Abstract
Paramutation is the epigenetic transfer of information between alleles that leads to the heritable change of expression of one allele. Paramutation at the b1 locus in maize requires seven noncoding tandem repeat (b1TR) sequences located ∼100 kb upstream of the transcription start site of b1, and mutations in several genes required for paramutation implicate an RNA-mediated mechanism. The mediator of paramutation (mop1) gene, which encodes a protein closely related to RNA-dependent RNA polymerases, is absolutely required for paramutation. Herein, we investigate the potential function of mop1 and the siRNAs that are produced from the b1TR sequences. Production of siRNAs from the b1TR sequences depends on a functional mop1 gene, but transcription of the repeats is not dependent on mop1. Further nuclear transcription assays suggest that the b1TR sequences are likely transcribed predominantly by RNA polymerase II. To address whether production of b1TR-siRNAs correlated with paramutation, we examined siRNA production in alleles that cannot undergo paramutation. Alleles that cannot participate in paramutation also produce b1TR-siRNAs, suggesting that b1TR-siRNAs are not sufficient for paramutation in the tissues analyzed. However, when b1TR-siRNAs are produced from a transgene expressing a hairpin RNA, b1 paramutation can be recapitulated. We hypothesize that either the b1TR-siRNAs or the dsRNA template mediates the trans-communication between the alleles that establishes paramutation. In addition, we uncovered a role for mop1 in the biogenesis of a subset of microRNAs (miRNAs) and show that it functions at the level of production of the primary miRNA transcripts.
Keywords: interchromosomal, transfer, epigenetic, information, trans-generational
Paramutation is an interaction between alleles that leads to a heritable change of expression of one allele. One of the most intensively studied examples of paramutation is at the b1 locus in maize (1), which encodes a transcription factor that activates the purple anthocyanin biosynthetic pathway (2). There are two alleles involved in b1 paramutation, the highly transcribed and darkly pigmented B-I allele and the lightly pigmented B′ allele that has much lower transcription. When B-I and B′ are crossed together, paramutation always occurs: B-I is always changed into B′ (3).
Several genes required for paramutation have been identified through forward genetic screens. The mediator of paramutation (mop) genes (1, 4–6) and the required to maintain repression (rmr) genes (6, 8–10) have been isolated using the b1 and pl1 systems, respectively. To date, all characterized genes required for paramutation identified through forward genetic screens encode proteins that have been associated with siRNA biogenesis in other species (1). Recently, a protein that binds to the b1 tandem repeat (b1TR) sequences was identified, and expression of this protein as a transgene can establish a paramutagenic state in B-I (11). The mop1 gene, which is the focus of this study, encodes a protein with high similarity to RNA-dependent RNA polymerases (RDRs) and is the predicted ortholog of RDR2 in Arabidopsis thaliana (Arabidopsis) (4, 5, 7). Activity of mop1 is required for paramutation at the b1 locus and other loci (5, 6, 12), and it is required to maintain the silent B′ state (5). Similar to Arabidopsis RDR2, mop1 is required for the accumulation of the vast majority of 24-nt siRNAs (13–15), and it is involved in regulating the expression of a subset of transposable elements (TEs), transgenes, and several non-TE genes (7, 13–18).
The b1 gene is one of only two genes for which the sequences mediating paramutation have been defined (12, 19, 20). The key sequences required for b1 paramutation are seven b1TR units (each of the b1TR units is 853 bp in length) of noncoding DNA located ∼100 kb upstream of the b1 transcription start site (20, 21). This sequence is unique to this location within the maize genome, and both B-I and B′ carry seven tandem repeats, whereas alleles that do not undergo paramutation have a single copy of the repeat unit (20, 21). B-I and B′ are epialleles; that is, they have identical DNA sequences but show distinct patterns of DNA methylation and chromatin structure within the tandem repeats (20, 22–24). Generation of an allelic series with different numbers of repeats demonstrated that multiple repeats are required for paramutation (20).
The tandem repeats mediate enhancer activity that functions in cis to increase transcription from the b1 gene when in the B-I state (20), potentially through a long-distance looping mechanism because the tandem repeats interact with the transcription start site of the b1 gene differentially in B-I vs. B′ (23). The molecular nature of the genes required for paramutation strongly suggests that an RNA-dependent mechanism is critical for paramutation. Consistent with this idea, transcription assays have demonstrated that the repeats are transcribed in B-I and B′ as well as in alleles that do not undergo paramutation (4). Bidirectional transcription potentially generates dsRNA, the trigger molecule in a number of transcriptional and posttranscriptional gene regulation mechanisms that involve the processing of dsRNA into different classes of regulatory small RNAs (25, 26). Recent experiments have shown that siRNAs are produced from the b1TR sequences in B′ (27).
In this study, we used transcription assays, deep sequencing of small RNA libraries, and Northern blot analysis to investigate the potential steps in siRNA biogenesis where mop1 may function and whether production of b1TR-siRNAs correlates with paramutation. We also test whether DNA-dependent RNA polymerases are mediating transcription from the b1TR sequences and investigate alterations in microRNAs (miRNAs) in a mop1 mutant.
Results
The mop1-1 Mutation Does Not Reduce Transcription of the b1TR Sequences.
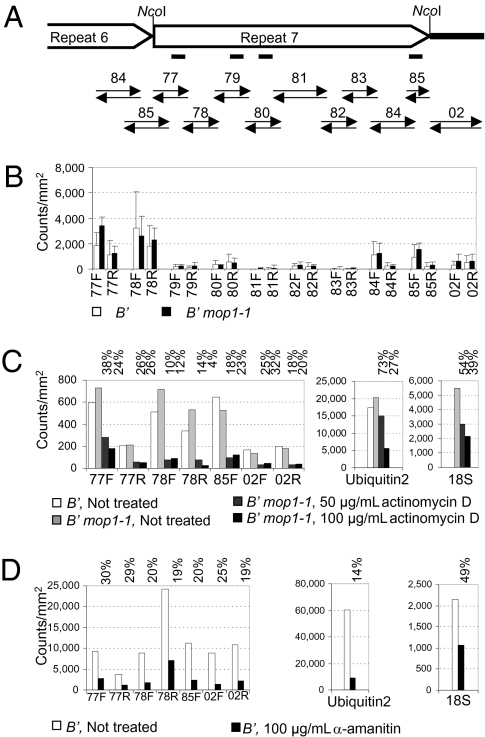
Previously, we showed that the b1TR sequences are transcribed on both strands using nuclear run-on experiments, which should monitor either DNA-templated or RNA-templated transcription in nuclei. To test whether the transcription we observe is carried out by mop1, a putative RDR, we performed nuclear run-on experiments using immature ears with B′ plants that were either wild type (WT) or homozygous for mop1-1. The results revealed that WT (B′) and B′ mop1-1 homozygous plants have very similar transcription levels from all the regions monitored (Fig. 1 A and B), demonstrating that the mop1-1 mutant did not reduce the b1TR transcription measured by nuclear run-on analysis.
Fig. 1.
Transcription from the b1TRs is not reduced in mop1-1 but is reduced by inhibitors of DNA-dependent RNA polymerases. (A) Map of the RNA probes used for nuclear run-on analyses. Open arrows depict parts of the sixth and seventh b1TRs required for paramutation. The black box indicates the sequence immediately downstream of the repeats. No transcription was detected upstream of the repeats (4); thus, that region was not tested in these experiments. Black paired arrows below the repeats indicate forward and reverse RNA probes used in relation to this drawing. The location of the four LNA probes used for Northern blot analysis (Fig. 2B) is indicated with four lines below repeat 7. (B) The b1 repeat transcription in B′ immature ears that are WT or homozygous for mop1-1. The results for the genotypes are indicated with open (WT) or solid (mop1-1) histograms for each forward (F) or reverse (R) probe, For each of the three biological replicates, raw counts were normalized to the Ubiquitin2 probe; SD is shown as bars within each histogram. (C) Transcription results after treatment with actinomycin D, a drug that inhibits all DNA-templated RNA synthesis. B′ and B′ mop1-1 samples not treated (no inhibitor) and B′ mop1-1 samples treated with 50 and 100 μg/mL actinomycin D are shown. The percent transcription from inhibitor-treated relative to no-inhibitor–treated control is indicated above each group of histograms. (D) Transcription results after treatment with α-amanitin, a drug that most strongly inhibits Pol-II transcription. The percent transcription from inhibitor-treated relative to no-inhibitor samples is indicated above each pair of histograms. For both C and D, young sheaths were used and transcription is shown for probes with the strongest signals in untreated samples. Transcription of control genes, Ubiquitin2 (transcribed by Pol-II) and 18S (transcribed by Pol-I), is shown separately to accommodate their high incorporation rates. The significance of the differences between control and treated samples was tested using an exact binomial probability calculation with the null hypothesis that drug treatments do not affect transcription and the alternative hypothesis that treatments reduce transcription (49). With either actinomycin treatment (P = 0.002) or α-amanitin treatment (P = 0.002), nine of nine probes demonstrated reduced transcription with drug treatment.
b1TR Sequences Are Transcribed by a DNA-Dependent RNA Polymerase.
To test whether DNA-dependent RNA polymerases might be contributing the majority of the transcription in nuclei, nuclear run-on reactions were performed with actinomycin D, an antibiotic that forms stable complexes with DNA, blocking all DNA-templated RNA synthesis. Results presented in Fig. 1C demonstrate that actinomycin D reduces transcription from the b1TR and downstream sequences to levels similar to those of control genes transcribed by RNA polymerase II (Pol II; Ubiquitin2) and Pol I (18S).
These results demonstrate that transcription from the b1TR and sequences immediately downstream is predominantly mediated by DNA-dependent RNA polymerases. Previous studies have shown that b1 repeat transcription is not altered in a mutation of mop2, which encodes the second largest subunit of a Pol IV/Pol V-related complex (27). This suggests that Pol II might be the major polymerase contributing to b1 repeat transcription. To test this hypothesis, nuclei were treated with α-amanitin, a small molecule that binds with high affinity within the Pol II active site, strongly inhibiting its transcription (Fig. 1D). At high α-amanitin concentrations, transcription from b1TR sequences was reduced to levels similar to that of the Pol II-transcribed Ubiqutin2 gene. As expected for α-amanitin (28, 29), transcription of the Pol I-transcribed 18S gene was less affected. These results suggest that the majority of the b1 repeat transcription measured in nuclei is mediated by Pol II.
Production of b1TR-siRNAs Depends on mop1 but also Occurs in Alleles That Cannot Participate in Paramutation.
Recent studies with mop2, which encodes the second largest subunit of Pol IV/PolV, demonstrated that the b1TR sequences generate siRNAs and that these are reduced in a mop2 mutant (27). To test if b1TR-siRNAs are also reduced in mop1, we examined deep sequencing data from small RNA libraries and performed Northern blots. We also used Northern blots to examine other genotypes to determine if b1TR-siRNA production correlated with the ability to undergo paramutation.
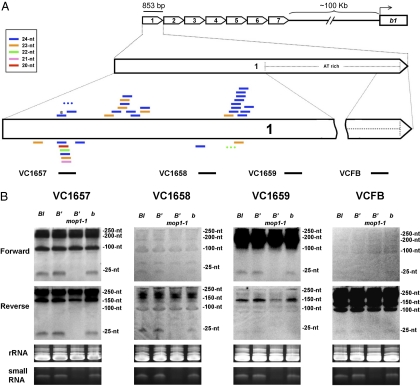
In the sequencing data from small RNA libraries from immature ears of WT (B′) and B′ mop1-1 homozygous mutant plants (one library from each genotype) (14), we identified a total of 35 unique small RNAs that perfectly and exclusively matched the b1TR over their entire length (Fig. 2A). A total of 33 and 3 b1TR-siRNAs were present in the WT and mop1-1 libraries, respectively, from a total of 13 million reads (14). These small RNAs had characteristics of siRNAs because they were predominantly 24 nt in size and examples were found that matched to both strands (Fig. 2A). The reduction in b1TR-siRNAs in mop1-1 was consistent with the global reduction in 24-nt siRNAs previously reported in the mop1-1 mutant (14). SNPs are present in four of the seven repeats, and they allowed us to map distinct siRNAs to more than one repeat (Table S1), indicating that multiple repeats are transcribed and processed into siRNAs in B′.
Fig. 2.
siRNAs associated with the b1TRs that mediate paramutation. (A) siRNAs matching the b1TRs (b1TR-siRNAs) from small RNA libraries. The seven tandem repeats found in B-I and B′ are depicted as open arrows relative to the b1 coding region (located ∼100 kb downstream of the repeats). Repeat 1 is expanded to show the location of the b1TR-siRNAs identified from deep sequencing [ref. 14; Gene Expression Omnibus (GEO) database accession nos. GSM306487 and GSM306488]. For simplicity, the mapping is shown only for the first repeat, with details of where each siRNA mapped summarized in Table S1. A dotted line inside the repeat unit indicates the AT-rich (72% AT-rich) region within the 3′-end of the tandem repeats. This region is not drawn to scale because no siRNAs were found in the libraries that matched the AT-rich region. The siRNAs are shown as bars above or below the repeat unit, representing the strand of DNA to which they match. Different colors represent different siRNA sizes, as indicated in the key. Two siRNAs observed only in the B′ mop1-1 libraries are shown as dotted bars. The siRNA observed in both libraries is labeled with an asterisk. All other siRNAs were observed only in the WT library. Locations of LNA probes VC1657, VC1658, VC1659, and VCFB (Materials and Methods) used for the Northern blot analysis (B) are shown below the repeat. (B) Northern blot analysis using 100 μg of small RNA-enriched fractions from immature ears. The genotypes are B-I and B′, each of which has seven tandem repeats and participates in paramutation (B-I is highly transcribed, and B′ has very low transcription) (50); B′ mop1-1, which does not participate in paramutation because of the mop1-1 mutation (5, 20); and the recessive b allele, which has a single copy of the repeat unit and does not participate in paramutation (21). Each b1TR probe used for hybridization is indicated above each panel. rRNA is shown as a loading control. Ethidium bromide staining of the small RNA-enriched fraction, which monitors global siRNA levels, is shown at the bottom of each panel.
Northern blots, combined with highly sensitive locked nucleic acid (LNA)-modified oligonucleotide probes, confirmed the deep sequencing results; the 24-nt siRNA signal, both globally and from the b1TR, was dramatically reduced in the B′ mop1-1 mutant relative to WT B′ (Fig. 2B). Also, as shown in Fig. 2B, the 24-nt b1TR-siRNA signal was detected from both strands with multiple probes in B-I, B′, and b. Despite the fact that all b1TR-siRNAs identified from the libraries mapped exclusively to the 5′-half of the repeats, we were able to detect faint levels of 24-nt siRNAs from the 3′-end of the repeats. These results demonstrated that most siRNAs produced from the b1TRs are dependent on mop1 function, which is absolutely required for paramutation. However, there is no correlation between the levels of b1TR-siRNAs detected in the blots and silencing (compare the active B-I vs. silent B′ alleles), nor is there a correlation with alleles that participate in paramutation (B-I and B′) vs. the b allele that does not.
In addition to the small RNA signal, the Northern blots revealed signals corresponding to larger RNA species ranging between 35 and 250 nt. These larger RNAs were consistently observed in a number of repetitions of these experiments with RNA from immature ears as well as from leaves and sheath tissues in all genotypes tested. These RNA species may represent transcripts synthesized directly as short molecules or intermediates from a stepwise processing of longer primary transcripts. The strong signal observed for these RNA populations suggests they may be highly transcribed, very stable, or both. The patterns were distinct with each probe, and the levels did not obviously correlate with the levels of the 24-nt b1TR-siRNAs observed with the same probe.
Transgenic Production of b1TR dsRNA and siRNAs Recapitulates Paramutation.
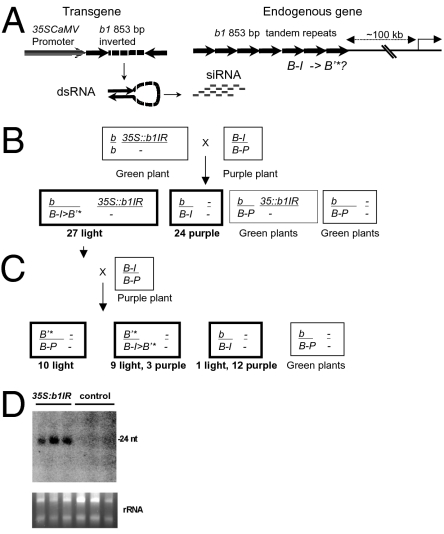
Northern blots revealed that both the active (B-I) and silenced (B′) alleles involved in paramutation (B-I and B′) and an allele (b) that does not participate in paramutation produce siRNAs, indicating that the b1TR-siRNAs are not sufficient for silencing in the tissues examined. Other experiments with a mop2 mutant, in which the b1TR-siRNAs are dramatically reduced but B′ silencing is not relieved, indicate that siRNAs are not required to maintain silencing (27). One possible explanation is that the b1TR-siRNAs are mediating the establishment of paramutation, which occurs very early in development, just after fertilization or in early embryogenesis (30). To test whether b1TR-siRNAs might be sufficient to establish paramutation, we used a transgenic approach to produce b1TR-siRNAs independent of b1 (Fig. 3A). Transgenic plants carrying the 35S:b1IR construct with a single b1TR unit cloned as an inverted repeat (IR) hairpin under the control of the strong constitutive 35SCaMV promoter (Fig. 3A) were crossed with a B-I/B-Peru (B-I/B-P) tester, and the progeny were screened for silencing of B-I (Fig. 3B). The B-I/B-P tester is particularly useful because B-I is less prone to spontaneous paramutation when heterozygous with an allele like B-P or b that cannot participate in paramutation. In addition, the purple seed (aleurone) pigmentation of B-P provides a useful marker for the allele, facilitating the crosses (5, 31, 32). The 35S:b1IR transgene was able to establish paramutation efficiently, because 100% of the B-I alleles were silenced in plants that inherited the transgene, a state indicated as B′* (Fig. 3B and Fig. S1). To determine if the silencing could be maintained in the absence of the transgene, B′* plants were crossed with the B-I/B-P tester and the progeny were examined (Fig. 3C). The newly developed B′* state was fully heritable in the absence of the transgene, and, importantly, the B′* state was able to paramutate naive B-I alleles (Fig. 3C), although the paramutagenic activity was reduced relative to B′, which is always 100% paramutagenic. Spontaneous paramutation of B-I occurred at a much lower frequency as determined by the b/B-I progeny (Fig. 3C). These results indicate that the key features of paramutation can be recapitulated by transcription of a hairpin of the b1 repeat unit that generates siRNAs (Fig. 3D).
Fig. 3.
Transgene-generated b1TR-siRNAs recapitulate key features of paramutation. (A) Diagram of the 35S:b1IR construct harboring a single tandem repeat unit (arrows) cloned as an inverted repeat that produces b1TR-siRNAs. (B and C) Crossing scheme used to test paramutation effects of 35S:b1IR on B-I. Progeny classes are diagrammed with the informative classes in bold, and the results are summarized below for each progeny class. (B) Test for establishment of paramutation. All B-I plants that inherited the transgene were silenced, as evidenced by reduced pigment levels. The phenotypes are illustrated in Fig. S1. We indicate this silent state as B′*. No spontaneous paramutation was observed in the nontransgenic B-I siblings. (C) Test for heritability of the B′* state and its ability to induce paramutation in the absence of the transgene. Only nontransgenic siblings are shown. (D) Northern blot to detect b1TR-siRNAs in three each 35S:b1IR transgenic and control nontransgenic siblings. Fifty micrograms of the small RNA-enriched fraction was probed with VC1658 (forward). Half of the amount of RNA and a 10-fold reduction of exposure time (12 h vs. 5 d) were used relative to results in Fig. 2B. rRNA is shown as a loading control.
In Arabidopsis, silencing induced by IR constructs is rarely heritable in the absence of the inducer transgene and is not paramutagenic. One possibility is that IR transgenes are more prone to paramutation in maize than in Arabidopsis. To test this hypothesis, we examined silencing of the anthocyaninless-1 gene, a1, by an a1 promoter IR (a1pIR). As diagrammed in Fig. S2, the 35S:a1pIR efficiently silenced the A1 allele, but the silencing was not paramutagenic in subsequent generations in the absence of the transgene. A similar result was observed with the pIR-induced silencing of a pollen-specific promoter, MS45 (33). Thus, we hypothesize that the ability of the 35S:b1IR to establish a heritable paramutagenic state is a property of the b1 repeats rather than a difference between the silencing machinery in maize and Arabidopsis.
Biogenesis of Several MiRNA Families Is Altered in the mop1-1 Mutant.
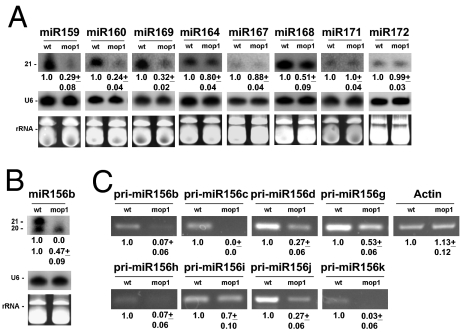
Mutations in mop1 often show developmental phenotypes, including delayed flowering, reduced height, feminized tassels (tasselseed phenotype), and reduced seed set (5). MiRNAs play key roles during plant development and mutations, altering the biogenesis or action of miRNAs result in numerous developmental defects (34). Based on the observation that raw abundances for perfect matches to known miRNAs were different in WT vs. mop1-1 small RNA libraries (14) and on a report showing that levels of miR156 were down-regulated in feminized tassels of mop1-1 (35), we hypothesized that other developmental phenotypes observed in the mop1-1 mutant might be related to alterations in levels of additional miRNAs. To investigate this, we examined the levels of nine highly conserved miRNAs (36) in mop1-1 homozygous and WT plants by RNA blot analysis. Results were highly reproducible in three independent experiments, with a representative experiment shown in Fig. 4 A and B. The levels of five of the miRNAs were reduced by 2- to 5-fold in mop1-1 relative to WT (Fig. 4 A and B), whereas miR164 exhibited a slight (1.25-fold) but reproducible reduction (Fig. 4A). In Arabidopsis, two miRNA species (20 and 21 nt) are observed for miR156 from shoot apices (37). In maize, a single miRNA species has been reported for miR156 (35, 38), but its specific size was not reported. We found that similar to Arabidopsis, two size classes of miR156 can be observed in maize. The 21-nt species is not detected in mop1-1, whereas the 20-nt species is reduced 2-fold relative to WT (Fig. 4B). To explore the step at which miRNA biogenesis might be compromised in mop1-1 mutants, we examined the levels of miR156 primary transcripts (pri-miRNAs) in WT and mop1-1 mutants. In maize, the miR156 family contains 11 members (a–k) (39) mapping to different genomic regions. We found EST supporting evidence for 8 of the 11 pri-miR156 members and designed a set of primers outside the precursor region (pre-miRNA) specifically to amplify each pri-miRNA (Table S2). Because there was no EST evidence for pri-miR156e and pri-miR156j, we used genomic sequences to design the primers. The levels of each pri-miR156 family member were determined using RT-PCR, with reduction observed for 8 pri-miR156 family members in mop1-1 relative to WT (1.25- to 10-fold; Fig. 4C). We could not detect pri-miR156a, pri-miR156e, or pri-miR156f in immature ears, despite increasing RNA concentration and PCR cycles. These results show that the levels of several miRNAs are affected in mop1-1 and, in the case of the miR156 family, that the reduction in miRNA levels correlates with a reduction in the levels of their corresponding pri-miRNAs.
Fig. 4.
The mop1-1 mutation influences biogenesis of certain miRNAs. (A and B) Levels of each indicated miRNA family were monitored by Northern blot analysis using 5′-end–labeled DNA oligonucleotides complementary to the mature miRNA and 20 μg of the small RNA fraction from B′ immature ears that were WT (wt) or homozygous for the mop1-1 (mop1) mutation. Levels of rRNA and U6 are shown as loading controls. The numbers indicate the mean abundance and SD of miRNAs in mop1-1 relative to the WT control after scanning and normalization for loading for three biological replicates. (C) Levels of pri-miRNAs were reduced in the mop1-1 mutant. RT-PCR analysis of pri-miR156 levels in WT and mop1-1. Total RNA was extracted from immature ears. Analysis of actin served as a loading control showing that equivalent amounts of RNA were tested in all reactions. The numbers indicate the mean abundance and SD of pri-miRNAs in mop1-1 relative to the WT control from three biological replicates.
Discussion
Here, we show that Pol II is likely to be the DNA polymerase contributing most significantly to the b1TR transcription, consistent with a role for mop1 functioning downstream of b1TR transcription. We found that b1TR-siRNAs are dramatically reduced in mop1-1 mutants, as are 24-nt siRNAs more globally, consistent with a role of mop1 in siRNA biogenesis (14). The previously described effects of mop1 mutants on transgene (17) and transposon (7, 16, 18) silencing are most easily explained by the global requirement of mop1 for siRNA biogenesis (14). Models for RNA-directed DNA methylation (RdDM) in Arabidopsis hypothesize that RDR2 (the likely ortholog of MOP1) is required to amplify the dsRNA signal (40). Studies in Tetrahymena suggest RDRs can also interact directly with Dicer proteins (41) and that this interaction is required for siRNA biogenesis. Our observations that mop1 is required to maintain the silent state (5) but that b1TR-siRNAs are not required to maintain silencing of B′ (27) suggest that mop1 may have a function in maintaining B′ silencing beyond generating b1TR-siRNAs.
Despite deep sequencing in WT stocks (6 million reads), not all b1TR-siRNAs were identified by this approach, as evidenced by the detection of 24-nt signals in regions where no siRNAs from the libraries mapped. This could represent the very low abundance of these siRNAs (every siRNA sequenced was detected only once), such that the level of sequencing was not saturating. It is also possible that there were library or sequencing biases. Larger RNA species are also observed on the blots, which may reflect processing intermediates, separate transcripts, or a combination of both. These larger RNAs were similarly produced in all genotypes, including mop1-1, and are thus unlikely to be directly involved in paramutation.
The presence of b1TR-siRNAs does not correlate with paramutation in the tissues examined, which include immature ears, sheaths, and leaves. The transcription of and siRNA presence in the single-repeat unit b allele indicate that the presence of siRNAs, at least in the tissues examined, is not sufficient for establishing silencing. The observation that the tandem repeats in the highly expressed B-I allele are transcribed at similar levels and have similar b1TR-siRNA levels as the silenced B′ allele raises the question as to how B-I remains active. One possibility is that the distinct chromatin structure of B-I relative to B′ provides “immunity” from silencing, potentially through a distinct nuclear localization or the binding of specific proteins. The initial silencing event happens early in development (30), and our studies of mop2 (27) indicate that b1TR-siRNAs are not required to maintain silencing in tissues such as immature ears and leaves, in which it is possible to examine siRNAs and transcription of the b1TR sequences. Thus, one possibility is that if we could examine siRNAs and transcription when paramutation is established, we might see a correlation between siRNAs or a larger RNA and establishment of silencing. There is precedence for a role of larger RNAs in silencing in Arabidopsis in that the heterochromatic silencing of certain 5S ribosomal DNA tandem repeats requires Pol V and most likely longer RNAs but not other RdDM components (42–44).
Despite the lack of correlation between b1TR-siRNAs and maintenance of silencing, our transgene results suggest that either b1TR-siRNAs or the hairpin template mediates the trans-communication that establishes silencing. These results are similar to transgene-induced silencing of FWA in Arabidopsis: The siRNA pathway is involved in establishing but not in maintaining silencing (45). A difference between FWA silencing in Arabidopsis and our observations is that the silenced B′* state is heritable and paramutagenic when the inducing transgene is segregated away. Although the B′* state established by the transgene is not as paramutagenic as “natural” paramutation, it is similar to that observed for B′ alleles with fewer repeats (20), the state established by the binding of the CBBP protein (11), and most other paramutation systems, which are often not 100% efficient (46). Thus, the generation of a hairpin dsRNA or siRNAs from the b1TR sequence can establish a heritable chromatin state in trans at the endogenous locus. Other RdDM constructs in maize, such as the a1 promoter described in this study, are not paramutagenic, similar to Arabidopsis RdDM, suggesting that there is something “special” about the b1TR sequences that enables them to mediate paramutation. Two possibilities that are not mutually exclusive are chromatin structure and self-generation of transcripts or siRNAs. Thus, although an RNA-based silencing mechanism is conserved among plant species, there are likely to be unique players that mediate the distinct characteristics of paramutation.
The reduction in miRNA levels observed for a subset of conserved miRNA families in the mop1-1 mutant indicates that mop1 also affects miRNA biogenesis. Further experiments are required to determine if mop1 is a multifunctional gene involved in several molecular processes, including siRNA biogenesis and miRNA biogenesis, or whether the observed phenotypes represent aspects of the same function. The observation of reduced levels of pri-miR156 transcripts is consistent with mop1 functioning at an early step, potentially through siRNA regulation of pri-miRNA expression. This finding is consistent with recent transcriptome analysis showing altered expression of a number of Pol II-transcribed genes (13). Independent of whether the mop1-1 effects on miRNA biogenesis are direct or indirect, the reduction in several miRNAs important for plant development provides an explanation for the developmental phenotypes frequently observed in mop1-1 mutants.
Materials and Methods
Plant Material.
Immature ears (3–5 cm in length) were harvested from plants grown outdoors in Tucson, AZ, between 68 and 75 d after germination.
RNA Analysis.
Nuclear run-on and Northern blot analyses were performed as described (27). LNA probes were synthesized by Sigma–Proligo. LNA-modified bases are preceded by a plus (+) sign: VC1657F (gCTg+CAgCCT+gTgCA+ggCTTAg+CCTCA+gCCTAT+CgTgg+CCCgA+CA), VC1657R (TgTC+gggCC+ACgATAg+gCTgA+ggCTAA+gCCTgC+ACAgg+CTgC+AgC), VC1658F (TgAA+CATCTT+gTCCA+gTTAAAT+CACTgg+ACACC+gTgAC+AgCC+ACA), VC1658R (TgT+ggCTgT+CACg+gTgTC+CAgT+gATTTAA+CTggA+CAAgAT+gTTCA), VC1659F (CAg+CATCAC+CCTCACA+CATgg+TCCg+CATgg+CTACg+CgTAT+CTATg), VC1659R (CATA+gATAC+gCgTAg+CCATg+CggAC+CATg+TgTgAg+ggTgATg+CTg), VCFBF (G+AGGGCTC+CAAGAGG+TCTATAA+AAATTTG+GTGTTTA+AAAATTC+ATG), and VCFBR (CA+TGAATTT+TTAAACA+CCAAATT+TTTATAG+ACCTCTT+GGAGCCC+TC). For the three miRNA Northern blot replicates, 20 μg of the small RNA-enriched fraction was loaded per lane and 5′-end–labeled oligonucleotides complementary to the mature miRNA were used as probes. For the replicate RT-PCR analysis of pri-miRNAs, cDNA was synthesized from 6 μg of total RNA using reverse transcriptase (Invitrogen) and oligo(dT). Specific primers for all miR156 family members are identified in Table S2. Hybridizations and image processing were performed using QuantityOne software (BioRad) and ImageJ software (National Center for Biotechnology Information).
Transgenic Construct, Plant Transformation, and Transgenic Plants.
An 853-bp b1 repeat unit was PCR-amplified using primers VC977A (ggTTggTTgCgATCgCCCTAggCCATgggTTTgCTgCATCCTTg with AvrII-SgfI tail) and VC977B (ggACTAgTggCgCgCCCCAAgTATTCggTATAAAAgTTgT with AscI-SpeI tail) and cloned in the pMCG161 vector to produce an inverted repeat construct similar to that described by McGinnis et al. (47). The resulting 35S:b1IR plasmid was introduced in the HiII genetic background by biolistic transformation, as described by Frame et al. (48). HiII carries a b allele that specifies no anthocyanin pigment and is neutral to paramutation. The primary transgenic line was crossed with a tester carrying B-I and B-P alleles. The B-P allele does not participate in paramutation and does not produce plant anthocyanin pigment, and its purple seed color allows preplanting identification of seeds carrying B-P. Resulting progeny were grown to assay the effect of 35S:b1IR transgene on B-I expression (Fig. 3B) using herbicide resistance to identify transgenic plants. In the paramutagenicity test (Fig. 3C), Southern blot analysis was used to distinguish between b/B-I→B′;−/− and B′*/B-I;−/− plants, taking advantage of restriction polymorphisms between b and B-I/B′.
Supplementary Material
Acknowledgments
We thank Cheng Lu for making the small RNA libraries under National Science Foundation Award 0638525 (to P.J.G. and B.C.M.). We also acknowledge the contributions of Devika Unnithan, Virginia O'Connell, and Vishwas Seshadri in generating constructs for the inverted repeat transgenes and Dean Billheimer for advice on statistical analyses. This work was supported primarily by National Institutes of Health Grant DPIOD575 (to V.L.C), with additional support from the National Science Foundation Plant Genome Research Program (PGRP) Award 0701745 (to B.C.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSM306487 and GSM306488).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007972107/-/DCSupplemental.
References
- 1.Arteaga-Vazquez MA, Chandler VL. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr Opin Genet Dev. 2010;20:156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goff SA, et al. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 1990;9:2517–2522. doi: 10.1002/j.1460-2075.1990.tb07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coe EH. A regular and continuing conversion-type phenomenon at the B locus in maize. Proc Natl Acad Sci USA. 1959;45:828–832. doi: 10.1073/pnas.45.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 5.Dorweiler JE, et al. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollick JB, Chandler VL. Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics. 2001;157:369–378. doi: 10.1093/genetics/157.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodhouse MR, Freeling M, Lisch D. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 2006;4:e339. doi: 10.1371/journal.pbio.0040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erhard KF, Jr, et al. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- 9.Hale CJ, Erhard KF, Jr, Lisch D, Hollick JB. Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet. 2009;5:e1000598. doi: 10.1371/journal.pgen.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stonaker JL, Lim JP, Erhard KF, Jr, Hollick JB. Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 2009;5:e1000706. doi: 10.1371/journal.pgen.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brzeska K, Brzeski J, Smith J. Chandler VL. Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proc Natl Acad Sci USA. 2010;107:5516–5521. doi: 10.1073/pnas.1001576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidorenko L, Chandler V. RNA-dependent RNA polymerase is required for enhancer-mediated transcriptional silencing associated with paramutation at the maize p1 gene. Genetics. 2008;180:1983–1993. doi: 10.1534/genetics.108.095281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia Y, et al. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5:e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nobuta K, et al. Distinct size distribution of endogenous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci USA. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lisch D, Carey CC, Dorweiler JE, Chandler VL. A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc Natl Acad Sci USA. 2002;99:6130–6135. doi: 10.1073/pnas.052152199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGinnis KM, Springer C, Lin Y, Carey CC, Chandler V. Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics. 2006;173:1637–1647. doi: 10.1534/genetics.106.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodhouse MR, Freeling M, Lisch D. The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics. 2006;172:579–592. doi: 10.1534/genetics.105.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidorenko LV, Peterson T. Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell. 2001;13:319–335. doi: 10.1105/tpc.13.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stam M, et al. The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandler VL, Eggleston WB, Dorweiler JE. Paramutation in maize. Plant Mol Biol. 2000;43:121–145. doi: 10.1023/a:1006499808317. [DOI] [PubMed] [Google Scholar]
- 23.Louwers M, et al. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell. 2009;21:832–842. doi: 10.1105/tpc.108.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haring M, et al. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04245.x. 10.1111/j.1365-313X.2010.04245.x. [DOI] [PubMed] [Google Scholar]
- 25.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 27.Sidorenko L, et al. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 2009;5:e1000725. doi: 10.1371/journal.pgen.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jendrisak J. The use of alpha-amanitin to inhibit in vivo RNA synthesis and germination in wheat embryos. J Biol Chem. 1980;255:8529–8533. [PubMed] [Google Scholar]
- 29.Strain GC, Mullinix KP, Bogorad L. RNA polymerases of maize: Nuclear RNA polymerases. Proc Natl Acad Sci USA. 1971;68:2647–2651. doi: 10.1073/pnas.68.11.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coe EH. The properties, origin, and mechanism of conversion-type inheritance at the B locus in maize. Genetics. 1966;53:1035–1063. doi: 10.1093/genetics/53.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson GI, Kubo KM, Shroyer T, Chandler VL. Sequences required for paramutation of the maize b gene map to a region containing the promoter and upstream sequences. Genetics. 1995;140:1389–1406. doi: 10.1093/genetics/140.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radicella JP, Brown D, Tolar LA, Chandler VL. Allelic diversity of the maize B regulatory gene: Different leader and promoter sequences of two B alleles determine distinct tissue specificities of anthocyanin production. Genes Dev. 1992;6:2152–2164. doi: 10.1101/gad.6.11.2152. [DOI] [PubMed] [Google Scholar]
- 33.Cigan AM, Unger-Wallace E, Haug-Collet K. Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J. 2005;43:929–940. doi: 10.1111/j.1365-313X.2005.02492.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hultquist JF, Dorweiler JE. Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta. 2008;229:99–113. doi: 10.1007/s00425-008-0813-2. [DOI] [PubMed] [Google Scholar]
- 36.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 42.Douet J, Tutois S, Tourmente S. A Pol V-mediated silencing, independent of RNA-directed DNA methylation, applies to 5S rDNA. PLoS Genet. 2009;5:e1000690. doi: 10.1371/journal.pgen.1000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pontes O, Costa-Nunes P, Vithayathil P, Pikaard CS. RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway. Mol Plant. 2009;2:700–710. doi: 10.1093/mp/ssp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daxinger L, et al. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 2009;28:48–57. doi: 10.1038/emboj.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandler VL, Stam M. Chromatin conversations: Mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 47.McGinnis K, et al. Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 2005;392:1–24. doi: 10.1016/S0076-6879(04)92001-0. [DOI] [PubMed] [Google Scholar]
- 48.Frame BR, et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 2002;129:13–22. doi: 10.1104/pp.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snedecor GW, Cochran WG. Statistical Methods. 7th Ed. Ames, Iowa: Iowa State Univ Press; 1980. p. 112. [Google Scholar]
- 50.Patterson GI, Thorpe CJ, Chandler VL. Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics. 1993;135:881–894. doi: 10.1093/genetics/135.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.