The discovery in 1991 by Fuerst and Webb that the chromosome of the bacterium Gemmata obscuriglobus (a member of Planctomycetales) is surrounded by a double membrane, mimicking a eukaryotic nucleus, challenged the traditional prokaryote/eukaryote classification of living organisms based on cell structure (1). Since then, such an unexpected observation has puzzled evolutionists, leading to different reactions. Most considered G. obscuriglobus a curiosity of the bacterial world. Until recently, this view has been supported by the absence of obvious eukaryotic protein signatures in the genomes of Planctomycetales and related bacteria (2). For a few others, the “nucleus” of G. obscuriglobus should, one way or another, testify as some sort of evolutionary link between Planctomycetes and modern eukaryotes (3). Two landmark papers now seem to strengthen this opinion.
In PNAS, Fuerst and colleagues show that G. obscuriglobus can perform a function previously thought to be restricted to eukaryotes: endocytosis (4). Whereas in traditional bacteria, such as Escherichia coli, the capture of proteins involves their transport through the cell membrane via pore-like structures, the data obtained by Fuerst and colleagues strongly suggest that, as in eukaryotes, proteins can be internalized by G. obscuriglobus via the formation of endosome-like structures. They show that imported proteins (GFP, Ig, or streptavidin) accumulate and are later degraded in the ribosome-free region of the G. obscuriglobus cytoplasm (the so-called paryphoplasm). Ultracentrifugation of a cell extract shows that internalized proteins copurify with a cell fraction containing both membrane debris and vesicles. These vesicles are visible within the paryphoplasm by electron microscopy, and some of them appear to be formed by invagination of the cytoplasmic membrane. Immunogold labeling revealed the presence of internalized GFP lining these vesicles and membrane invagination, suggesting that vesicle formation is indeed linked to protein uptake. Although genetic experiments lack definitive proof (there are presently no available genetic tools for Planctomycetales), these data strongly suggest that this is indeed a demonstration of endocytosis in a bacterium.
Of course, endocytosis by G. obscuriglobus could be considered another curiosity of this bacterium with no link to “true” endocytosis, a hallmark of eukaryotes. However, a recent work, by Devos and colleagues from the European Molecular Biology Laboratory (Heidelberg), nicely complements the results of Fuerst and colleagues (5). Using sensitive structure-based in silico search approaches, these authors screened all available archaeal and bacterial proteomes for the presence of structural analogs of eukaryotic membrane coat (MC) proteins. In eukaryotes, MC proteins are involved in both vesicle-trafficking systems and in the formation of the nuclear pore. Strikingly, Devos and colleagues only detected eukaryotic MC-like proteins (from 5 to 16 copies) in the proteomes of G. obscuriglobus and several related bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae (PVC) superphylum. This superphylum groups Planctomycetes, Verrucomicrobia, Chlamydiae, Lentispherae, and Poribacteria based on rRNA phylogeny (2). Importantly, it includes all known bacteria with cell plans involving compartmentalization of the cytoplasm by an intracytoplasmic membrane (ICM). The double membrane of the G. obscuriglobus nucleus is formed by invagination of this ICM, much like the double membrane of the eukaryotic nucleus is formed by invagination of the endoplasmic reticulum (ER) (3). Significantly, Devos and colleagues detected MC proteins in compartmentalized PVC bacteria only. In particular, they did not find any MC proteins in Chlamydiae, which in fact do not have an ICM. Using antibodies prepared by Devos and colleagues against one of the eight MC proteins of G. obscuriglobus, gp4978, Fuerst and colleagues found that this protein colocalizes with vesicles containing proteins imported by endocytosis (4). They also obtained images showing that gp4978 associates with the cytoplasmic membrane in the first stage of membrane invagination. These data strongly suggest that gp4978 is involved in the membrane-remodeling process that triggers endocytosis. It is tempting to think that, more generally, the multiple MC proteins present in several PVC bacteria are required to manipulate their complex endomembrane systems. In G. obscuriglobus, the nuclear double membrane segregates the nucleoid from a large fraction of ribosomes (3). This supposes the existence of nuclear pores allowing the transport of messenger RNA from the nucleus to these ribosomes. By analogy with nucleoporin (one of the eukaryotic MC proteins), it is possible that some of the MC proteins encoded by G. obscuriglobus could be used to build these pores.
The analogies between the membrane-trafficking systems of PVC bacteria and Eukarya, both at the cytological and protein structure levels, are thus strikingly evident. It now seems impossible to ignore these data when discussing the origin of the eukaryotic nucleus. A major objective of future research should now be to determine whether bacterial MC proteins are only structural analogs of eukaryotic ones (a case of convergent evolution) or whether instead they are homologous. This cannot be tested through sequence similarity (even between eukaryotic MC proteins), because these proteins evolve too rapidly at the sequence level. However, MC proteins have retained their core architecture during evolution, which is formed by a unique combination of two protein domains composed of long stretches of β-strands in the N terminus and α-helices in the C terminus. The respective lengths of the two domains, as well as the respective numbers of β-strands and α-helices in each domain, are strikingly similar in PVC bacteria and Eukarya (5). In their paper, Devos and colleagues thus argue in favor of homology instead of convergence. The final proof will probably require solving the structures of several MCs from Bacteria and Eukarya and reconstructing (if possible) the structure of the ancestral MC protein in each domain. Preliminary results have nevertheless already provided important information, suggesting in fact an ancient origin of these proteins in both PVC bacteria and in Eukarya, because several copies of MC proteins were probably already present in their respective last common ancestors (5).
If we assume that bacterial and eukaryotic MC proteins have a common origin, how can this information be fitted with current theories on the origin of eukaryotes? Three scenarios can be imagined. First, in models suggesting that eukaryotes originated from the association of a bacterium and an archaeon (symbiotic hypotheses), it appears reasonable now, in light of the data by Fuerst and Devos and colleagues, to suggest a compartmentalized PVC bacterium as the host. The archaeal symbiont would have lost its cell membrane, and its chromosome would have been surrounded by a nuclear membrane coming from the ICM of the bacterial host. In this scenario, it is tempting to suggest that the archaeal symbiont belonged to the newly recognized phylum Thaumarchaeota, because these Archaea are mostly mesophiles and exhibit eukaryotic features that are missing in other archaeal phyla (6). However, symbiotic hypotheses for the origin of Eukarya remain difficult to understand in terms of known biological mechanisms. For example, they imply a specific association between a bacterium and an archaeon for which there are no examples in nature, and assume a very unlikely process where all of the genes of the bacterial host coding for informational proteins would have been replaced by those of the archaeal symbiont. Alternatively, under the evolutionary scenarios where modern Eukarya originated from an ancestral protoeukaryotic lineage [urkaryote, sensu
Endocytosis of proteins might well be an ancient trait.
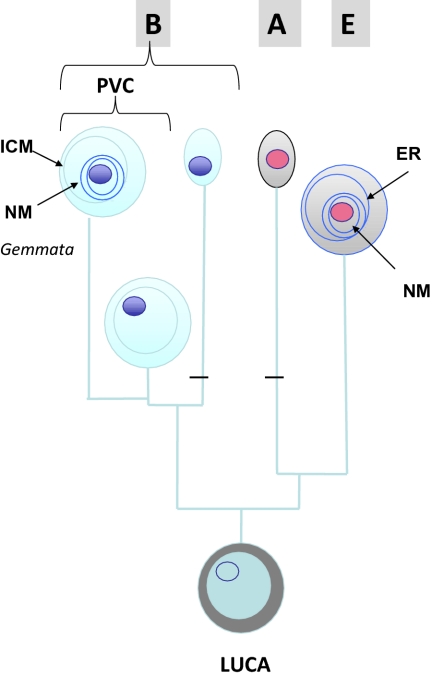
Woese and Fox (7)], two hypotheses can be proposed. In the first one, MC proteins appeared in the lineage leading to PVC bacteria and were subsequently transferred to the lineage leading to Eukarya, or the reverse. In the second one, MC proteins would have already been present in the last universal common ancestor (LUCA) and were inherited in Eukarya and PVC bacteria, whereas they were lost in all other bacterial phyla and in Archaea. The second hypothesis (favored by Fuerst and Webb) is supported by the fact that the loss of MC proteins appears to have occurred recurrently in the PVC superphylum, whereas lateral gene transfer of MC proteins to other bacteria or Eukarya (and vice versa) has not been observed. Moreover, Brochier and Philippe reported several years ago that Planctomycetales are the earliest diverging bacterial lineage in a 16S rRNA phylogeny based on the slowest evolving positions (8). If this is so, only two losses of MC proteins (one in Bacteria after the divergence of the PVC lineage and another in the branch leading to Archaea) would be necessary to explain the present distribution of MC proteins in the three domains of life (Fig. 1).
Fig. 1.
A scenario for the origin of modern compartmentalized cells (Eukarya and PVC bacteria) from a compartmentalized LUCA. A, Archaea; B, Bacteria; E, Eukarya. The cytoplasm and chromosomes of Archaea and Eukarya are shown with the same color in reference to the high similarity of these two domains for informational proteins and several membrane-bound processes (e.g., ATP synthesis). For the sake of simplicity, eukaryotic organelles have not been indicated. Also for simplicity, the ICM of PVC bacteria and the ER of Eukarya have been schematically drawn as intracellular circles. The horizontal black bars indicate loss of MC proteins. NM, nuclear membrane.
If the LUCA already harbored MC proteins, it was probably compartmentalized. This idea can appear odd to many biologists who use to think of the LUCA and all its contemporaries as very primitive entities. However, the formation of vesicles and membrane manipulation may be very ancient features of life. For instance, eukaryotic RNA viruses encode proteins that can manipulate the host ER to produce viral factories surrounded by membranes (9), suggesting, by analogy, that even ancient cells with RNA genomes could have had such capacity and therefore already be compartmentalized. If MC proteins were already around at the time of the LUCA, the ancient biosphere might have been more diversified than usually suspected, with various lineages of compartmentalized cells, some of them with nuclei (which could be named synkaryotes) and others without (akaryotes), thriving in various environments. Endocytosis of proteins might well be an ancient trait that was lost in bacteria with rigid cell walls. Although PVC bacteria are bona fide members of the bacterial domain, they might therefore have conserved some ancestral features in terms of cellular structure and function that open up new avenues of thinking about the nature of our cellular ancestors. Further exploration of microbial diversity will most likely bring surprises. Other compartmentalized cells could in fact exist among the vast numbers of still uncultivated archaeal and bacterial lineages. In any case, the results of Fuerst and Devos and colleagues remind us that we should definitely stop thinking of bacteria in terms of simple “lower” organisms.
Footnotes
The authors declare no conflict of interest.
See companion article on page 12883.
References
- 1.Fuerst JA, Webb RI. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA. 1991;88:8184–8188. doi: 10.1073/pnas.88.18.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Fuerst JA. Intracellular compartmentation in Planctomycetes. Annu Rev Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 4.Lonhienne TGA, et al. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA. 2010;107:12883–12888. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santarella-Mellwig R, et al. The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 2010;8:e10000281. doi: 10.1371/journal.pbio.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brochier-Armanet C, Gribaldo S, Forterre P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol Direct. 2008;3:54. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brochier C, Philippe H. Phylogeny: A non-hyperthermophilic ancestor for bacteria. Nature. 2002;417:244. doi: 10.1038/417244a. [DOI] [PubMed] [Google Scholar]
- 9.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]