Abstract
Cellulose synthase-interactive protein 1 (CSI1) was identified in a two-hybrid screen for proteins that interact with cellulose synthase (CESA) isoforms involved in primary plant cell wall synthesis. CSI1 encodes a 2,150-amino acid protein that contains 10 predicted Armadillo repeats and a C2 domain. Mutations in CSI1 cause defective cell elongation in hypocotyls and roots and reduce cellulose content. CSI1 is associated with CESA complexes, and csi1 mutants affect the distribution and movement of CESA complexes in the plasma membrane.
Keywords: cell walls, CESA complexes, cell expansion, microtubules, polysaccharides
Cellulose is synthesized at the plasma membrane by hexameric protein complexes with a diameter of 25–30 nm when observed by freeze-fracture electron microscopy in algae, moss, and vascular plants (1–4). The only known component of the complexes is cellulose synthase (CESA), which is represented by 10 isoforms in Arabidopsis (5). Genetic studies indicate at least three Arabidopsis CESA isoforms are required for primary cell wall synthesis (6, 7). Lesions in CESA1 (rsw1), CESA3 (cev1), or CESA6 (prc1) lead to a deficiency in elongation in dark-grown seedlings (8–10). Genetic and biochemical studies demonstrating interactions between CESAs led to a heteromeric model of cellulose synthesis (11, 12) in which the complexes are composed of at least three functionally nonredundant CESA isoforms. It generally is accepted that in Arabidopsis CESA1, CESA3, and CESA6 or CESA6-like proteins are required for functional primary cell wall complexes, whereas CESA4, CESA7, and CESA8 are required for functional secondary cell wall complexes. However, the exact number of CESA proteins contained within the complex, their stoichiometry, and their specific interactions are unknown, and no other components of the complex have been reported.
Recent advances in cell biology and microscopy allow imaging of CESA complexes in live tissues. At least two of the three primary CESAs (CESA3 and CESA6) are functional when labeled with GFP and its derivatives. Both GFP-CESA3 and YFP-CESA6 were observed at the plasma membrane as discreet particles that move along linear trajectories coincident with underlying cortical microtubules (7, 13). CESA particles move bidirectionally with an average velocity of about 350 nm/min corresponding to the addition of ~700 glucose residues per glucan chain per minute (13). CESA particle dynamics are sensitive to osmotic stress and to several drugs that affect cytoskeleton and cellulose synthesis. The observation that perturbation of microtubule polymerization by oryzalin affects the overall distribution and motility of CESA particles supports models in which the microtubules guide the deposition of cellulose. However, CESA particles appear to have an intrinsic level of organization that is evident when microtubules are completely depleted (13).
Genetic screens for mutants deficient in cellulose have implicated a number of proteins in the overall process of cellulose biosynthesis. Mutations in KORRIGAN (KOR), which encodes an endo-β-1,4-glucanase, exhibit deficiencies in cell elongation and reduced cellulose production (14). KOR-like proteins from Brassica napus and poplar exhibit cellulase activity in vitro. However, the exact role of cellulase in cellulose synthesis is unknown. Additional cellulose-deficient mutants include cobra, kobito, pom1, rsw3, fragile fiber1, and fragile fiber2, none of which has been assigned a clear mechanistic function in cellulose synthesis. Here we report the identification of a protein involved in cellulose synthesis that appears to be associated with primary CESA complexes. Identification of this protein opens an avenue in ongoing efforts to understand the mechanism of cellulose synthesis.
Results
Cellulose Synthase-Interactive Protein 1 Interacts with Multiple Primary CESAs.
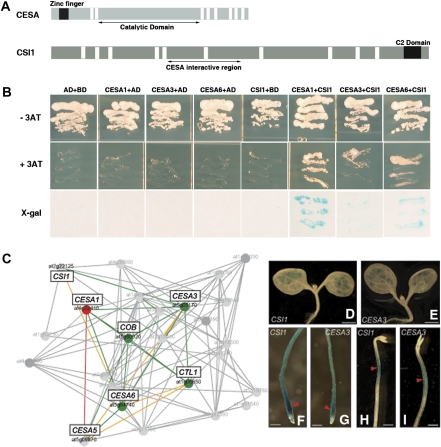
To explore whether additional proteins may be required for cellulose biosynthesis, we performed yeast two-hybrid screens to identify proteins that physically interact with CESA1, -3, and -6. Using 541 amino acids of the putative catalytic domain of CESA6 as bait, we identified a protein referred to as “cellulose synthase-interactive protein 1” (CSI1; At2g22125) (Fig. 1A). To confirm the interaction of CESA6 and CSI1, we subcloned the prey into a GAL4 activation domain (GAL4-AD) and fused the putative catalytic domain of CESA6 with GAL4 binding domain (GAL4-BD). Coexpression of these constructs in yeast resulted in the appearance of β-galactosidase (GUS) activity, confirming the interaction between CESA6 and CSI1. CESA1 and CESA3 also showed positive interactions with CSI1, although the CESA3 interaction appeared to be weaker than the CESA1 or CESA6 interaction (Fig. 1B).
Fig. 1.
Identification of CSI1. (A) Schematic representation of CESA and CSI1 proteins. White bars represent predicted transmembrane domains and ARM repeats in CESA and CSI1, respectively. The regions of the proteins used for the two-hybrid interaction tests are shown as arrows below the bars representing the proteins. (B) CSI1 interacts with three primary CESA proteins in yeast. CSI1 fused with GAL4-AD specifically interacted with the central catalytic domain of three primary CESAs fused with GAL4-BD. (C) Truncated coexpression network for primary wall cellulose-related genes using the AraGenNet at http://aranet.mpimp-golm.mpg.de/aranet/AraGenNet (29). Colored lines indicate strength of transcriptional coordination: green, mutual rank ≤10; orange, mutual rank ≤20; red, mutual rank ≤30. Connections of moderate interest for the study are shown in gray. Low mutual rank indicates stronger coexpression relationships. (D–I) Promoter GUS analysis of CSI1::GUS (D, F, and H) and CESA3::GUS (E, G, and I). GUS staining pattern is shown in light-grown seedlings (D and E) and etiolated seedlings (F–I). Red arrows indicate strong GUS activity. (Scale bars: 750 μm in D and E and 200 μm in F–I.)
CSI1 was identified previously as one of the genes that is coregulated transcriptionally with the primary CESAs (15). In addition to CSI1, several genes that affect cellulose deposition, including COBRA and CTL1/POM1, also are coexpressed with the CESA genes (Fig. 1C). Consistent with coexpression analysis, transgenic plants in which GUS was placed under the control of the 1.4-kb promoter region upstream of the CSI1 gene exhibited a pattern of GUS activity (Fig. 1 D, F, and H and Fig. S1) similar to that seen with promoter:GUS fusions of primary wall CESA genes (Fig. 1 E, G, and I). In addition, the CSI1 promoter drives GUS expression in floral tissues, rosette leaves, roots, and pollen (Fig. S1), indicating that the CSI1 expression pattern is similar to that of CESAs throughout development.
CSI1 Encodes an Armadillo Repeat-Containing Protein.
Protein sequence homology searches identified CSI1-related sequences in a variety of dicots, monocots, and the moss Physomitrella patens (Fig. S2). The Arabidopsis genome contains two closely related genes, which we refer to as “CSI2” and “CSI3” and which share about 55% sequence similarity with CSI1. No CSI1-like proteins were identified outside the land plants. The CSI protein contains multiple tandem copies of a degenerate protein sequence motif, the armadillo (ARM) repeat. The ARM repeat is an ≈40-aa long, tandemly repeated sequence first identified in the Drosophila segment polarity gene, armadillo (16). ARM repeats are found in more than 240 proteins which are predicted to share a conserved 3D structure and often participate in protein–protein interactions (17–19). CSI1 also contains a C2 domain at its C terminus. Some C2 domains have been shown to bind phospholipids in a calcium-dependent or -independent manner and are involved in targeting proteins to cell membranes (20, 21). Other C2 domains have been shown to mediate protein–protein interactions (22).
csi1 Mutants Have Defects in Expansion.
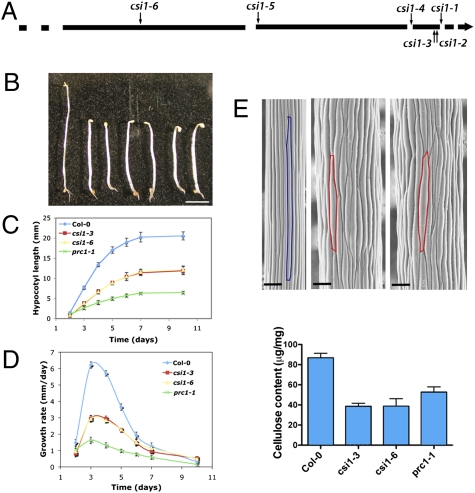
To investigate the biological function of CSI1, we analyzed six independent homozygous transfer DNA (T-DNA) insertion lines with insertions in either exons or introns of CSI1 from the Salk Institute Genomic Analysis Laboratory (SIGnAL) collection (Fig. 2A) (23). At least five of the lines probably were null alleles for CSI1, because no CSI1 mRNA was detectable by RT-PCR (Fig. S3A). Hypocotyls in etiolated csi1 mutants were ≈30% shorter and ≈80% wider than in wild-type plants (Fig. 2 B, C, and E) and elongated less rapidly than in wild-type but more quickly than in prc1-1 (a CESA6 mutant) plants (Fig. 2D). The reduced hypocotyl length and increased diameter indicate that csi1 mutants have defects in the control of anisotropic expansion (highlighted cells in Fig. 2E). The etiolated seedlings of csi1 alleles had a 50% reduction in crystalline cellulose (Fig. 2F). Several cellulose-deficient mutants, such as cob-6, ctl1/pom1, and kor1, display similar cell elongation phenotypes (14, 24, 25).
Fig. 2.
CSI1 is required for normal cell expansion. (A) Schematic representation of six T-DNA insertion sites in csi1. Exons are represented by black lines, and introns are shown by breaks. (B) Morphology of 4-d-old dark-grown seedlings: (Left to Right) Col-0 (wild-type) and csi1-1, csi1-2, csi1-3, csi1-4, csi1-5, and csi1-6 mutants. (Scale bar: 2 mm.) (C and D) Hypocotyl length (C) and growth rate (D) of dark-grown wild-type (Col-0) plants and csi1-3, csi1-6, and prc1-1 mutants. Data were collected from the measurement of ~50 seedlings for each genotype. Error bars represent SE (absent error bars were obscured by symbols). (E) SEM of dark-grown hypocotyls in wild-type plants and csi1 mutants: (Left to Right) Arabidopsis thaliana Columbia (Col-0), csi1-3, and csi1-6 mutants. Colors outline one epidermal cell. (Scale bar: 100 μM.) (F) Cellulose content was reduced in csi1 mutants. n = 5. Error bars represent SE.
csi1 mutants also exhibited short, and swollen, seedling roots. In 8-d-old light-grown seedlings, roots in csi1 mutants were ≈25% shorter than in wild-type plants (Fig. S3 B and C). The roots in csi1 mutants also exhibited epidermal cell swelling and were 80% wider than wild-type roots (Fig. S3 D–F). Additionally, adult csi1 mutants were dwarfed and had shorter siliques than wild-type plants (Fig. S3G). To test whether the smaller siliques were caused by partial sterility, we conducted reciprocal backcrosses to wild-type plants using heterozygous plants. The progeny of these crosses clearly showed that the transmission of the csi1 allele occurred at much lower frequency from the male gametophytes than from the female, indicating that the reduced fertility of csi1 mutants was caused mainly by pollen defects (Table S1). Therefore, we examined the surface structure of the mature pollen grains by SEM. Although all wild-type pollen grains displayed typical morphology, very few typical pollen grains were observed in the csi1 alleles. Instead, the majority of the csi1 pollen grains displayed irregular or collapsed cell wall morphologies (Fig. S3 I and K). Similar observations also have been reported for null alleles of CESA1 and CESA3 (6).
CSI1 Is Associated with CESA Complexes.
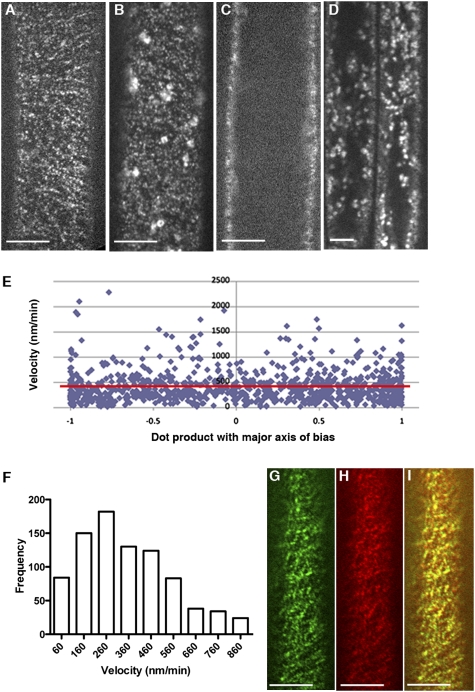
To investigate the subcellular localization of CSI1, we generated both N- and C-terminal translational fusions of red fluorescent protein (RFP) to the CSI1 protein under the native CSI1 promoter in the csi1-6–mutant background. Both N- and C-terminal constructs complemented the mutant phenotypes of reduced hypocotyl elongation and reduced expansion anisotropy, indicating that the fusion protein was functional (Fig S4). Observation of epidermal cells in dark-grown hypocotyls by spinning disk confocal microscopy revealed that RFP-CSI1 was detected as distinct particles at the plasma membrane (Fig. 3A and Movie S1). Similar to YFP-CESA6, RFP-CSI1 particles were organized into linear arrays. To assess whether RFP-CSI1 particles move bidirectionally like those of YFP-CESA6, directional bias was analyzed by calculating the dot product of each particle relative to the dominant axis of potential particle bias and correlating this dot product with the velocity of each particle (Fig. 3E). Greater differences between particle velocities going with and against the major axis result in a larger slope in a linear regression of the plot. As seen in Fig. 3E, RFP-CSI1 particles (n = 917) travel bidirectionally with no bias relative to the major axis. The average velocity of RFP-CSI1 particles in epidermal cells in dark-grown hypocotyls was 416 nm/min (range 63–860 nm/min; Fig. 3F). The velocity of RFP-CSI1 is similar to that reported for YFP-CESA6 (13). Unlike YFP-CESA6, which is associated with the Golgi complex in addition to the plasma membrane (Fig. 3 B and D) (13), RFP-CSI1 was not detected in the cytoplasmic compartments (Fig. 3C). In plants containing both GFP-CESA3 and RFP-CSI1, the RFP-CSI1 particles were substantially colocalized with GFP-CESA3 (Fig. 3 G–I).
Fig. 3.
CSI1 is localized to CESA-like particles in dark-grown hypocotyls cells. (A–D) Optical sections of epidermal cells in 3-d-old dark-grown hypocotyls expressing RFP-CSI1 (A and C) and YFP-CESA6 (B and D). Focal planes of the plasma membrane (A and B) and lower cortex (C and D) are shown. (Scale bar: 5 μm.) (E) Plot of RFP-CSI1 particle velocity vs. its dot product (i.e., scalar product; see Methods) with the direction of bias. Red line indicates the linear regression of the plot. A slope of 0 indicates no bias in direction. (F) Histogram of measured RFP-CSI1 particle velocities. The mean is 416 nm/min (n = 917). (G–I) Localization of GFP-CESA3 (G), RFP-CSI1 (H), and merge (I). (Scale bar: 5 μm.)
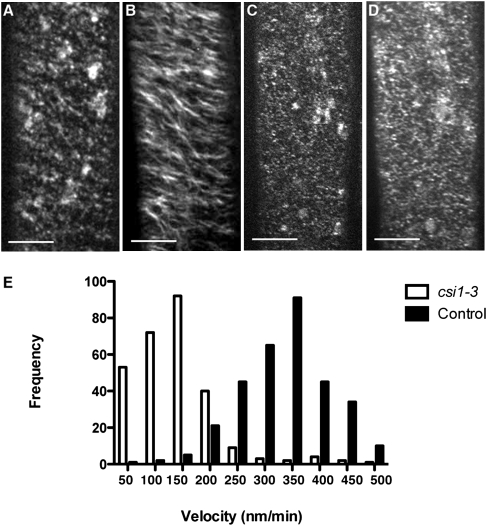
Lesions in csi1 Affect the Distribution and Motility of YFP-CESA6.
To monitor the dynamics of CESA complexes in csi1 mutants directly, we introduced a homozygous csi1-3 allele into a YFP-CESA6 line (13). Using the YFP-CESA6 marker, CESA complexes can be observed at the plasma membrane as distinct punctuate particles that move at constant rates along linear tracks (13). Although the CESA particles in the plasma membranes of wild-type (control) epidermal cells were organized into linear arrays, the distribution of the CESA particles appeared to be disorganized in plasma membranes of the csi1-3 background (Fig. 4C and Movie S2). In contrast, CESA particles in the internal cell layers were not affected in csi1-3 mutants. We did not observe any significant differences in the distribution or motility of CESA particles associated with the Golgi complex in csi1-3 mutants compared with control lines (Golgi in the csi1-3 mutants moved at 6021 ± 405 nm/min vs. 5214 ± 982 nm/min in wild-type plants). In control cells, CESA particles associated with the plasma membrane migrated with an average velocity of 365 ± 45 nm/min (n = 318). In csi1-3 mutants, however, the average velocity of CESA particles was reduced to 132 ± 52 nm/min (n = 225; Fig. 4E). These observations are reflected clearly in time-averaged projections of the CESA particles (Fig. 4 B and D). In control cells, motile particles form linear trajectories along the axes of the particle arrays. Although CESA particles still move bidirectionally in csi1-3 mutants (as assessed by Imaris software), linear tracks in time-averaged images were much shorter.
Fig. 4.
YFP-CESA6 dynamics are altered in csi1-3 mutants. YFP-CESA6 localization in dark-grown hypocotyls cells is shown in wild-type plants (A and B) and csi1-3 mutants (C and D). (A) A single optical section acquired at the plane of plasma membrane in wild-type plants. (B) Average of 61 frames (duration: 2 min; 2-s interval) showing movement of labeled particles along linear trajectories. (C) Single optical section of YFP-CESA6 in csi1-3 mutant. (D) Average of 61 frames (duration: 2 min; 2-s interval) in csi1-3 mutant. (Scale bars: 5 μm.) (E) Histogram of measured particle velocities. The mean velocity is 365 nm/min in control plants (n = 318) and 132 nm/min in csi1-3 mutants (n = 225).
Uniformity of Cellulose Microfibrils Is Affected in csi1 Mutants.
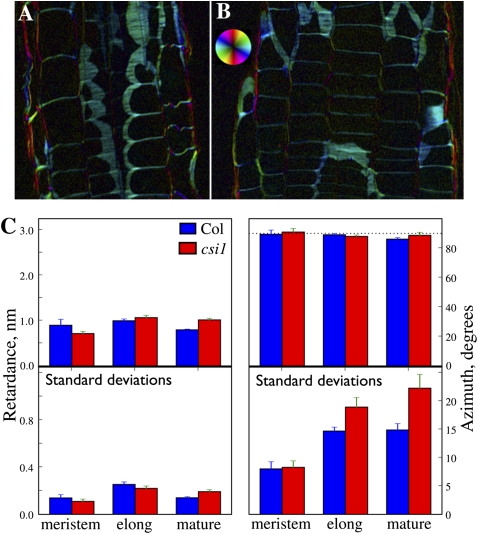
To investigate effects of the csi mutations on the arrangement of cellulose microfibrils, we observed the longitudinal cell walls of wild-type and csi1-1 roots using circularly polarized light enhanced with the universal compensator (26). Polarized light can be used to assay the abundance and orientation of cellulose microfibrils because of their partial crystallinity. Overall, the cell walls appeared similar in both genotypes (Fig. 5 A and B). To determine whether there was a quantitative difference in microfibril abundance or orientation, we quantified the retardance of the cell walls and the azimuth of the crystalline elements. The average retardance was about 1 nm, similar to that reported previously for Arabidopsis cell walls (27), and there was no significant difference between the genotypes in the three delineated zones of the root (Fig. 5C). Similarly, in the three root zones of both genotypes, the azimuths averaged to 90° transverse to the long axis of the root. However, for the elongation and mature zones, the variability among azimuth measurements made in individual roots was significantly greater in csi1-1 mutants than in wild-type roots, suggesting that microfibril alignment in these cell walls had become less uniform.
Fig. 5.
Polarized light analysis of csi1 mutants. (A and B) Polarized-light micrographs of (A) wild-type and (B) csi1-1-mutant roots. Images are of longitudinal-tangential sections through cortex and epidermis. Intensity is proportional to retardance, and color represents optical axis (azimuth) of the crystalline elements, as shown by the color wheel. Note the similar intensity and color for the two genotypes. (C) Quantification of retardance and azimuth (90° is transverse to the root's long axis). Top panels show means ± SEM (n = 3 roots); bottom panels show the average SD (n ~ 200 measurements) ± SEM for each of the roots. Note the larger SDs for csi1-1 mutants in the elongation and mature zones.
Discussion
The CSI1 protein is the first non-CESA protein associated with CESA complexes. Several lines of evidence led us to hypothesize that CSI1 exerts a direct effect on cellulose synthesis through its association with CESA complexes. First, CSI1 physically interacts with multiple primary CESAs in yeast two-hybrid assays, and CSI1 is transcriptionally coregulated with several of the CESA genes involved in primary cell wall synthesis, but there was no obvious association with CESAs involved in secondary cell wall synthesis. Additionally, CSI1 colocalizes with primary CESA complexes, and csi1 mutations affect the distribution and movement of CESA complexes, resulting in strongly reduced rates of CESA complex movement. If we assume that the length of cellulose microfibrils is affected by the velocity and lifetimes of CESA particles, the cellulose deficiency and the associated swelling phenotype can be attributed to the effect of the csi1 mutations on the activity of the CESA complexes. Additionally, the csi1 mutations appeared to decrease the degree to which cellulose microfibrils are coaligned. Polarized light is sensitive to the azimuth at which the optical axis of the crystalline sample is oriented. In both csi1 mutants and wild-type roots the average azimuth of cellulose microfibrils was transverse to the long axis of the root. However, the SD of the azimuth measurements was larger in csi1 mutants than in wild-type roots. In other words, microfibril alignment, on a scale greater than that of a wavelength, is noisier in csi1 mutants than in wild-type roots. A similar reduction in the uniformity of microfibril alignment across the root has been reported for treatment with low concentrations of the microtubule inhibitor oryzalin (27). The decrease in cellulose organization in csi1 mutants indicates that CSI1 may participate in the mechanisms responsible for organizing the deployment of cellulose microfibrils in primary walls.
CSI1 belongs to a family of highly conserved land plant-specific proteins. CSI1 contains multiple predicted ARM repeats and a single C2 domain. Three-dimensional structures of previously characterized ARM repeats comprise three α helices. For example, yeast importin-α contains a central region of 442 aa that contains 10 ARM repeats of 42 aa, forming a right-handed superhelix of helices that creates a surface for protein–protein interactions (18). By comparison, CSI1 has 10 predicted ARM repeats distributed unevenly across the entire protein (2,151 aa). We are not able to draw direct structural comparisons between CSI1 and other proteins containing ARM repeats.
Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana Columbia (Col-0) seeds were sterilized and germinated on Murashinge and Skoog plates (1/2 × MS salts, 0.8% agar, 0.05% monohydrate 2-(N-Morpholino) ethanesulfonic acid, pH 5.7). Seedlings were grown vertically on the agar at 22 °C in darkness for 3 d before imaging. For soil-grown plants, seedlings were germinated on MS plates containing 1% sucrose and then were transferred to pots in a greenhouse at 22 °C under 16-h light and 8-h dark.
Yeast Two-Hybrid Assay.
The yeast two-hybrid screen was carried out by Hybrigenics. CSI1 was identified as a CESA6-interactive protein. To confirm the interaction between CSI1 and CESA6, we subcloned the prey fragment into pACTII (28). The catalytic domains of CESA1, CESA3, and CESA6 were cloned into pAS1-CYH2 (28) using primers indicated in Table S2. The resulting constructs were cotransformed into yeast strain Y190 as pairs (Fig. 1B). Transformants were selected on SC-Trp-Leu-His plates. Positive interactions were tested by their ability to grow on SC-Trp-Leu-His plates supplemented with 100 mM 3-aminotriazole (Sigma) and for GUS activity using a filter assay.
Coexpression Analysis.
A coexpression network for CSI1 (Cluster 86) was obtained from AraGenNet at http://aranet.mpimp-golm.mpg.de/aranet/AraGenNet (29) and trimmed to facilitate readability.
GUS Construct and Staining.
A genomic DNA fragment (1.4 kb) upstream from the ATG start codon of CSI1 was cloned into pCAMBIA1305 GUS-Plus (Table S2) using BamHI and NcoI. The construct was transformed into Arabidopsis using Agrobacterium-mediated transformation. Transgenic plants were selected on hygromycin, stained for GUS activity in 100 mM sodium phosphate (pH 7.0), 10 mM EDTA, 1 mM ferricyanide, 1 mM ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl β-D-glucuronic acid at 37 °C, cleared in 70% ethanol, and observed under a Leica MZ12.5 stereomicroscope (Leica DFC420 digital camera).
Construction of Transgenic Lines.
A 35S promoter in pH7WGR2 and pH7RWG2 (30) was replaced with a 3-kb promoter of CSI1 using Spe I and SacI to create construct pYG104 and pYG105, respectively. The full-length coding sequence of CSI1 (accession # NM_12781.4) synthesized by DNA 2.0 was introduced into pYG104 and pYG105 using Gateway LR Clonase II (Invitrogen). The verified constructs (pYG106 and pYG107) then were introduced into csi1-6 by Agrobacterium-mediated transformation. Complemented lines were selected for further analysis.
Cellulose Measurement.
Cellulose was measured from 4-d-old etiolated seedlings using the Updegraff method (31). Data were collected from five technical replicates for each tissue sample.
Microscopy.
Anthers from 5-wk-old plants or hypocotyls of 4-d-old dark-grown seedlings were mounted on stubs under a pressure of 50 Pa in an Hitachi TM-1000 scanning electron microscope. Roots of 8-d-old light-grown seedlings were wet-mounted on slides and viewed and photographed with a Leica DM5000B epifluorescence microscope (JH Technology). ImageJ software (W. Rasband, National Institute of Health, Bethesda, MD) was used for measurement of hypocotyls and root length. Whole 5-d-old seedlings were viewed on MS plates using a Leica MZ12.5 stereomicroscope (Leica DFC420 digital camera).
Polarized Light Analysis.
Roots were prepared for polarized light analysis as described previously (27). Briefly, root tips were cryofixed in liquid propane, freeze-substituted in ethanol, embedded in butyl-methyl-methacrylate, and sectioned at 2-μm thickness. Sections were imaged on an Interphako polarized-light microscope (Zeiss) equipped with an LC Polscope quantification system (Cambridge Research Instruments) implementing the universal compensator (26). This instrument operates in circularly polarized light and generates two images. The intensity of each pixel is proportional to birefringent retardance in the first image and to the azimuth of the optical axis of the crystalline elements in the second image. For display (Fig. 5) the two images are superimposed with pixel intensity giving retardance and color giving azimuth. Measurements were taken from subcellular areas of cell wall in cortex and epidermis, as viewed in longitudinal sections. Approximately 20 sections per root and three roots per genotype were measured. For analysis, the root was divided into meristem, elongation, and mature zone based on cell length. Azimuth was defined with respect to the local midline (longitudinal axis) of the root.
To assess the statistical significance of the polarized light observations, circular statistical techniques were used (32). Specifically, microfibril orientation was expressed as a unit vector corresponding to the doubled azimuth angle; to obtain the mean orientation, this unit vector was averaged over each observation zone, and the result was halved. The angular deviation was calculated as half the (nonnegative) angular distance between the unit vectors for an individual patch, a, and the mean, b: arccos [cos (a − b)]. Zone-wise mean angular deviations of microfibrils were compared between genotypes using a Mann–Whitney U test, similar to the approach used to compare sample angular deviations reported by Wallraff (32). The distribution of zone mean angular deviations was skewed right, but the distribution became approximately normal after log transformation. Therefore, logarithms of mean angular deviations also were compared using an independent sample t test. The results of the t test on log-transformed data and the u test were the same.
Isolation of T-DNA Insertion Line.
The identification of csi1 knockout lines from the SIGnAL (Salk Institute Genomic Analysis Laboratory; http://signal.salk.edu/cgi-bin/tdnaexpress) collection was based on a combination of database searches and PCR amplification of T-DNA flanking regions. For T-DNA lines identified from the SIGnAL collection, seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University; http://www.biosci.ohio-state.edu/~plantbio/Facilities/abrc/abrchome.htm). PCR reactions were carried out to identify single plants for the T-DNA insertion. Primers used for T-DNA genotyping of csi1 alleles are listed in Table S2.
Confocal Microscopy and Image Analysis.
For analyses of microtubule dynamics, seeds were germinated on MS agar plates and grown vertically in darkness for 3 d at 22 °C. Seedlings were mounted between two coverslips in water. Imaging was performed on a Yokogawa CSUX1spinning disk system featuring the DMI6000 Leica motorized microscope (13) and a Leica 100×/1.4 NA oil objective. YFP was excited at 488 nm, and a band-pass filter (520/50 nm) was used for emission filtering. Image analysis was performed using Metamorph (Molecular Devices) and Imaris (Bitplane) software.
For Imaris analysis, the contrast was enhanced and normalized for each slice within a movie using ImageJ. The enhanced movie was processed in Imaris 6.2.1 from Bitplane. Automated particle detection was performed to find particles with a diameter of ~230 nm, and tracks were generated over the lifetime of the particle. To filter noise in particle detection, only particles detected for 14 s (seven frames) were analyzed. The data for the total displacement and duration for each track in a movie were exported. Directional bias was analyzed by summing the vectors of all particles to determine the direction of greatest particle flux. The dot product of each particle's trajectory against this direction was calculated, and the velocity of each particle was plotted against its dot product. Greater differences between particle velocities going with and against the major axis result in a larger slope in a linear regression of this plot.
Database Search and Sequence Alignment.
The predicted amino acid sequence of CSI1 (At2g22125) was retrieved from the Arabidopsis Information Resource (TAIR) database (www.arabidopsis.org). This protein sequence was used to identify full-length CSI1-like proteins in the National Center for Biotechnology Information GenBank protein database using BLASTP (www.ncbi.nlm.nih.gov/BLAST). CSI1-like proteins (Table S3) were aligned using ClustalW implemented in MegAlign (DNASTAR); protein alignments then were used to generate the phylogenetic tree of CSI1-like proteins (MegAlign; DNASTAR).
Supplementary Material
Acknowledgments
We thank H. Höfte (INRA, Versailles, France) for providing GFPCESA3 line and S. Li and K. Hematy for helpful discussions. This work was supported in part by an award from The Balzan Foundation, Grants DE-FG02-03ER15421 (to T.I.B.) and DOE-FG02-03ER20133 (to C.R.S.) from the US Department of Energy, and the Energy Biosciences Institute. M.B., A.S., and S.P. were funded through the Max-Planck Gesellschaft.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007092107/-/DCSupplemental.
References
- 1.Brown RM, Jr, Willison JH, Richardson CL. Cellulose biosynthesis in Acetobacter xylinum: Visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci USA. 1976;73:4565–4569. doi: 10.1073/pnas.73.12.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller SC, Brown RM, Jr, Scott TK. Cellulosic microfibrils: Nascent stages of synthesis in a higher plant cell. Science. 1976;194:949–951. doi: 10.1126/science.194.4268.949. [DOI] [PubMed] [Google Scholar]
- 3.Giddings TH, Jr, Brower DL, Staehelin LA. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol. 1980;84:327–339. doi: 10.1083/jcb.84.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller SC, Brown RM., Jr Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol. 1980;84:315–326. doi: 10.1083/jcb.84.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 6.Persson S, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desprez T, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagard M, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arioli T, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 11.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 14.Nicol F, et al. A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 17.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 18.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 19.Hatzfeld M. The armadillo family of structural proteins. Int Rev Cytol. 1999;186:179–224. doi: 10.1016/s0074-7696(08)61054-2. [DOI] [PubMed] [Google Scholar]
- 20.Davletov BA, Südhof TC. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 21.Ochoa WF, et al. Structure of the C2 domain from novel protein kinase Cepsilon. A membrane binding model for Ca(2+)-independent C2 domains. J Mol Biol. 2001;311:837–849. doi: 10.1006/jmbi.2001.4910. [DOI] [PubMed] [Google Scholar]
- 22.Benes CH, et al. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 24.Schindelman G, et al. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong R, Kays SJ, Schroeder BP, Ye ZH. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell. 2002;14:165–179. doi: 10.1105/tpc.010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oldenbourg R, Mei G. New polarized light microscope with precision universal compensator. J Microsc. 1995;180:140–147. doi: 10.1111/j.1365-2818.1995.tb03669.x. [DOI] [PubMed] [Google Scholar]
- 27.Baskin TI, Beemster GT, Judy-March JE, Marga F. Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 2004;135:2279–2290. doi: 10.1104/pp.104.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mutwil M, et al. Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 2010;152:29–43. doi: 10.1104/pp.109.145318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 31.Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 32.Batschelet E. Circular Statistics in Biology. London: Academic; 1981. p. 371. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.