Abstract
The ESCRT pathway helps mediate the final abscission step of cytokinesis in mammals and archaea. In mammals, two early acting proteins of the ESCRT pathway, ALIX and TSG101, are recruited to the midbody through direct interactions with the phosphoprotein CEP55. CEP55 resides at the centrosome through most of the cell cycle but then migrates to the midbody at the start of cytokinesis, suggesting that the ESCRT pathway may also have centrosomal links. Here, we have systematically analyzed the requirements for late-acting mammalian ESCRT-III and VPS4 proteins at different stages of mitosis and cell division. We found that depletion of VPS4A, VPS4B, or any of the 11 different human ESCRT-III (CHMP) proteins inhibited abscission. Remarkably, depletion of individual ESCRT-III and VPS4 proteins also altered centrosome and spindle pole numbers, producing multipolar spindles (most ESCRT-III/VPS4 proteins) or monopolar spindles (CHMP2A or CHMP5) and causing defects in chromosome segregation and nuclear morphology. VPS4 proteins concentrated at spindle poles during mitosis and then at midbodies during cytokinesis, implying that these proteins function directly at both sites. We conclude that ESCRT-III/VPS4 proteins function at centrosomes to help regulate their maintenance or proliferation and then at midbodies during abscission, thereby helping ensure the ordered progression through the different stages of cell division.
Keywords: ESCRT, CHMP, mitosis, cytokinesis, abscission
The ESCRT pathway functions across eukaryotes and many archaeal species, where it helps mediate (i) vesicle formation at multivesicular bodies (MVB) (1), (ii) enveloped virus budding (2), and (iii) the abscission stage of cytokinesis (3–7). These seemingly disparate biological processes all involve the resolution of thin, cytoplasm-filled membrane tubules, implying that ESCRT machinery can be recruited to different biological membranes to mediate topologically similar membrane fission events. Most ESCRT pathway proteins function as subunits of five multiprotein complexes, termed the ESCRT-0, -I, -II, -III, and VPS4 complexes. Other ESCRT factors, such as ALIX, function as discrete proteins. Classic studies in yeast have established that the ESCRT components are recruited sequentially to endosomal membranes where they assemble into higher order complexes that mediate protein sorting, membrane remodeling, and fission (8). Initially, early-acting factors such as ESCRT-I, ESCRT-II, and ALIX interact with upstream recruiting factors, concentrate protein cargoes, and help deform membranes (9). These early-acting factors recruit subunits of the ESCRT-III complex, which form filaments within the necks of membrane tubules and mediate membrane fission (10–16). ESCRT-III assemblies, in turn, recruit VPS4 ATPases, which use the energy of ATP hydrolysis to disassemble the ESCRT complexes (10–14, 17).
ESCRT-III and VPS4 homologs mediate abscission in hyperthermophilic crenarchaeal species that diverged from eukaryotes several billion years ago, suggesting that cell division may have been the primordial function of the ESCRT pathway (6, 7). The ESCRT pathway also helps mediate abscission in mammals (3), where its activities must be integrated with a number of other important processes, including centrosome function, mitosis, and contractile ring formation and constriction. As cells exit mitosis, the phosphoprotein CEP55 leaves centrosomes and concentrates at the midbody (18), where it helps to organize factors required for abscission. This translocation event is one of many emerging links between centrosomes and cytokinesis (reviewed in ref. 19). Indeed, it has been proposed that centrosomes may regulate mammalian pathway(s) that help control the transition from mitosis to cytokinesis, as is the case for the “mitotic exit network,” which is organized by yeast spindle pole bodies (20). CEP55 binds and recruits the early-acting ALIX and ESCRT-I proteins to the midbody, and these interactions are required for efficient abscission (3, 4, 21). The roles of late-acting ESCRT factors in cytokinesis are less well characterized, but several recent reports indicate that VPS4, the ESCRT-III protein CHMP1B, and the ESCRT-III-like protein IST1 can also function at midbodies (3, 4, 21–24).
In this study, we tested the requirements for each human ESCRT-III and VPS4 protein during different stages of cell division. Cells lacking each of the late-acting ESCRT-III/VPS4 proteins exhibited abscission defects. Remarkably, cells lacking ESCRT-III/VPS4 proteins also exhibited profound defects earlier in mitosis, including aberrations in centrosome number, morphology, and function, implying that ESCRT-III/VPS4 proteins perform important centrosomal functions and may also function during other stages of mitosis.
Results
ESCRT-III and VPS4 Proteins Are Required for Normal Cell Division.
To test for roles in cell division, each of the 11 different human ESCRT-III/CHMP proteins and both VPS4 proteins were individually depleted from HeLa cells using siRNA treatments. At least four siRNAs against each ESCRT-III/VPS4 protein were tested for efficacy using antibodies against the endogenous proteins, except in the case of CHMP4C, where suitable antibodies were not available and protein depletion was therefore tested using exogenously expressed CHMP4C. At least two effective siRNAs against each target protein were identified, and their specificities were confirmed by testing for cross-reactivity with related ESCRT-III/VPS4 family members (Fig. S1 and Table S1). This set of ESCRT-III/VPS4 siRNAs was then used to test the roles of each ESCRT-III and VPS4 protein in cell division. Measurements of cellular DNA content and nuclear morphology were confirmed with at least two different siRNAs against each ESCRT-III/VPS4 protein, but for clarity the results are reported for a single siRNA (except where noted).
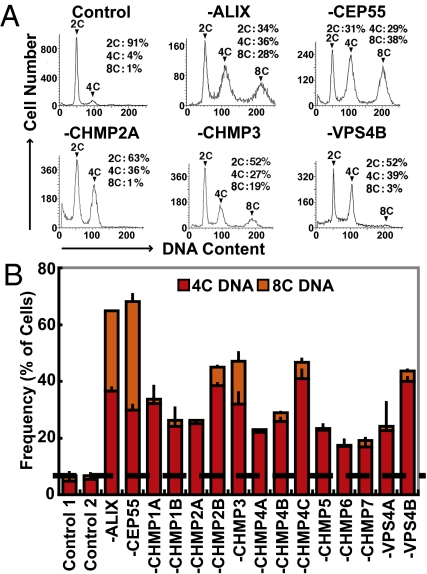
As an initial screen for cell cycle defects, HeLa cells were depleted of individual ESCRT-III/VPS4 proteins and assayed for changes in DNA copy number using flow cytometry measurements (Fig. 1). Cells received two siRNA treatments at 24-h intervals, and the resulting phenotypes were measured 24 h after the second treatment. Irrelevant siRNAs were used as negative controls, and siRNAs that targeted ALIX or CEP55 were used as positive controls for abscission defects (3, 4). As expected, depletion of ALIX or CEP55 dramatically increased the frequencies of cells with four copy (4C) DNA content (>6-fold increases) and 8C DNA (>16-fold increases) compared with the negative controls. These profiles arose because the cells went through multiple rounds of mitosis without completing cytokinesis, and therefore ended up with multiple nuclei and multiple copies of their DNA (Figs. 1 and 2). Depletion of individual ESCRT-III/VPS4 proteins similarly increased the frequency of cells with 4C DNA content (3- to 8-fold increases), implying nonredundant roles for all of the human ESCRT-III/VPS4 proteins in cell division. CHMP3 depletion additionally increased the fraction of cells with 8C DNA content (approximately 9-fold increase), and this phenotype therefore resembled the ALIX and CEP55 controls. In contrast, depletion of other ESCRT-III/VPS4 proteins caused a significant increase in the fraction of cells with 4C but not 8C DNA, suggesting that these ESCRT-III/VPS4 proteins might perform different or additional functions in cell division.
Fig. 1.
Aberrant DNA contents of cells lacking ESCRT-III and VPS4 proteins. (A) Flow cytometry analyses showing the DNA content of HeLa cells treated with a control siRNA or with siRNAs to deplete ALIX (control), CEP55 (control), CHMP2A, CHMP3, or VPS4B. The 2C, 4C, and 8C DNA peaks are labeled, and cell percentages are provided. (B) Quantified percentages of siRNA-treated cells with 4C (red) and 8C (orange) DNA (n = 2 ± range, dotted line denotes the average total percent of control cells with 4C and 8C DNA).
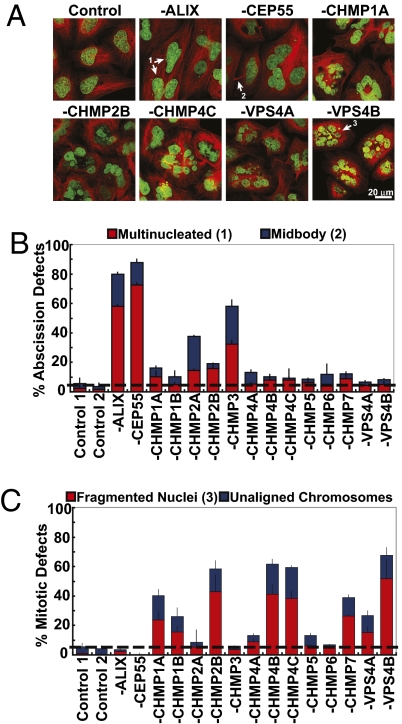
Fig. 2.
Mitosis and cytokinesis defects in cells lacking ESCRT-III and VPS4 proteins. (A) Confocal fluorescent slices from HeLa cells depleted of the designated proteins. Fluorescent staining shows microtubules (red, anti-α-Tubulin) and nuclei (green, SYTOX Green). Multinucleated cells (1), visible midbodies (2), and fragmented nuclei (3) are highlighted with arrows. (B) Percentages of cells with multiple nuclei (red), or visible midbodies (blue) (A), resulting from abscission defects following depletion of the designated proteins (n = 3 ± SD). (C) Percentages of cells with fragmented nuclei (red) (A) or unaligned chromosomes (blue) (Fig. 3A), following depletion of the designated proteins (n = 3 ± SD).
ESCRT-III and VPS4 Proteins Are Required for Mitosis.
To identify other potential ESCRT-III/VPS4 protein functions, cells were depleted of individual ESCRT-III/VPS4 proteins, fixed, stained with fluorescent markers for DNA (green) and microtubules (red), and examined by confocal microscopy (Fig. 2A). As expected, depletion of CEP55 or ALIX dramatically increased the frequencies of cells with visible midbodies (>5-fold increases, reflecting delayed abscission) and multiple nuclei (>25-fold increases, reflecting failed abscission) (Fig. 2B). Depletion of CHMP3 similarly increased the frequencies of cells with visible midbodies and multiple nuclei (10- and 16-fold increases, respectively), again indicating that this ESCRT-III protein, like ALIX and CEP55, functions primarily during abscission. Cells lacking CHMP2A also had similar phenotypes, but these cells exhibited other mitotic defects (described below). Depletion of the remaining ESCRT-III/VPS4 proteins also modestly increased the numbers of cells with visible midbodies and multiple nuclei, but these cells also exhibited two striking new phenotypes: (i) Many cells had condensed but poorly aligned chromosomes, suggesting prometaphase or metaphase defects (Fig. 3A), (ii) Many cells had aberrant nuclei that appeared to be composed of fragmented or interconnected micronuclei or that adopted unusual crescent shapes (Fig. 2A). Strong phenotypes of this type were induced by depletion of CHMP1A, CHMP1B, CHMP2B, CHMP4B, CHMP4C, CHMP7, VPS4A, and VPS4B (Fig. 2C). Similar, but weaker phenotypes were induced by depletion of the remaining ESCRT-III/VPS4 proteins (with the exception of CHMP3). Strong mitotic phenotypes often correlated with weaker abscission defects, possibly because the prevalence of earlier mitotic defects masked downstream abscission defects (Fig. 2 B and C). These observations indicate that most ESCRT-III/VPS4 proteins are required both for efficient abscission and also for normal mitotic chromosome alignment and nuclear morphology.
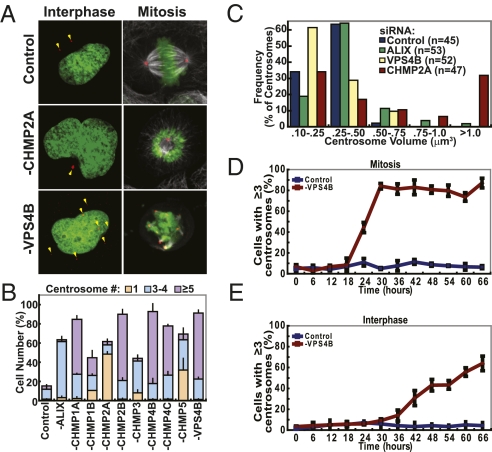
Fig. 3.
Centrosomal defects in cells lacking ESCRT-III and VPS4 proteins. (A) Confocal fluorescent images of HeLa cells stained for DNA (SYTOX Green) and centrosomes (red, anti-Pericentrin), following treatment with a control, CHMP2A, or VPS4B siRNA. Cells are shown during interphase (Left) and mitosis (Right, with microtubules in white, anti-α-Tubulin). Arrows highlight subsets of visible centrosomes (Figs. S2 and S3). (B) Percentages of cells with abnormal centrosome numbers following siRNA treatment (Color key above, n = 3 ± SD). (C) Centrosome volumes measured in 3D reconstructions of fixed HeLa cells depleted of the designated protein, stained for Pericentrin and binned into the designated volume ranges. Color key and total cell numbers from three different experiments are given within the graph. (D and E) Percentages of HeLa cells with three or more centrosomes following treatment with control or VPS4B siRNA. Individual samples were harvested every 6 h and stained with antibodies against Pericentrin and γ-Tubulin. Only centrosomes that stained with both antibodies were counted (n = 3 ± SD). Mitotic (D) and interphase (E) cells from the same samples were counted separately.
ESCRT-III and VPS4 Proteins Maintain Normal Centrosome and Spindle Numbers and Morphologies.
To characterize the ESCRT-III/VPS4 mitotic defects further, we analyzed centrosome, chromosome, and spindle organization in HeLa cells depleted of a subset of the ESCRT-III/VPS4 proteins with the strongest mitotic phenotypes. Cells were treated with an irrelevant siRNA (negative control), an siRNA against ALIX (control), or siRNAs against ESCRT-III/VPS4 proteins and then fixed and stained for centrosomes (red), DNA (green), and microtubules (white) (Fig. 3A). As expected, interphase cells treated with control siRNAs typically had two distinguishable centrosomes (85 ± 3% of cells, Fig. 3A, column 1). Cells with one, three to four, or more than five centrosomes were also present, but were infrequent (1.5 ± 0.5%, 10 ± 4%, and 3 ± 1%, respectively) (Fig. 3B). During metaphase, control cells typically formed normal bipolar spindles with two centrosomes (red), canonical spindle microtubules (white), and aligned chromosomes (green) (Fig. 3A, column 2). Spindles were also normal in cells that lacked ALIX and CHMP3, but centrosome numbers were elevated modestly. These similarities further support the idea that this particular ESCRT-III protein functions primarily during abscission, where failed abscission allowed subsequent round(s) of DNA and centrosome duplication in the next cell cycle, resulting in 8C DNA and four centrosomes per cell.
In contrast, cells that lacked most other ESCRT-III/VPS4 proteins had abnormally large numbers of centrosomes and spindles. This phenotype is illustrated for interphase cells that lacked VPS4B (Fig. 3A Lower Left) and was quantified for each of the nine different ESCRT-III/VPS4 proteins tested (Fig. 3B). Similarly, mitotic cells that lacked most ESCRT-III/VPS4 proteins exhibited multipolar spindles and aberrant chromosome alignment during metaphase (Fig. 3A Lower Right). Strong phenotypes of this type were observed in cells that lacked CHMP1A, CHMP2B, CHMP4B, CHMP4C, or VPS4B (Fig. 3B and Fig. S2). In each of these cases, most cells had at least five discernable centrosomes, and some cells had large numbers of centrosomes (>20).
As shown in Fig. 3 A and B, CHMP2A and CHMP5 differed from most other ESCRT-III proteins, as their depletion resulted in abnormally high percentages of interphase cells with only a single discernable centrosome (49 ± 3% of CHMP2A-depleted cells, vs. 1.5 ± 0.5% of control cells; Fig. S3). During mitosis, cells that lacked CHMP2A frequently exhibited monopolar spindles that appeared to pull the condensed, aligned chromosomes around themselves into characteristic “C” or “O” shapes (Fig. 3A Middle Right). The lone centrosomes in CHMP2A-depleted cells often appeared unusually large and highly fluorescent, suggesting that they might have abnormal organization and/or increased recruitment of pericentriolar material. This phenotype was quantified by measuring centrosome volumes in 3D reconstructions from thin confocal sections of cells depleted of two representative ESCRT-III/VPS4 proteins (CHMP2A and VPS4B). As shown in Fig. 3C, the sizes of centrosomes stained with anti-Pericentrin antibodies were relatively homogenous in control cells, with reconstructed volumes typically ranging between 0.25 and 0.50 μm3 (64% of centrosomes) or 0.1 and 0.25 μm3 (34% of centrosomes). Centrosomes in cells lacking ALIX were of similar size, albeit with small pools of larger centrosomes. In contrast, centrosomes in cells that lacked VPS4B were typically smaller than control centrosomes (62% between 0.1 and 0.25 μm3), but also exhibited a slightly expanded size range (e.g., 10% were between 0.50 and 0.75 μm3). Most notably, a significant subset of the centrosomes from cells lacking CHMP2A were much larger than control centrosomes, with 32% >1 μm3. Thus, depletion of either VPS4B or CHMP2A altered the overall size of centrosomes and associated pericentriolar material, but in opposite directions.
A series of control experiments were performed to confirm the specificity of the anti-Pericentrin antibody, test whether the aberrant centrosomes contained the expected array of other centrosomal proteins, determine whether VPS4B depletion induced similar phenotypes in multiple different human cell lines, and examine cell viability. As shown in Fig. S4, the aberrant centrosomes in VPS4B- and CHMP2A-depleted cells stained with a second anti-Pericentrin antibody, with anti-γ-Tubulin antibodies, and with anti-aurora A antibodies during mitosis, as would be expected for functional centrosomes. We also tested the effects of depleting VPS4B from four different human cell types: HeLa, GHD-1, 293T, and HOS cells. HeLa and GHD-1 cells exhibited highly penetrant mitotic and centrosomal phenotypes, whereas these defects were weaker but measurable in 293T and HOS cells (Fig. S5 A–C). Thus, VPS4B is required to maintain normal centrosome numbers in multiple human cell types, although the degree to which different cell types depend upon VPS4B may vary. Cell death was also increased in cells depleted of CHMP2A, CHMP4B, and VPS4B, consistent with roles for these proteins in cell division (Fig. S5D).
Centrosome Amplification in VPS4-Depleted Cells Occurs During Mitosis.
Time courses were performed to determine the timing of centrosome amplification within the cell cycle. Cells were treated with either a control siRNA or an siRNA against VPS4B (at t = 0 h and t = 24 h) and analyzed at 6-h intervals by counting the numbers of centrosomes in fixed, stained interphase or mitotic cells. As expected, centrosome numbers remained constant in cells treated with control siRNAs. In contrast, centrosome numbers in VPS4B-depleted mitotic cells began to increase significantly at t = 24 h and continued to rise until t = 30 h (Fig. 3D), consistent with amplification during the first round of mitosis following protein depletion. Remarkably, approximately 70% of the mitotic cells with supernumerary spindles had at least five centrosomes/spindles, implying that their numbers increased rapidly from two to more than five during the first mitosis.
As shown in Fig. 3 D and E, the rise in mitotic cells with supernumerary centrosomes was followed by a rise in the number of interphase cells with elevated centrosome numbers, implying that centrosome amplification occurred primarily during mitosis, resulting in amplified numbers of centrosomes in the subsequent interphase. The fraction of cells with elevated centrosome numbers was higher in mitotic cells than in interphase cells, possibly because mitotic cells with high spindle numbers had greater probabilities of undergoing apoptosis (or cell cycle delays) and/or because amplified centrosomes clustered during interphase.
Live Cell Imaging of Cells Lacking VPS4B and CHMP2A.
The dynamic processes of spindle formation, chromosome alignment, chromosome segregation and cytokinesis were analyzed by imaging live HeLa cells that stably expressed fluorescently labeled chromatin (H2B-mCherry) and microtubules (YFP-α-Tubulin). Time lapse images, schematic illustrations, and movies of dividing cells lacking ALIX, VPS4B or CHMP2A are provided in Fig. 4 A and B and Movies S1, S2, S3, S4, S5, S6, S7, and S8. Movie images were quantified to determine the duration of mitosis and the frequencies of mitosis and abscission failure. As expected, control cells proceeded normally through the cell cycle (Fig. 4 A and B and Movies S1 and S2). The average duration of mitosis was 71 ± 51 min (Fig. 4C), and mitosis and abscission were almost always successful [6 ± 2 and 2 ± 1% failure rates, respectively (Fig. 4 D and E)]. Cells that lacked ALIX exhibited minor mitotic defects (58 ± 18 min, 14 ± 2% failure) but then arrested during abscission (91 ± 3% abscission failure) (Fig. 4 and Movies S3 and S4). Eventually, abscission failure resulted in dissolution of the midbody and coalescence into single cells with multiple nuclei.
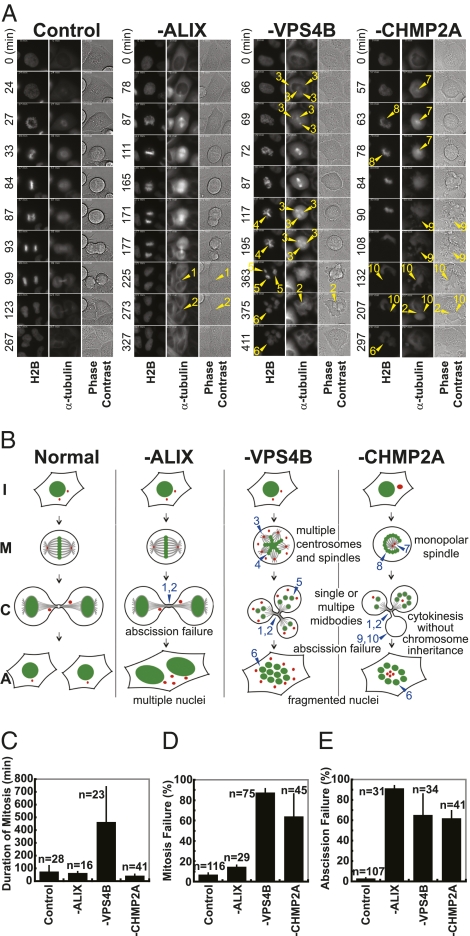
Fig. 4.
Time-lapse analyses of mitosis and cytokinesis defects in living cells lacking VPS4B or CHMP2A. (A) Time-lapse images of dividing HeLa cells treated with a control siRNA or depleted of the designated proteins. Stably transduced HeLa cells expressed fluorescent chromatin (H2B-mCherry, first columns in each panel) and microtubules (YFP-α-Tubulin, second columns). Phase contrast images are provided in the third columns, and elapsed times are given (Left). Numerically coded event key: (1) arrested abscission, (2) dissolution of midbody and coalescence, (3) multiple spindle poles/centrosomes, (4) failed chromosome alignment, (5) asymmetric chromosome segregation into three nascent cells, (6) formation of multiple small nuclei, (7) monopolar spindles/single centrosomes or clusters, (8) “C”-shaped chromosome clusters, (9) emerging daughter cells, and (10) failed chromosome segregation into daughter cells. Annotated movies of these single cells and of fields of cells are provided in Movies S1, S2, S3, S4, S5, S6, S7, and S8. (B) Schematic illustrations of the different stages of mitosis and cytokinesis in cells lacking the designated proteins. I, interphase; M, metaphase; C, cytokinesis; A, abscission. Specific defects are coded numerically as in A. (C–E) Duration of mitosis (C) and percentages of cells with mitotic (D) and abscission (E) defects in cells lacking the designated proteins (SI Materials and Methods).
In cells that lacked VPS4B, bipolar spindles typically formed initially and the condensed chromosomes began to align on the metaphase plate. However, additional spindles soon formed, and the chromosomes became increasingly incoherent until they ultimately dispersed into small clusters (Fig. 4A panel 3, 117–363 min, and Movies S5 and S6). Cells remained in this aberrant prometaphase stage for extended periods of time (458 ± 283 min vs. 71 ± 51 min for control cells, see Fig. 4C for quantification). Eventually, these cells usually initiated cytokinesis, but defects in chromosomal segregation (87 ± 5% failure) and/or abscission (65 ± 21% failure) typically ensued (Fig. 4 D and E). Ultimately, multiple small discrete or interconnected nuclei appeared, apparently because nuclear membranes formed around the dispersed chromosomes (Fig. 4A, panel 3, 375 min, and Movies S5 and S6). Thus, our live cell imaging experiments confirmed that cells lacking VPS4B formed aberrantly large numbers of spindles, arrested early in mitosis, failed to segregate chromosomes normally, failed to complete abscission, and ultimately assembled aberrant nuclei.
In contrast, cells that lacked CHMP2A typically formed monopolar spindles early in mitosis. Chromosomes initially condensed and began to align, but were then drawn in about the monopole, forming characteristic C- or O-shaped arrays (Fig. 4A, panel 4, 63–78 min, Movies S7 and S8, and Fig. 3A). We surmise that the chromosome arrays curved about the monopole owing to the lack of opposing forces normally generated by a bipolar spindle. However, CHMP2A-depleted cells subsequently progressed into stages that resembled metaphase and anaphase and proceeded through mitosis at normal rates (37 ± 16 min, Fig. 4C). During these stages, aberrant bipolar spindles formed and the chromosomes separated (e.g., Fig. 4A, panel 4, 207 min, and Movies S7 and S8). Chromosome segregation was often asymmetric (63 ± 23%, Fig. 4D), contractile rings formed at multiple sites (Fig. 4A, panel 4, 84–207 min, and Movies S7 and S8), and the “daughter cells” often received no chromosomes. Finally, abscission typically failed (62 ± 8%, Fig. 4E) and small fragmented nuclei sometimes formed, although this phenotype was less severe than for VPS4B-depleted cells (Fig. 2C). These live cell imaging experiments therefore revealed that cells lacking CHMP2A formed transient monopolar spindles, failed to segregate chromosomes normally, failed to complete abscission, and ultimately assembled aberrant nuclei.
Human VPS4 Proteins Concentrate at Centrosomes.
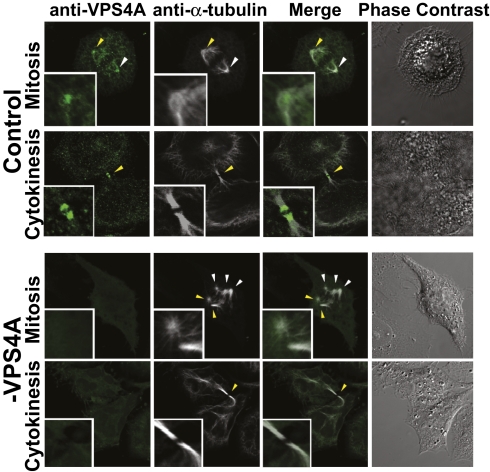
The severity of the centrosomal defects in cells lacking ESCRT-III/VPS4 proteins suggested that these proteins might function at centrosomes before concentrating at midbodies to facilitate abscission. To test this idea, we examined the localization of endogenous VPS4A (Fig. 5) and VPS4B (Fig. S6) at different stages of the cell cycle using immunofluorescence microscopy. During interphase, prophase, and metaphase, both VPS4 proteins were distributed in puncta throughout the cytoplasm (and sometimes also in the nucleus) but with visible concentrations at centrosomes and spindle poles (Fig. 5, row 1 and Fig. S6, row 1). Antibody specificity was confirmed by the loss of staining in cells depleted of VPS4A or VPS4B (Fig. 5 and Fig. S6, rows 3 and 4) under conditions where centrosome and spindle numbers were amplified (Fig. 5 and Fig. S6, row 3).
Fig. 5.
Localization of endogenous VPS4A during metaphase and cytokinesis. Representative images showing localization of endogenous VPS4A (green, anti-VPS4A, columns 1 and 3) and microtubules (white, anti-α-Tubulin, columns 2 and 3) during mitosis (row 1) or cytokinesis (row 2) in cells treated with a control siRNA. Phase contrast images are shown in column 4. Note that VPS4A concentrates at the spindle pole during metaphase (row 1) and at the midbody during cytokinesis (row 2). Images in rows 3 and 4 show cells depleted of VPS4A, demonstrating the specificity of antibody staining and showing multipolar spindles, characteristic of the depletion-dependent phenotype (row 3). Centrosomes and midbodies are highlighted with arrows, and magnified Insets correspond to regions highlighted by yellow arrows.
VPS4 protein localizations changed dramatically during cytokinesis, when both VPS4 proteins concentrated at the midbody and formed discrete bands on either side of the Flemming body (row 2). This midbody localization pattern is consistent with previous reports for VPS4 and other ESCRT proteins (3, 4, 21, 24). Moreover, we recently reported that the ESCRT-III-like protein IST1, which binds both ESCRT-III and VPS4 proteins, also localizes to centrosomes and midbodies in a cell cycle–dependent manner (22, 23). Thus these three late-acting ESCRT proteins are positioned to function directly at centrosomes and spindle poles during mitosis and at midbodies during abscission.
Discussion
Recent studies have shown that the ESCRT pathway functions in abscission and that this is an ancestral function that predates the divergence of archaea and eukaryotes (6, 7, 25). We now report that, in addition to their anticipated roles in abscission, most ESCRT-III/VPS4 proteins are also required for centrosome and spindle maintenance. The centrosomal defects in cells depleted of ESCRT-III/VPS4 proteins were often striking and highly penetrant. For example, approximately 80% of HeLa cells lacking VPS4B exhibited multiple centrosomes during the first mitosis after protein depletion (Fig. 3D) and most had five or more centrosomes.
Our study demonstrates a functional role for late-acting ESCRT-III/VPS4 proteins at mammalian centrosomes/spindles. However, a series of previous observations have suggested possible links between other ESCRT proteins and centrosomes and/or mitotic functions. (i) We and others have previously reported that a series of different ESCRT proteins can localize to centrosomes, microtubule organizing centers, and/or mitotic spindles, including ESCRT-I (4, 26, 27), ALIX (4), ESCRT-II (28), and IST1 (22). (ii) The TSG101 subunit of ESCRT-I was originally identified as a tumor susceptibility gene whose reduction induced mitotic abnormalities and aneuploidy (26). (iii) Cell division defects observed in Arabidopsis elc/tsg101 mutants were attributed to possible microtubule misregulation (29). (iv) The Drosophila homolog of CHMP5 was identified in a genome-wide siRNA screen for suppressors of multipolar mitoses (30). (v) S. pombe dot2/eap30/vps22 (ESCRT-II) mutants were reported to overamplify their spindle pole bodies (equivalent to mammalian centrosomes) during meiosis, although not during mitosis (31). (vi) Finally, a study focused on possible CHMP1A nuclear functions demonstrated that overexpression of this ESCRT-III protein caused cells to arrest with 4C DNA content (32). Together with the results from this study, these observations collectively suggest that the entire ESCRT pathway probably exerts centrosomal and possibly other functions that are conserved across fission yeast, plants, and mammals. The late-acting ESCRT-III/VPS4 proteins appear to play the most critical roles in centrosome maintenance, however, because their depletion induced stronger mitotic defects than did depletion of early-acting ESCRT factors such as TSG101 and ALIX (e.g., Figs. 1–3).
At least a subset of ESCRT pathway components appear to follow a recurring protein trafficking pathway in which proteins localize to centrosomes for most of the cell cycle, and then relocalize to the midbody to function during cytokinesis. This behavior is well documented for CEP55, which migrates from the centrosome to the midbody upon phosphorylation by Cdk1 and Erk2 (18, 33, 34). Cell cycle–dependent centrosomal and midbody localization patterns have now also been reported for IST1 (22) and VPS4 (this study) and for the CHMP4-binding protein, TTC19, which was recently show to translocate together with its binding partner, FYVE-CENT, from centrosomes to midbodies (35). Furthermore, our current study shows that ESCRT-III and VPS4 proteins are required for proper functioning of both centrosomes and midbodies, supporting the idea that the ESCRT pathway and its accessory factors help control multiple steps in the transition from mitosis to cytokinesis, perhaps with parallels to the well-characterized mitotic exit and septation initiation networks in yeast and plants (36). Localization of VPS4 to both centrosomes and midbodies seems to imply that the protein functions directly at both sites. At present, however, we cannot rule out indirect effects because VPS4 activity is required for normal endosomal trafficking (1, 5, 8), and endosomes participate in cleavage furrow ingression and cytokinesis (37).
Defining the mechanism(s) by which ESCRT-III/VPS4 proteins help maintain proper centrosome numbers is complicated because centrosome amplification and fragmentation can occur via several different mechanisms (reviewed in ref. 38) and because the centrosomal phenotypes we observed were quite complex (e.g., CHMP2A and VPS4B depletion altered the size and abundance of the pericentriolar matrix in different ways). One attractive possibility is that ESCRT-III/VPS4 proteins may help control centrosome numbers via interactions with machinery that regulates centriole disengagement and licenses centrosomes for duplication. In particular, the protein CC2D1A(Aki1) was recently shown to help control centrosome disengagement and duplication (39) and has also been identified as a potential CHMP4 binding protein in yeast two hybrid screens (40). Alternatively, it is possible that changes in centrosome numbers could reflect centrosome fragmentation and/or alterations in centrosome clustering. Indeed, the “single” centrosomes in CHMP2A-depleted cells sometimes appeared to be clustered (e.g., see the leftmost image in row 3 of Fig. S3), and the Drosophila homolog of CHMP5 (termed CG6259) was identified in a global siRNA screen for proteins involved in centrosomal clustering (30). In that case, the supernumerary centrosomes of Drosophila S2 cells reportedly formed multipolar spindles in the absence of CHMP5, suggesting that CHMP5 can help cluster multiple centrosomes into bipolar spindles.
It remains to be determined whether known ESCRT-III/VPS4 activities like ring formation, force generation, and/or membrane constriction and fission could function in centrosome duplication. One possibility is that polymeric centrosomal ESCRT-III/VPS4 assemblies could serve as scaffolds that recruit or modulate important activities of the centrosome. For example, the ESCRT-III protein CHMP1B binds specifically to the AAA ATPase spastin and thereby recruits this microtubule severing protein to the midbody (24). It is conceivable that analogous microtubule remodeling activities might be used at mitotic spindles, consistent with the suggestion that spindle microtubule misregulation is responsible for cell division defects in Arabidopsis cells lacking TSG101 (29).
Finally, it is intriguing that two ESCRT-III proteins, CHMP3 and CHMP4, were recently reported to localize to kinetochores in a global screen for proteins with possible mitotic functions (41). Thus, it appears possible that the ESCRT pathway is highly integrated into the biology of centrosomes, spindles, and possibly other mitotic machinery. This integration could allow ESCRT proteins to help coordinate three fundamental processes that must occur during cell division: chromosome segregation, centrosome duplication, and cytokinesis. We speculate that the evolution of centrosomal functions for the primordial ESCRT abscission machinery may have helped to ensure the ordered coupling of these essential steps of cell division.
Materials and Methods
SI Materials and Methods provides additional experimental detail. siRNA constructs, expression vectors, and antibodies are summarized in Tables S1–S4.
siRNA Treatments.
At least four siRNAs against each ESCRT-III protein were tested for efficacy, specificity, and lack of toxicity. HeLa cells were transfected twice with 20 nM siRNA (Lipofectamine RNAi MAX) at 24-h intervals and analyzed 24 h after the second transfection (except where noted).
Flow Cytometry Analyses of Cellular DNA Contents.
Following siRNA treatment, cells were treated with trypsin, collected, and resuspended in 0.3 mL propidium iodide (PI) solution (50 μg PI/mL in PBS, 0.1% Triton X100, 0.25 mg RNase/mL, 30 min, 4 °C). 10,000 PI-positive cells were counted with a FACScan fluorescence-activated cell sorter (FACScan; BD Bioscience), and peak volumes were analyzed using Modifit LT v2.0 software.
Immunofluorescence Imaging.
HeLa cells were seeded onto coverslips and transfected with siRNA at 30–50% confluence. Cells were then fixed with 3% paraformaldehyde in PBS or 100% methanol at −20 °C and stained with SYTOX Green (1 μM) and/or with antibodies against Pericentrin, α-Tubulin, γ-Tubulin, Aurora A, or VPS4 proteins. Confocal immunofluorescence images were acquired using Fluoview software on a FV300 IX81 Olympus microscope.
Time-Lapse Microscopy.
HeLa cells stably transduced with retroviruses that expressed YFP-α-Tubulin and H2B-mCherry were cultured in a 37 °C microscope chamber (Oko-Lab) with 5% CO2. 24 h after siRNA transfection, cells were imaged for YFP, mCherry, and by phase contrast on an Olympus IX81 microscope (UPLANSAPO 40× NA 0.90 objective, filters (Semrock) YFP (2427A), mCherry (TRITC-A). Images were acquired every 3 min with MetaMorph v7.02 software (Molecular Devices Corp.).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant AI051174 (to W.I.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005938107/-/DCSupplemental.
References
- 1.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 4.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghazi-Tabatabai S, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajorek M, et al. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbro M, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Doxsey SJ. Molecular links between centrosome and midbody. Mol Cell. 2005;20:170–172. doi: 10.1016/j.molcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 20.McCollum D, Gould KL. Timing is everything: Regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajorek M, et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agromayor M, et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlton JG, Martin-Serrano J. The ESCRT machinery: New functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, Li L, Cohen SN. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc Natl Acad Sci USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita E, et al. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langelier C, et al. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spitzer C, et al. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- 30.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y, Mancuso JJ, Uzawa S, Cronembold D, Cande WZ. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell. 2005;9:63–73. doi: 10.1016/j.devcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- 33.Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17:3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Garay I, Rustom A, Gerdes HH, Kutsche K. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics. 2006;87:243–253. doi: 10.1016/j.ygeno.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Sagona AP, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 36.Bedhomme M, Jouannic S, Champion A, Simanis V, Henry Y. Plants, MEN and SIN. Plant Physiol Biochem. 2008;46:1–10. doi: 10.1016/j.plaphy.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 37.van Ijzendoorn SC. Recycling endosomes. J Cell Sci. 2006;119:1679–1681. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
- 38.Srsen V, Merdes A. The centrosome and cell proliferation. Cell Div. 2006;1:26. doi: 10.1186/1747-1028-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol. 2009;187:607–614. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang HT, et al. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.