Abstract
Pigmentation patterning has long interested biologists, integrating topics in ecology, development, genetics, and physiology. Wild-type neonatal larvae of the silkworm, Bombyx mori, are completely black. By contrast, the epidermis and head of larvae of the homozygous recessive sex-linked chocolate (sch) mutant are reddish brown. When incubated at 30 °C, mutants with the sch allele fail to hatch; moreover, homozygous mutants carrying the allele sch lethal (schl) do not hatch even at room temperature (25 °C). By positional cloning, we narrowed a region containing sch to 239,622 bp on chromosome 1 using 4,501 backcross (BC1) individuals. Based on expression analyses, the best sch candidate gene was shown to be tyrosine hydroxylase (BmTh). BmTh coding sequences were identical among sch, schl, and wild-type. However, in sch the ∼70-kb sequence was replaced with ∼4.6 kb of a Tc1-mariner type transposon located ∼6 kb upstream of BmTh, and in schl, a large fragment of an L1Bm retrotransposon was inserted just in front of the transcription start site of BmTh. In both cases, we observed a drastic reduction of BmTh expression. Use of RNAi with BmTh prevented pigmentation and hatching, and feeding of a tyrosine hydroxylase inhibitor also suppressed larval pigmentation in the wild-type strain, pnd+ and in a pS (black-striped) heterozygote. Feeding L-dopa to sch neonate larvae rescued the mutant phenotype from chocolate to black. Our results indicate the BmTh gene is responsible for the sch mutation, which plays an important role in melanin synthesis producing neonatal larval color.
Pigmentation patterning has long interested biologists (1–3) and its study integrates diverse classical branches of biology, including ecology, development, genetics, and physiology (4). A variety of body color and pattern reflects natural biodiversity, the biology of visual communication, and a number of physiological processes (5–7). In insects, body-color pigment precursors and pigments are synthesized in the epidermal cells, and subsequently become an integral part of the body. Insect pigmentation not only varies among individuals, species, and populations, but also changes in time and space during the lifetime of a single individual. The intricacies of color pattern are important in investigating the reciprocal interactions that shape phenotypic variation between evolutionary and developmental processes (6). Genetic analyses show that the development of pigmentation depends on many pleiotropic regulatory factors, such as sex determination genes, HOX genes, and signaling pathways, which also regulate many other traits (8–11). In most Drosophila species, nonsex-specific striped pigmentation is controlled by the transcription factor, optomotor-blind (omb) (12). In many butterfly species, differences in engrailed, spalt, and Distal-less expression are associated with the diversification of color rings. The same transcription factors are expressed in eyespot fields, but in different relative spatial domains, suggesting that they correlate with divergent eyespot color schemes (13). Among different Drosophila species, the divergent expression of the yellow gene correlates with the distribution of black melanin, but other genes are also involved in epistatic interactions to generate the evolutionary variations observed in Yellow expression (14–17). The known candidate genes are generally conserved among insects and are responsible for pigmentation diversity in other insect lineages (6). Nevertheless, it is a challenge to identify additional novel genes that specify other color pattern polymorphisms or mutations in insects.
Diverse color mutants of Bombyx mori show differing pigment patterns of the egg, eye, wing, and body (18–21). B. mori has therefore been regarded as the major lepidopteran model for the study of color mutants, particularly because of the publication of its complete genome sequence (22–24) and a high-density linkage map (25, 26). Mutations in the sepiapterin reductase (SPR) gene (BmSpr) are responsible for the yellow body coloration of the lemon and lemon lethal mutations during the larval developmental stages (20). Mutations at the chocolate (ch) locus produce larvae that are reddish brown in later instars, and a mutation at the sooty (so) locus, an ortholog of ebony, leads to smoky larvae and black pupae (19).
Wild-type neonate larvae have black bodies. However, in recessive homozygotes of the sex-linked chocolate (sch) mutant, the epidermis and head of neonate larvae are chocolate colored (Fig. 1). Genetic analysis reveals that the chocolate phenotype is controlled by a single recessive gene located at 21.5 cM on chromosome 1. Pan and colleagues found that sch reduces hatchability and becomes lethal in the late-embryo stage when sch eggs are incubated at 30 °C (27). In 2001, Ohnuma obtained a new sex-linked chocolate mutant from a polyphagous silkworm strain, Sj, named schl (28). Linkage analysis showed it was allelic to sch, sharing the same mutant phenotype, except that it was unable to hatch even at room temperature (25 °C). Apart from attempts to investigate the pigment mechanism of the sch mutation, there have been only sporadic studies on sch and schl, limited to the analysis of linked molecular markers (29–31). Consequently, the gene responsible for the chocolate pigment patterns in neonatal sch and its relationship to temperature sensitivity have not been determined. In this study, we examined the genetic basis of the chocolate color mutation by positional cloning.
Fig. 1.
Phenotypes of neonatal larvae of p50T wild-type and sch mutant. The sch neonate larvae show a chocolate color in the epidermis and head (Lower) compared with the black color of the wild-type p50T (Upper). (Scale bar, 0.5 mm.)
Results
Mapping of the sch Mutation.
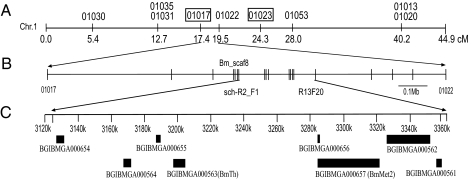
To identify a candidate region for the sch gene, we carried out a genetic linkage analysis using primer sets designed with reference to a high resolution SNP linkage map (26) and information from the silkworm genome sequence (22–24) (Table S1). First, the sch mutation was roughly mapped using 67 backcross (BC1) individuals completely linked to the SNP marker 01022, so that the linked region was narrowed to the area between markers 01017 and 01023. This region was 6.9 cM, according to the SNP map, and contained a genome sequence of about 3.45 Mb (Fig. 2A). Second, the sch gene was further localized to the area between the markers 01017 and 01022 using 4,501 BC1 individuals. Finally, with newly developed markers between 01017 and 01022, a region of 239,622 bp tightly linked to the sch phenotype was located on Bm_scaf8 from 3,125,075 to 3,364,696 bp between the markers sch-R2_F1 and R13F20 (Fig. 2B) (Table S2). We predicted eight genes within this particular region using gene-prediction models (Fig. 2C) (24).
Fig. 2.
Mapping of the sch mutation on linkage group 1. (A) SNP markers used for approximate mapping to define the region linked to the sch phenotype. The approximate region linked to the sch phenotype was narrowed down with the boxed markers, 01017 and 01023, using 67 BC1 individuals. The distance is shown in centiMorgan units (cM). (B) Newly developed SNP markers used for fine mapping to narrow the region tightly linked to the sch phenotype using 4,501 BC1 individuals. (C) Gene annotation in the sch linked region. Eight genes were predicted in this region, including BGIBMGA000654, BGIBMGA000564, BGIBMGA000655, BGIBMGA000563, BGIBMGA000657, BGIBMGA000656, BGIBMGA000562 and BGIBMGA000561. Transcriptional direction: Left to Right for BGIBMGA000654, 000655, 000656, and 000657 (BmMet2); Right to Left for BGIBMGA000561, 000562, 000563 (BmTh), and 000564.
Identification of an sch Candidate Gene.
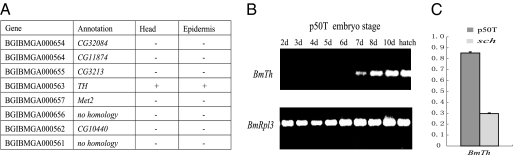
Because the pigmentation of the head and larval epidermis is suppressed in the sch mutant (Fig. 1), the gene responsible for sch should be expressed in both of these regions. We analyzed the expression profiles of the eight initial candidate genes based on microarray data for different larval tissues on day 3 of the fifth instar (32). Only one gene was expressed in both head and epidermis (Fig. 3A). This gene was BGIBMGA000563, which encodes a tyrosine hydroxylase (TH), called BmTh (NM_001145322.1) in the present study. TH is the first enzyme of the melanin synthesis pathway and plays an important role in dark pigment production (6). Interestingly, the expression profile of BmTh increased as the process of body pigmentation progressed during the embryonic stage of the wild-type strain p50T (Fig. 3B). We therefore speculated that BmTh was probably the gene responsible for the sch mutation and focused on it in our subsequent experiments.
Fig. 3.
Expression profiles of sch candidate genes. (A) List of expression patterns of eight candidate genes in the head and epidermis of fifth instar day 3 larvae using microarray data (32). (B) Expression profile of BmTh during the embryonic stage of p50T with semiquantitative RT-PCR. (C) The relative expression level of BmTh between p50T and sch on the day before hatching as shown by real-time PCR, plotted as the mean ± SD (n = 3). The relative expression level of BmTh was determined by comparing the signal intensity with BmRpl3.
We first obtained the complete coding sequences (CDS) of BmTh from embryonic cDNA from the sch mutant and wild-type strain p50T using primer sets designed for the CDS of BmTh (Table S3), and found that they were identical. However, semiquantitative RT-PCR analysis showed that the mRNA transcript level of BmTh during embryogenesis was much lower in the sch mutant than in p50T (Fig. S1A). We therefore hypothesized that a change in the regulatory region of the BmTh gene was probably involved in the mutation.
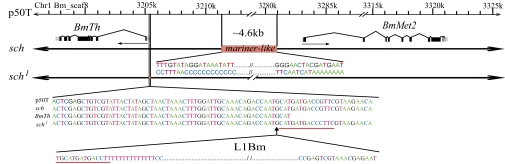
When making the fine linkage map, we found that many primers would not amplify within a large region of the sch mutant DNA lying just upstream of the BmTh gene (Table S4), although they worked well in p50T. Upon designing more primers for PCR comparisons between the mutant and wild-type strains, we finally found that a 69,464-bp region from ∼6 kb upstream of the BmTh gene was replaced by a ∼4.6-kb Tc-1 mariner type DNA transposon in the sch mutant (Fig. 4 and Fig. S1B). The other end of the mutant site was ∼2 kb from the BmMet2 gene, which codes for juvenile hormone resistance protein II in silkworm. To determine which gene was affected by the deletion in sch, we compared the expression of BmTh and BmMet2 between neonatal larvae of p50T and sch, using real-time PCR. The expression of BmTh in the sch mutant was only one-third of that in p50T (Fig. 3C), whereas BmMet2 showed almost no expression in either wild-type or mutant strains. We therefore concluded that the BmTh gene was the most likely candidate for the sch mutation.
Fig. 4.
Schematic structure of the mutant sites in sch and schl. In the sch mutant a sequence of ∼70 kb was replaced with a ∼4.6-kb mariner-like transposon located between ∼6 kb upstream of the BmTh gene and ∼2 kb upstream of the BmMet2 gene (Middle), whereas the schl mutant had a large insertion in front of the transcription start site of BmTh (Bottom). Red underlines indicate a repeated sequence found in the insertion sequence. Arrows indicate the direction of transcription.
Identification of the Mutant Site in the schl Genome.
To test this conclusion, we characterized the allelic mutation, schl. As occurred previously, sequence analysis showed that the coding regions of BmTh were identical among p50T, sch, and schl. However, analysis of the schl genomic DNA using the same primer sets that detected the deleted region in the sch mutant showed no evidence for a similar deletion in the schl mutant. Instead, we found a large fragment inserted in the promoter region between the TATA box region and the transcription start site of the BmTh gene (Fig. 4). Using primer sets near both ends of the insertion, we were not able to amplify a PCR band from the schl mutant, even using long-range PCR. We therefore speculated that the insertion fragment was a long complex retrotransposon containing many other transposons and retrotransposons, like those present in W chromosomes (33, 34). Using reverse PCR with primer sets near the insertion sites (Table S5), we identified and sequenced amplicons from both ends of the insertion fragment. As proposed, the insertion sequence was a non-LTR retrotransposon, L1Bm (Fig. 4) (35).
We then compared the expression of BmTh in p50T, sch, and schl using semiquantitative RT-PCR. In the schl mutant, the BmTh gene was expressed at a lower level than in sch, and it was much lower than in p50T (Fig. S1C). These results showed that the insertion event led to a significant decrease in the BmTh transcript in the schl mutant, similar to the decrease of the BmTh transcript in the sch mutant caused by the deletion event. The expression profiles were consistent with the hypothesis that changes in the transcriptional regulatory region of the BmTh gene were responsible for the sch and schl mutations, but that in one case the mutant phenotype was caused by a deletion, and in the other by an insertion.
Functional Validation of BmTh.
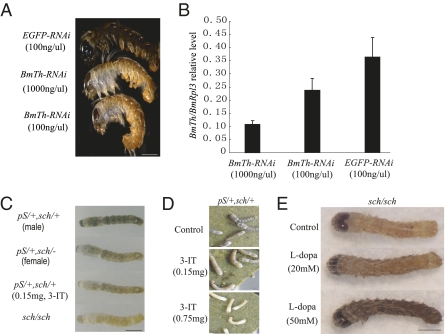
To determine whether decreasing the amount of BmTh mRNA caused the chocolate color or altered the hatching success of neonatal larvae, we synthesized dsRNA corresponding to two different fragments of the BmTh CDS and injected them into early silkworm eggs at doses of 0.3 ng and 3 ng per egg within 4 h after laying. Most of the control samples, which we injected with EGFP dsRNA under the same conditions, hatched after incubation, whereas none of the eggs injected with BmTh dsRNA hatched successfully. Interestingly, upon microscopic inspection and dissection, we found that most of the unhatched embryos were still alive and had attempted to bite through the egg shell to emerge, but had failed. All of the individuals injected with BmTh dsRNA showed evidence of chocolate coloration on the head and epidermis compared with the black color of individuals injected with EGFP dsRNA (Fig. 5A). After 2 d, when the control samples had finished hatching, none of the larvae injected with BmTh dsRNA had hatched, and stereomicroscopic examination showed that most of them had died. This phenotype was very similar to the schl mutant. Details of the BmTh RNAi results are shown in Table S6. Using real-time PCR, we compared the expression of BmTh between individuals injected with BmTh and EGFP dsRNA. The level of BmTh mRNA in the embryos injected with BmTh dsRNA was less than half that of the controls (Fig. 5B).
Fig. 5.
Functional analysis of the BmTh gene. (A) RNAi effects on coloration in neonate larvae. Double-stranded RNA for EGFP (0.3 ng per individual, Top) or BmTh (3 ng per individual, Middle, and 0.3 ng per individual, Bottom) were injected into eggs within 4 h after laying. (Scale bar, 0.5 mm.) (B) Dose-dependent RNAi-induced suppression of targeted gene transcripts. EGFP-RNAi (EGFP) is shown as control. (C) Effects of the TH inhibitor, 3-iodo-tyrosine (3-IT), on the cuticular pattern of second instar day 0 larvae. (Scale bar, 1.5 mm.) (D) Dosage-dependence of treatment with 3-IT on the black stripe of the larval cuticle on second instar day 0 larvae. (E) Rescue of the sch mutant phenotype from chocolate to black color by feeding L-dopa to sch neonate larvae (Scale bar 0.5 mm.)
To determine whether down-regulation of BmTh mRNA was equivalent to affecting pigmentation by depression of BmTH activity, we generated F1 hybrids with sch using pS (striped, 2–3.0), a dominant mutation that produces black stripes in second instar and later stages, which could be used to monitor melanin formation. We fed neonatal larvae of pS(heterozygous)-sch(hetero) with an artificial diet containing 3-iodo-tyrosine (3-IT), an inhibitor of TH. The dark stripes changed to colorless in the second instar larvae of the hybrid mutants fed with TH inhibitor, similar to the pS(hetero)-sch(homo) and sch/sch phenotypes (Fig. 5 C and D). These results, combined with the results of the RNAi experiments, clearly demonstrated that decreasing BmTh mRNA was equivalent to depressing the activity of BmTH. When fed with a higher amount of TH inhibitor, most second instar larvae died after the first molt. The surviving larvae fed with a lower amount of TH inhibitor recovered the black-colored stripes in the third instar so long as they were not fed additional TH inhibitor during the second instar.
TH (EC 1.14.16.2) catalyzes the oxidation of tyrosine to produce dopa, which is necessary for melanin formation (36). Our results suggested that the low expression level of BmTh might limit dopa synthesis, leading to chocolate coloration in the sch mutant. Therefore, we fed L-dopa to sch mutant neonate larvae and successfully rescued the mutant phenotype from chocolate to black coloration within 16 h. Furthermore, the extent of this rescue was dependent on the L-dopa dosage (Fig. 5E).
In conclusion, our results indicated that the sch and schl mutations correspond to the BmTh gene. Each insertion event clearly interrupted the BmTh regulatory region and directly caused the low expression of BmTh, although to varying degrees in the sch and schl mutants. A decrease of BmTh mRNA resulted in a decrease in melanin, leading to the chocolate embryonic color and an inability to hatch, but the lethal effect was dependent on the dosage.
Discussion
In the present study, we identified the gene corresponding to the sch mutation in the silkworm. This conclusion was validated by the following evidence: (i) the BmTh gene was located within a ∼240 kb region perfectly linked with the sch phenotype on a sample of 4,501 BC1 individuals; (ii) the BmTh gene was the only one of eight predicted genes in the candidate region that had an expression profile consistent with the sch phenotype; (iii) BmTh expression was significantly reduced in the sch and schl mutants, accompanied by transposon-mediated disruption of the BmTh promoter regions; (iv) knockdown of BmTh by RNAi and reduction of TH activity by feeding an enzyme inhibitor produced the same phenotypes as the sch and schl mutants; and (v) feeding L-dopa to sch neonate larvae successfully rescued the mutant phenotype from chocolate to black coloration. These data strongly support the conclusion that BmTh corresponds to the Bmsch gene and that the decreased expression of BmTh prevents melanin synthesis, resulting in the reddish-brown coloration in the head and epidermis of the sch and schl mutants.
TH catalyzes the oxidation of tyrosine to dopa in the presence of tetrahydrobiopterin (BH4) as a cofactor; dopa decarboxylase then converts dopa to dopamine, which is then processed by phenoloxidases and cofactors to form melanin during cuticle development (36). THs are highly conserved enzymes found in both vertebrates and invertebrates. We found no difference in the amino acid sequence among p50T, sch, or schl mutants, nor even a single nucleotide polymorphism, implying that conservation of TH is important in silkworms. Silkworms have a single copy BmTh gene and no isoforms have been reported. This finding is in contrast to Drosophila melanogaster, which has two alternatively spliced isoforms of DTH, a minor one expressed specifically in neural tissue and a major one expressed predominantly in the late larval and adult hypodermis (37).
In Drosophila, the pale (ple) gene encodes TH (38), which shows 70% homology with the amino acid sequence of BmTH. Null mutants of ple result in unpigmented embryos that are unable to hatch. Furthermore, a temperature-sensitive mutant (plets1) induced by ethyl methanesulfonate mutagenesis shows a low permissive temperature (18 °C) and a high restrictive temperature (29 °C) (39). However, the precise mutant site and molecular mechanism have not been identified. In the silkworm, the schl and sch mutants have very similar phenotypes to the Drosophila ple and plets1 mutants, respectively. The sch mutant is lethal in the late embryonic stage only at high temperatures (>30 °C), whereas the schl mutant is lethal even at room temperatures (27, 28). We also showed that the expression of BmTh was up-regulated after heat treatment in both p50T and sch embryos, but that the up-regulated quantity in p50T was higher than that of the sch mutant (Fig. S2). Considering all our results and the findings in Drosophila, we suggest that BmTh is not only a lethal gene but also a temperature-regulated gene. In Drosophila, both isoforms of TH have their highest enzymatic activity at 25 to 29 °C and are only 30% active at 37 °C (40). Candidate heat-shock protein (HSP) binding sites were predicted within the 70-kb upstream region of BmTh in the wild-type strain p50T using TFSEARCH (41). However, this region was deleted in the sch mutant, consistent with the possibility that HSP may play a role in normal and wild-type expression of BmTh.
Based on our results, we hypothesize a possible explanation for the inability of sch mutants to hatch, and their subsequent death under high-temperature regimes (>30 °C). At temperatures higher than 30 °C, the enzymatic activity of BmTH decreases. In p50T, the decrease in enzymatic activity may be compensated by an increase in the expression level of BmTh, so that the neonate is still able to develop a black color and hatch normally. In the sch mutant, however, the large deletion of the upstream sequence containing HSP factor binding sites leads to a partial loss of BmTh regulatory function when the temperature is increased. The up-regulated expression level of BmTh is not sufficient to compensate for loss of BmTH enzymatic activity. Lower amounts of the downstream products, such as dopa and dopamine, are therefore produced at 30 °C rather than at 25 °C, resulting in insufficient sclerotization of sch larval mandibles to bite through the egg shell. Compared with sch, the schl mutant completely loses the ability to hatch, even at room temperature. The most likely explanation for schl lethality is that the insertion is positioned just in front of the transcription start site of BmTh and seriously interrupts its basic core promoter, which leads to a lower expression of BmTh in the schl mutant than in sch, where the insertion occurs far from the core promoter.
The sch and schl mutants are typical examples of loss-of-gene function caused by transposable elements (TEs). Interestingly, two different TE insertion events interrupted the function of the BmTh regulatory region, resulting in low transcriptional levels of BmTh. A Tc1-mariner type transposon replaced a ∼70-kb region upstream of BmTh in the sch mutant, whereas a non-LTR retrotransposon L1Bm was inserted just in front of the transcription start site of the BmTh gene in the schl mutant. Nevertheless, we observed a barely detectable expression in the schl mutant, resulting in the lethality of schl at room temperature. Among B. mori mutants, TE-mediated gene disruption has also been reported for the Yellow blood and distinct translucent genes (42, 43). In the revised B. mori genome datasets, Tc1-mariner type transposons and non-LTR retrotransposons comprise about 13.8 and 2.7% of the genome sequence, respectively (44). Moreover, there are indications that the size of silkworm genes and its genome has been increased by the insertion of large numbers of TEs in relatively recent evolutionary time. As with the insertion of TEs in the BmTh regulatory region, TEs play a crucial role in affecting gene function and evolution.
Materials and Methods
Silkworm Strains.
The sch mutant was obtained from the silkworm stock pool of the Southwest University of China. The p50T wild-type was kindly supplied by the University of Tokyo. The schl mutant was obtained from the Institute of Sericulture, Ibaraki, Japan (27). The pS mutant was kindly provided by the Institute of Genetic Resources of Kyushu University. The pnd+ eggs were provided by the National Institute of Agrobiological Sciences. The silkworms were reared on mulberry leaves or artificial diet at 25 °C.
Positional Cloning.
Silkworm strains and crosses.
The sch and p50T strains were used as parent strains for the mapping panel. For the linkage analysis, 4,501 segregants of a single-pair backcross (BC1) between an sch female × an F1 male (sch female × p50T male) were used.
Genomic DNA extraction.
Genomic DNA of parental strains and F1 individuals was isolated from adult legs, and genomic DNA of individual BC1 segregants was isolated from whole neonatal larvae using DNAzol (Invitrogen) (26). BC1 genomic DNA samples were purified with PI-1100 (Kurabo).
The sch linkage map construction.
To construct the sch linkage map, SNP markers on chromosome 1 and newly developed markers from BAC end sequences (25, 26) were used to survey the segregation patterns in 4,501 BC1 individuals from the cross sch female × F1 male (sch female × p50T male). The primers for the SNP markers used in the linkage analysis are listed in Table S1.
Annotation Analysis.
The candidate genes in the region narrowed by the linkage analysis were annotated using the silkworm genome database SilkDB (http://silkworm.swu.edu.cn/silkdb/) and BLASTX in National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
RNAi Reduction of BmTh Gene Expression.
To produce BmTh dsRNAs, two different PCR fragments (402 bp and 637 bp) of BmTh cDNA were cloned into a pTA2 plasmid (Ambion). The two fragments were amplified using the silkworm full-length cDNA clone fcaL30O04 containing BmTh as a template with the following two pairs of primers: BmTh PCR 1 for the 402-bp fragment, forward: GTACAATGGCAGTCGCAGCA, reverse: CCGCTTCAGAAGATTCACTAGCAG; BmTh PCR2 for the 637-bp fragment, forward: CGCCACGCTTCTGATCTCGATAAC, reverse: TGCACAGTCCGAACTCAACCCTAA. Plasmids with PCR fragments for the BmTh gene inserted in both directions were isolated for the synthesis of the dsRNAs. The plasmids were linearized by digestion with the restriction enzymes BamHI and PstI to use as templates for RNA synthesis. Sense and antisense RNAs were synthesized for each fragment using a MEGAscript RNAi kit (Ambion). RNA synthesis and purification were carried out according to the manufacturer’s instructions and the integrity of dsRNA was confirmed by nondenaturing agarose gel electrophoresis. The purity and concentration of dsRNA were measured using a BECKMAN DU-600 spectrophotometer and adjusted to final concentrations of 100 ng/μL and 1,000 ng/μL with an injection buffer. EGFP dsRNA was synthesized using the method of Quan et al. (45).
Silkworm Strains and Microinjection.
Strain pnd+ larvae were reared on an artificial diet at 25 °C. The strain has dark-brown eggs and the larval skin color is black brown immediately after hatching. The eggs were used for the microinjection of dsRNA following the method of Quan et al. (45). About 3 nL of the solution at a dose of 0.3 ng or 3.0 ng were injected into each egg followed by incubation at 25 °C in a moist Petri dish. The embryo color was investigated after the control eggs hatched and photos were taken with an MZ16FA microscope (Leica).
RNA Extraction and Semiquantitative RT-PCR.
Total RNA from different embryonic stages and neonates was extracted using TRIzol reagent (Roche) and reverse transcribed with random primer (N6) and an oligo (dT) of Ready-To-Go RT-PCR Beads (Amersham Biosciences). Primer sets for RT-PCR of BmTh are listed in Table S3. The semiquantitative RT-PCR conditions were 94 °C for 30 s, followed by 28 (or 30) cycles of 94 °C for 10 s, 60 °C for 15 s, and 72 °C for 90 s. The reactions were kept at 72 °C for 7 min after the last cycle. The silkworm housekeeping gene ribosomal protein L3 (BmRpl3) was used as an internal control for normalization of sample loading.
Real-Time RT-PCR.
Total RNA was isolated as described above. Reverse transcription was carried out using 500 ng of total RNA in 10-μL reactions using a PrimeScript RT reagent Kit (Takara) by an oligo (dT) and (N6) priming method according to the manufacturer’s recommendations. Real-time PCR was performed with a LightCycler 480 (Roche) using the manufacturer’s recommended procedure. The real-time PCR condition was denaturation at 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The primers for the BmRpl3 and BmTh genes are listed in Table S7. Reactions were run in triplicate using 1 μL of cDNA per reaction. Standard curves to quantify relative gene copy number were made from 10-fold serial dilutions from 10 ng to 0.001 ng of PCR products of BmRpl3 and BmTh. BmTh transcript levels were compared with BmRpl3 transcript levels in the same sample.
3-IT and L-Dopa Feeding Experiments.
The TH inhibitor (3-IT) (Sigma) solution was added to artificial diet at concentrations of 0.75 mg/g and 0.15 mg/g (3-IT/artificial diet). After the artificial diet had completely absorbed the solution, it was fed to neonate larvae of pS(heterozygous)-sch(hetero) until they finished the first instar. For the rescue experiment, 20 mM and 50 mM L-dopa (Sigma) solutions were painted uniformly onto both surfaces of soft mulberry leaves and then fed to sch neonate larvae. PBS solutions were applied in the same way to form control groups in both feeding experiments. Photos were taken with a DXM 1200F system (Nikon).
Supplementary Material
Acknowledgments
We thank Dr. Masahiro Ajimura for his advice and help throughout the experiments, Dr. Yuki Nakamura for her guidance on real-time PCR, and all the members of the groups of Dr. Kazuei Mita and Dr Kimiko Yamamoto for their support. This work was supported by Grant 2005CB121000 from the National Basic Research Program of China (to Q.-Y.X.), the Ministry of Education of the People’s Republic of China (Program for Changjiang Scholars and Innovative Research Team in University, IRT0750) (to Q.-Y.X.), the 111 Project (B07045) (to Q.-Y.X.), and grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Integrated research project for plant, insect and animal using genome technology) (to K. M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001725107/-/DCSupplemental.
References
- 1.Beadle GW, Ephrussi B. Development of eye colors in Drosophila: Transplantation experiments with suppressor of vermilion. Proc Natl Acad Sci USA. 1936;22:536–540. doi: 10.1073/pnas.22.9.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beadle GW. Development of eye colors in Drosophila: Fat bodies and malpighian tubes in relation to diffusible substances. Genetics. 1937;22:587–611. doi: 10.1093/genetics/22.6.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ephrussi B, Chevais S. Development of eye colors in Drosophila: Relation between pigmentation and release of the diffusible substances. Proc Natl Acad Sci USA. 1937;23:428–434. doi: 10.1073/pnas.23.8.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- 5.True JR. Insect melanism: The molecules matter. Trends Ecol Evol. 2003;18:640–647. [Google Scholar]
- 6.Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: Genetic mechanisms and the potential consequences of pleiotropy. Semin Cell Dev Biol. 2009;20:65–71. doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: Genetic control of pigment patterns in Drosophila. Trends Genet. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 9.Weatherbee SD, et al. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr Biol. 1999;9:109–115. doi: 10.1016/s0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 10.Brakefield PM, et al. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-Herrero E, Vernós I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- 12.Kopp A, Duncan I. Control of cell fate and polarity in the adult abdominal segments of Drosophila by optomotor-blind. Development. 1997;124:3715–3726. doi: 10.1242/dev.124.19.3715. [DOI] [PubMed] [Google Scholar]
- 13.Brunetti CR, et al. The generation and diversification of butterfly eyespot color patterns. Curr Biol. 2001;11:1578–1585. doi: 10.1016/s0960-9822(01)00502-4. [DOI] [PubMed] [Google Scholar]
- 14.Biessmann H. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc Natl Acad Sci USA. 1985;82:7369–7373. doi: 10.1073/pnas.82.21.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter MF, et al. Temporal and spatial expression of the yellow gene in correlation with cuticle formation and dopa decarboxylase activity in Drosophila development. Dev Biol. 1991;147:32–45. doi: 10.1016/s0012-1606(05)80005-3. [DOI] [PubMed] [Google Scholar]
- 16.Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol. 2002;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- 17.Munté A, Aguadé M, Segarra C. Divergence of the yellow gene between Drosophila melanogaster and D. subobscura: recombination rate, codon bias and synonymous substitutions. Genetics. 1997;147:165–175. doi: 10.1093/genetics/147.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikkawa H. Mechanism of pigment formation in Bombyx and Drosophila. Genetics. 1941;26:587–607. doi: 10.1093/genetics/26.6.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futahashi R, et al. yellow and ebony are the responsible genes for the larval color mutants of the silkworm Bombyx mori. Genetics. 2008;180:1995–2005. doi: 10.1534/genetics.108.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Y, et al. The silkworm mutant lemon (lemon lethal) is a potential insect model for human sepiapterin reductase deficiency. J Biol Chem. 2009;284:11698–11705. doi: 10.1074/jbc.M900485200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banno Y, Fujii H, Kawaguchi Y, Yamamoto K, Nishikawa K. A Guide to the Silkworm Mutants: 2005 Gene Name and Gene Symbol. Fukuoka, Japan: Kyusyu University; 2005. [Google Scholar]
- 22.Xia Q, et al. Biology Analysis Group. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 23.Mita K, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- 24.International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto K, et al. Construction of a single nucleotide polymorphism linkage map for the silkworm, Bombyx mori, based on bacterial artificial chromosome end sequences. Genetics. 2006;173:151–161. doi: 10.1534/genetics.105.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto K, et al. A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol. 2008;9:R21. doi: 10.1186/gb-2008-9-1-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Q, Chen Y, Chen J, Lin J, Huang Z. Using temperature- and humidity-sensitivity traits to control silkworm sex. Chin Sci Bull. 1992;37:1133–1136. [Google Scholar]
- 28.Ohnuma A. A new type of sex-linked chocolate mutant, schl. Journal of Dainippon Silk Foundation. 2001;49:1–7. [Google Scholar]
- 29.Miao XX, et al. Inheritance and linkage analysis of co-dominant SSR markers on the Z chromosome of the silkworm (Bombyx mori L.) Genet Res. 2008;90:151–156. doi: 10.1017/S0016672308009221. [DOI] [PubMed] [Google Scholar]
- 30.Gao G, Zhu Y, Chen P, Dai F, Lu C. Studies on the molecular marker of the sex-linkage chocolate gene (sch) in silkworm with RAPD. Newsletter of Sericultural Science. 2000;20:1–3. [Google Scholar]
- 31.Yasukochi Y. A dense genetic map of the silkworm, Bombyx mori, covering all chromosomes based on 1018 molecular markers. Genetics. 1998;150:1513–1525. doi: 10.1093/genetics/150.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia Q, et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8:R162. doi: 10.1186/gb-2007-8-8-r162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe H, Mita K, Yasukochi Y, Oshiki T, Shimada T. Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet Genome Res. 2005;110:144–151. doi: 10.1159/000084946. [DOI] [PubMed] [Google Scholar]
- 34.Abe H, et al. Nested retrotransposons on the W chromosome of the wild silkworm Bombyx mandarina. Insect Mol Biol. 2002;11:307–314. doi: 10.1046/j.1365-2583.2002.00339.x. [DOI] [PubMed] [Google Scholar]
- 35.Ichimura S, Mita K, Sugaya K. A major non-LTR retrotransposon of Bombyx mori, L1Bm. J Mol Evol. 1997;45:253–264. doi: 10.1007/pl00006228. [DOI] [PubMed] [Google Scholar]
- 36.Wright TR. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv Genet. 1987;24:127–222. [PubMed] [Google Scholar]
- 37.Friggi-Grelin F, Iché M, Birman S. Tissue-specific developmental requirements of Drosophila tyrosine hydroxylase isoforms. Genesis. 2003;35:260–269. doi: 10.1002/gene.1082. [DOI] [PubMed] [Google Scholar]
- 38.Neckameyer WS, Quinn WG. Isolation and characterization of the gene for Drosophila tyrosine hydroxylase. Neuron. 1989;2:1167–1175. doi: 10.1016/0896-6273(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 39.Pendleton RG, Rasheed A, Sardina T, Tully T, Hillman R. Effects of tyrosine hydroxylase mutants on locomotor activity in Drosophila: A study in functional genomics. Behav Genet. 2002;32:89–94. doi: 10.1023/a:1015279221600. [DOI] [PubMed] [Google Scholar]
- 40.Vié A, Cigna M, Toci R, Birman S. Differential regulation of Drosophila tyrosine hydroxylase isoforms by dopamine binding and cAMP-dependent phosphorylation. J Biol Chem. 1999;274:16788–16795. doi: 10.1074/jbc.274.24.16788. [DOI] [PubMed] [Google Scholar]
- 41.Heinemeyer T, et al. Databases on Transcriptional Regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:364–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakudoh T, et al. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc Natl Acad Sci USA. 2007;104:8941–8946. doi: 10.1073/pnas.0702860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujii T, Abe H, Katsuma S, Mita K, Shimada T. Mapping of sex-linked genes onto the genome sequence using various aberrations of the Z chromosome in Bombyx mori. Insect Biochem Mol Biol. 2008;38:1072–1079. doi: 10.1016/j.ibmb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1046–1057. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Quan GX, Kanda T, Tamura T. Induction of the white egg 3 mutant phenotype by injection of the double-stranded RNA of the silkworm white gene. Insect Mol Biol. 2002;11:217–222. doi: 10.1046/j.1365-2583.2002.00328.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.