 13, 17, 20.
Literature Comparisons
Approximately one-half of CHD risk appears to be genetically transmitted1. However, the proportion of variation among markers of human traits and diseases (including CAD) attributed to individual SNPs has been modest 21–22, leading to the “common disease, common variant hypothesis”23. Combining these SNPs to form a more powerful risk predictor underlies the GRS concept. GRS modeling for CHD is in its infancy, but a few reports have appeared, mostly modeling plasma lipids5, 7, 13. CorGen extends these models to multiple additional genes and pathways identified by either candidate-gene or GWAS methods and introduces new markers.
Accounting for Genetic Susceptibility
Uncovering the genetic basis of CHD is an unrealized goal1, 10. The pathophysiology of CHD is a complex interaction of genetic and environmental factors acting directly and indirectly on multiple disease stages from preclinical to clinical CAD to acute coronary syndromes, each stage with a distinct set of risk factors1. Despite methodological advances such as GWAS, progress has been slow, and well-validated associations remain few, with modest impact. This emphasizes the need for broadening the genetic search, more precisely defining the coronary phenotype, and aggregating individual genetic risk markers into an overall metric.
A major success in this effort has been the discovery and replication of a risk locus at chromosome 9p21.3 (Supplementary references). Although its mechanism remains to be precisely defined, it appears to be involved in vascular development and function, increasing susceptibility to CAD rather than precipitating MI10, 24–25. Some but not other studies have suggested that knowledge of 9p21 status may improve individual risk classification26–28. In CorGen, 9p21 emerged as a major contributor to the multivariant GRS.
Three other contributors to GRS5 are involved with lipoprotein metabolism, but in contrast to a previous report7, these were not restricted to LDL-C–related genes. In addition, a less common variant from the thrombosis pathway (F2) with a relatively large effect was selected. Such uncommon SNPs have been suggested as a focus for future discovery efforts12, 21. A variant representing vascular inflammation (CRP rs2794520) demonstrated a preliminary association but was eliminated in the final model. Future research should aim to discover additional genetic risk contributors, both universal and population-specific, including those from vascular inflammation and thrombosis pathways.
Study Implications
Here we show that a multivariant GRS can discriminate CAD as well as or better than many standard non-genetic tests and may improve individual risk assessment. In contrast, GRS5 added little to the AUC for the ROC curve, i.e., did not importantly improve risk prediction over the Framingham Risk Score at a population level. This dichotomy also has been reported for other risk predictors7, 13, 18, 26.
We view CorGen as a proof-of-concept study. Very recent29 and future genetic studies may identify SNPs that add to and refine the CorGen GRS. Nevertheless, of the many high-profile literature SNPs associated with lipids, other disease markers, or even CAD already reported that we tested, only 5 of 28 leading candidates contributed independently to CAD prediction in multivariable GRS modeling. Similarly, of 38 tSNPs discovered internally by extensive scanning to characterize variation in the 6 key lipoprotein metabolic genes, only 2 contributed to the final GRS model. Hence, the goal to account for the greater part of the genetic basis of CAD remains challenging.
GRS may be of particular value in younger cohorts in whom traditional risk factors have not developed and who may benefit from closer surveillance and more aggressive preventive measures. GRS, as other novel risk predictors, may be of greatest incremental value in those at intermediate pre-test probability18–19, 30. Finally, by interfacing genetic loci from multiple pathways, additional insights into disease pathogenesis and treatment targets may be expected.
Strengths and Limitations
The study though prospective in enrollment and hypothesis possesses the limitations of observational studies, including the possibility of uncorrected confounding. The predictive value of the GRS is restricted to angiographic CAD and not myocardial infarction per se. This study focused on a younger population of Euro-Americans, minimizing population stratification and heterogeneity but potentially limiting applicability to other racial/ethnic groups (i.e., African-Americans) and older ages. Risk-associated SNPs while valuable for disease discrimination may not represent casual variants. Although moderately large, CorGen has limited power to discern associations with small effect sizes. However, small-effect variants are unlikely to have important clinical impact or be cost effective for clinical application. Although the GRS5 was validated internally, external validation in geographically distinct populations is needed, as is further testing of the novel and rare apoF variant. Here we test GRS in the context of the commonly used ATP-III version of the Framingham Risk Score; other risk stratification methods (e.g., FRS including diabetics; Diamond Forrester score) may deserve testing. Given our case-control study design, we are limited to discriminatory analysis and technically cannot precisely estimate the predictive power of GRS for CAD risk. Finally, we acknowledge that our highlighting of a pathway-based approach is primarily conceptual; this effort represents an initial, selective rather than a thorough, definitive application of a pathway approach.
Conclusions
Using a multiple step study design, CorGen has validated the ability of a GRS derived from 5 SNPs to predict premature CAD and has demonstrated the complementary nature of candidate-gene and GWAS approaches. The GRS5 model provided greater discrimination than any single variant, predicted a 24% risk increment per allele, identified a highest quartile GRS with a two-fold increase in CAD risk, and improved net risk classification in 28% of intermediate-risk individuals. Thus, CorGen demonstrates proof-of-concept that genetic risk scores are a feasible and promising approach to account for the genetic basis of CAD and identify individuals at increased CAD risk.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Samera Khan, John A. Huntinghouse, Matthew J. Kolek, Nathan Hull, and Brianna S. Ronnow.
This study was funded by the Heart, Lung, and Blood Institute, grant R01- HL071878.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hopkins PN, Hunt SC, Wu LL. Family history and genetic factors. In: Wong ND, Black HR, Gardin JM, editors. Preventive cardiology, a practical approach. New York: McGraw-Hill; 2005. pp. 92–148. [Google Scholar]
- 2.Anderson JL, Carlquist JF, Horne BD, Hopkins PN. Progress in unraveling the genetics of coronary artery disease and myocardial infarction. Current Atherosclerosis Reports. 2007;9:179–86. doi: 10.1007/s11883-007-0017-4. [DOI] [PubMed] [Google Scholar]
- 3.Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009;360:1699–701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 4.Topol EJ, Smith J, Plow EF, Wang QK. Genetic susceptibility to myocardial infarction and coronary artery disease. Hum Molec Genet. 2006;15:R117–23. doi: 10.1093/hmg/ddl183. [DOI] [PubMed] [Google Scholar]
- 5.Horne BD, Anderson JL, Carlquist JF, et al. Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Ann Human Genetics. 2005;69:176–86. doi: 10.1046/j.1529-8817.2005.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piccolo SR, Abo RP, Allen-Brady BK, et al. Evaluation of genetic risk scores for lipid levels using genome wide markers in the Framingham Heart Study. BMC. 2009 doi: 10.1186/1753-6561-3-s7-s46. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 8.Taylor GS, Muhlestein JB, Wagner GS, Bair TL, Li P, Anderson JL. Implementation of a computerized cardiovascular information system in a private hospital setting. Am Heart J. 1998;136:792–803. doi: 10.1016/s0002-8703(98)70123-1. [DOI] [PubMed] [Google Scholar]
- 9.Horne BD, Camp NJ, Carlquist JF, et al. Multiple-polymorphism associations of seven matrix metalloproteinase and tissue inhibitor metalloproteinase genes with myocardial infarction and angiographic coronary artery disease. Am Heart J. 2007;154:751–8. doi: 10.1016/j.ahj.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horne BD, Carlquist JF, Muhlestein JB, Bair TL, Anderson JL. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circulation Cardiovascular Genetics. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horne BD, Carlquist JF, Cannon-Albright LA, et al. High-Resolution Characterization of Linkage Disequilibrium Structure and Selection of Tagging SNPs for the Cholesteryl Ester Transfer Protein Gene. Ann Human Genetics. 2006;70:524–34. doi: 10.1111/j.1469-1809.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 12.Horne BD, Camp NJ, Anderson JL, et al. Multiple less common genetic variants explain the association of the cholesteryl ester transfer protein gene with coronary artery disease. J Am Coll Cardiol. 2007;49:2053–60. doi: 10.1016/j.jacc.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk of type 2 diabetes in U. S. men and women of European ancestry. Ann Intern Med. 2009;150:541–50. doi: 10.7326/0003-4819-150-8-200904210-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horne BD, Camp NJ, Carlquist JF, Anderson JL. Modeling multiplicative SNP interactions in the presence of an additive genetic risk score. Genet Epidemiol. 2009;33:771. [Google Scholar]
- 15.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 16.Pencina JJ, D’Agostino RBS, D’Agostino RBJ, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statist Med. 2008;27:157–72. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 17.Hlatky MA, Greenland P, Arnett DK, et al. Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119:2408–16. doi: 10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridker PM, Buring JE, Rifai N, Cook N. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women. JAMA. 2007;297:611–19. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- 19.Cook NR. Use and misuse of receiver operating characteristic curves in risk prediction. Circulation. 2007;115:928–35. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 20.Cook NR, Ridker PM. Advances in measuring the effect of individual predictors of cardiovascular risk: the role of reclassification measures. Ann Intern Med. 2009;150:795–802. doi: 10.7326/0003-4819-150-11-200906020-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–98. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 22.Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360:1701–03. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 23.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–9. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 24.Anderson JL, Horne BD, Kolek MJ, et al. Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J. 2008;156:1155–62. doi: 10.1016/j.ahj.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Dandona S, Stewart AFR, Chen L, et al. Gene dosage of the common variant 9p21 predicts severity of coronary artery disesae. J Am Coll Cardiol. 2010;55 doi: 10.1016/j.jacc.2009.10.092. (in press) [DOI] [PubMed] [Google Scholar]
- 26.Talmud PJ, Cooper JA, Palmen J, et al. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54:467–74. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 27.Brautbar A, Ballantyne CM, Lawson K, et al. Impact of adding a single allele in the 9p21 locus to traditional risk factors on reclassification of coronary heart disease risk and implications for lipid-modifiying therapy in Atherosclerosis Risk in Communities study. Clin Cardiovasc Genet. 2009;2:279–85. doi: 10.1161/CIRCGENETICS.108.817338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coronary Artery Disease Consortium. Large scale association analysis of novel genetic loci for coronary artery disease. Arterioscl Thromb Vasc Biol. 2009;29:775–80. doi: 10.1161/ATVBAHA.108.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106:3148–21. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.