Abstract
p73 is a p53-related transcription factor with fundamental roles in development and tumor suppression. Transcription from two different promoters on the p73 gene results in generation of transcriptionally active TAp73 isoforms and dominant negative ΔNp73 isoforms with opposing pro- and anti-apoptotic functions. Therefore, the relative ratio of each isoform is an important determinant of the cell fate. Proteasomal degradation of p73 is mediated by polyubiquitination-dependent and -independent processes both of which appear, thus far, to lack selectivity for the TAp73 and ΔNp73 isoforms. Here, we describe the characterization of another transcriptional target of TAp73; a ring finger domain ubiquitin ligase p73 Induced RING 2 protein (PIR2). Although PIR2 was initially identified a p53-induced gene (p53RFP), low abundance of PIR2 transcript in mouse embryonic fibroblasts of TAp73 KO mice compared with WT mice and comparison of PIR2 mRNA and protein levels following TAp73 or p53 overexpression substantiate TAp73 isoforms as strong inducers of PIR2. Although PIR2 expression was induced by DNA damage, its expression did not alter apoptotic response or cell cycle profile per se. However, coexpression of PIR2 with TAp73 or ΔNp73 resulted in an increase of the TA/ΔNp73 ratio, due to preferential degradation of ΔNp73. Finally, PIR2 was able to relieve the inhibitory effect of ΔNp73 on TAp73 induced apoptosis following DNA damage. These results suggest that PIR2, by being induced by TAp73 and degrading ΔNp73, differentially regulates TAp73/ΔNp73 stability, and, hence, it may offer a therapeutic approach to enhance the chemosensitivity of tumor cells.
Keywords: p73, RNF144b, ubiquitylation, DNA damage
TP73 is a member of the p53 family of transcription factors implicated in development and tumor suppression (1). In addition to the presence of alternative splicing at the 3′ end of mRNA, leading to generation of several different isoforms, the TRP73 gene contains two promoters coding for isoforms with or without the transactivation domain, TAp73 and ΔNp73, respectively (2). The ΔNp73 isoforms have a very important regulatory role as, for example, they exhibit dominant negative activity toward the tumor suppressor functions of both TAp73 and p53. This inhibitory function depends on either dominant negative competition for DNA binding sites (3, 4) or on oligomerization of ΔNp73 with TAp73 isoforms and p53 (5, 6). Moreover, overexpression of ΔNp73 confers chemoresistance to cancer cell lines (7, 8) and increased levels of ΔNp73 in primary tumors has been shown to correlate with poor prognosis (6, 9). The regulatory loop is completed by transcriptional activation of ΔNp73 by both TAp73 and p53 (10–12). Therefore, accumulating evidence supports the fact that an imbalance between TAp73 and ΔNp73 protein levels may be of great importance in both tumorigenesis and resistance to chemotherapy.
In response to genotoxic stress, TAp73 rapidly accumulates due to reduced degradation along with increased stabilization due to acetylation by p300, tyrosine phosphorylation by c-abl, and PML interaction (13–16). Following accumulation, and owing to its structural similarity to p53, TAp73 binds to p53-responsive elements to activate target genes to induce cell cycle arrest, senescence, or apoptosis (2, 17, 18). The apoptotic function of p73 is further enhanced through a transcription-independent pathway, which involves proteolytic cleavage by activated caspases and mitochondrial localization (19). In contrast, in response to DNA damage, ΔNp73 isoforms are preferentially degraded by an unknown mechanism, thus eliminating their dominant negative effects and allowing TAp73 and p53 to exert their proapoptotic activities (20). Even though TAp73 and ΔNp73 show a clear differential degradation, the molecular mechanism underlying this difference is still unknown. A better understanding of these mechanisms will be crucial to the design of other cancer therapies.
We previously performed a microarray analysis to identify the transcriptional targets of p73 (21). Among these, we identified a ring finger domain ubiquitin ligase variously named as in between ring finger domain containing protein 2 (IBRDC2), ring finger protein 144b (RNF144b), and p53 induced ring finger protein (p53RFP). This protein was originally shown to regulate the stability of p21, and the human sequence is located on 6p22.3 with the accession number AK096832. The sequence is shown in Fig. S1, together with the high degree of sequence conservation of the orthologs from Drosophila and zebrafish to man. Fig. S1 also shows the structure of the protein as well as cellular localization. Because, in our hands, the protein is not induced by p53, we refer to it as p73-Induced Ring Protein 2 (PIR2), and, in the present study, we characterize the functional consequences of its p73-mediated transactivation.
Consistent with PIR2 expression being regulated by TAp73, MEF's from TAp73 KO mice have significantly lower steady state levels of PIR2. We have generated a PIR2-specific antibody and shown that PIR2 protein expression is induced in response to DNA damage without a significant change in the apoptotic response or cell cycle profile. Coexpression of PIR2 with TAp73 or ΔNp73 resulted in preferential degradation of the ΔNp73 isoform in an ubiquitin-dependent manner, with a resulting increase in the TA/ΔNp73 protein ratio. Correspondingly, coexpression of PIR2 together with ΔNp73 relieved the inhibitory effect of ΔNp73 on TAp73 mediated apoptosis. These results suggest that PIR2, by differentially regulating TAp73 and ΔNp73 protein stability, may both promote apoptosis of tumor cells and enhance their chemosensitivity. PIR2 is therefore a therapeutic target, and agents that enhance its activity may usefully complement existing therapies particularly in chemoresistant tumors expressing high endogenous levels of ΔNp73.
Results
TAp73 Induces Expression of the Ring Finger Protein PIR2.
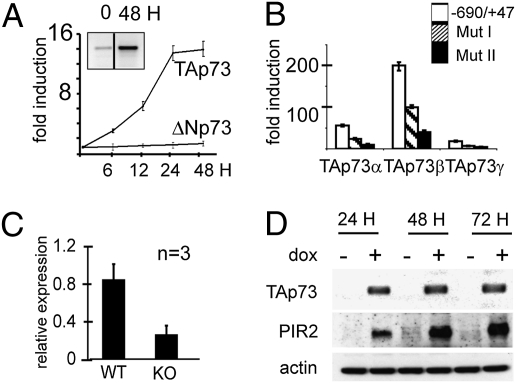
To test the possibility that p73 is able to transcriptionally induce PIR2, as originally suggested by the microarray analysis, we used SaOS-2 cells expressing TAp73 or ΔNp73 under the control of a tet-inducible promoter (22). Under normal conditions these cells express only low levels of PIR2 transcripts. Following induction of TAp73, but not ΔNp73, expression, PIR2 levels increased significantly as detected by real-time and semiquantitave–RT-PCR (Fig. 1A and Fig. S2A). By real-time PCR, PIR2 mRNA was increased 14-fold within 24 h of TAp73 induction (Fig. 1A). Induction of p53 expression in the SaOS-2-p53-inducible cell line did not cause any change in the PIR2 mRNA level (Fig. S2 B and C). To further evaluate transcriptional activation of PIR2 expression by TAp73, we cloned regions −690/+ 47 and −1068/+47 of the PIR2 promoter into the pGL3 basic reporter plasmid. TAp73 expression resulted in a 60-fold induction of the short promoter region, whereas the longer promoter showed less activity, suggesting the presence of an inhibitory site/sites between −1068 and −690. ΔNp73 failed to activate both the long and the short promoter regions as detected by luciferase assays (Fig. S2D).
Fig. 1.
p73 induces PIR2 expression. (A) Induction of PIR2 by TAp73. TAp73-SaOS-2 tet-on cells were treated with doxycyclin (dox) and cells were collected at the indicated time points. Quantitative analysis of PIR2 induction was performed by real-time PCR. The PIR2 mRNA level was up-regulated approximately 14-fold compared with basal level following 24–48 h of TAp73 expression. Induction of PIR2 detected by semiquantitative PCR is shown in the box. (B) TAp73 transactivates PIR2 promoter. Point mutations targeting the −211/−148 region caused a marked reduction of transcriptional activity of p73 on PIR2 promoter. (C) Expression of PIR2 in TAp73 KO MEFs. TAp73 KO MEFs have reduced PIR2 transcript compared with WT counterparts. cDNA from WT and TAp73 KO MEFs were used to quantitate PIR2 mRNA level by real-time PCR. (D) TAp73 induces PIR2 protein expression. Induction of PIR2 was also evident at protein level in TAp73 inducible SaOS-2 cells. PIR2 protein was detected in TAp73 expressing cells, whereas the uninduced cells had no detectable PIR2 protein.
Serial deletions from −690 to −211 resulted in a decrease of p73 transcriptional activity on the PIR2 promoter, probably due to a progressive deletion of responsive promoter elements (Fig. 2E). A further deletion up to −148 almost totally abolished transcriptional activity, suggesting that the region spanning nucleotide −211/−148 harbored the p73 responsive elements. Nucleotide sequence analysis of this region identified a full p73 responsive element located at position −169. Mutating the CAGG residues at position −163 to TTAA and CAAG residues at position −153 to TTAA (Mut I and II, respectively) by site directed mutagenesis produced a profound reduction of p73 transactivation activity indicating this region as the major p73 binding site on the PIR2 promoter (Fig. 1B). To confirm that PIR2 is a bona fide target of TAp73, we measured PIR2 levels in the embryonic fibroblasts (MEF) of TAp73 WT and KO mice by real-time PCR. The 70% reduction in PIR2 levels in TAp73 KO MEFs compared with WT MEFs, suggested that TAp73 is an important regulator of PIR2, at least in MEFs (Fig. 1C).
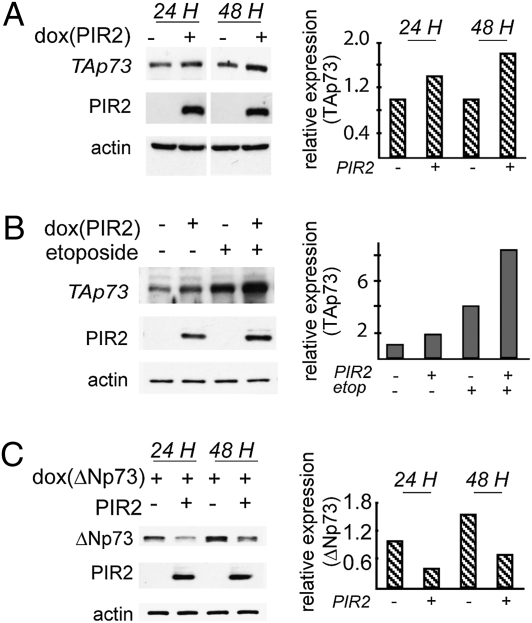
Fig. 2.
PIR2 expression modulates TAp73α and ΔNp73α protein levels. (A) Effect of PIR2 expression on endogenous TAp73α was analyzed by using HCT116 cells stably transfected with the plasmid harboring PIR2 cDNA under the control of a doxycylin-regulated promoter (tet-on PIR2-HCT116). PIR2 expression resulted in twofold induction of TAp73α expression. (B) TAp73α protein levels during DNA damage. Tet-on PIR2-HCT116 cells were treated with 100 μM etoposide for 24 h in the presence or absence of PIR2. Endogenous TAp73α accumulation was more evident in cells expressing PIR2. (C) ΔNp73α-SaOS-2 tet-on cells were transfected with the myc-PIR2 plasmid, and 24 h after transfection cells were induced to express ΔNp73. Cells were harvested 24 or 48 h after induction, and total cell lysate was used to assess p73 levels by Western blotting. Less ΔNp73α protein was detected in cells that express PIR2. All Western blots were subjected to densitometric analysis and results were normalized based on actin expression levels and reported in graphical form (Right).
To study PIR2 and its induction by p73 at the protein level, we generated an antibody against PIR2. The antibody was reactive with HA-tagged overexpressed PIR2 protein (Fig. S3A). To confirm the specificity of this antibody, we used various approaches. First, we performed semiquantitative–RT-PCR to analyze PIR2 expression levels in different lines and compared this with the presence or absence of PIR2 antibody reactive proteins by Western blotting (Fig. S3A). High levels of PIR2 transcripts in MDA-MB-231 and MDA-MB-468 cells correlated well with the presence of a protein migrating at a similar rate to HA-tagged PIR2. Likewise no protein band was detected in SHSY5Y and K562 cells, which do not have detectable levels of PIR2 transcripts. Furthermore, transfection of both MDA-MB-231 and MDA-MB-468 cells with siRNA against PIR2 resulted in reduction of the PIR2-antibody reactive protein level (Fig. S3B). The size of the PIR2 antibody responsive band was calculated to be 40 kDa on a semilogarithmic graph and this correlated with the expected size of the PIR2 protein (Fig. S3C). Together these results clearly demonstrated that the antibody generated against PIR2 is highly specific.
Next, we investigated if induction of TAp73 expression would result in accumulation of PIR2 protein in SaOS-2 cells. During the course of 72 h of TAp73 induction, PIR2 protein levels progressively increased (Fig. 1D). We also investigated the relative effects of TAp73 and ΔNp73 on PIR2 expression at the protein level. To this end, we transfected H1299 cells with the indicated plasmids and assessed PIR2 protein levels by Western blotting. As expected, ΔNp73 failed to induce PIR2 expression, whereas TAp73 (both α and β isoforms) induced PIR2 expression efficiently (Fig. S3D), further supporting the hypothesis that TAp73 is a strong inducer of PIR2.
To demonstrate the in vivo efficiency of TAp73 to induce PIR2 at protein level, we first determined the specificity of our antibody in mouse by silencing PIR2 in MEFs with siRNA. As shown in Fig. S3E, silencing of PIR2 resulted in reduction of the antibody reactive protein, indicating that the antibody is reactive with mouse PIR2. Mouse PIR2 migrated at a slightly slower rate compared with human PIR2 (Fig. S3E). Comparison of PIR2 levels in different mouse tissues revealed that PIR2 is highly expressed in brain (Fig. S3F). We compared PIR2 levels in the cortex of TAp73 WT and KO mice and showed that the absence of TAp73 results in reduction of PIR2 protein levels (Fig.S3G).
PIR2 Expression Modulates p73 Protein Stability.
To assess functional consequences of PIR2 expression, we generated PIR2-inducible HCT116 cells. These cells express TAp73α as the predominant endogenous p73 isoform (19). Following induction of PIR2 expression for 48 h, we detected a slight increase in TAp73α levels (Fig. 2A). However, accumulation of TAp73α became much more evident following induction of DNA damage by etoposide and was further increased in HCT116 cells expressing PIR2 (Fig. 2B). This result suggested the presence of a positive feedback loop where, following DNA damage, TAp73 increases its stability via a transcriptional target. To analyze the effect of PIR2 expression on ΔNp73, we used ΔNp73-inducible SaOS-2 cells. Expression of PIR2 resulted in a reduction in ΔNp73 levels, compared with control cells, both 24 or 48 h after induction (Fig. 2C). Indeed, coexpression of PIR2 and TAp73 in H1299 cells resulted in an increase in TAp73 protein levels (Fig. S4A), whereas coexpression of PIR2 and ΔNp73 resulted in a striking decrease in ΔNp73 levels (Fig. S4B). This suggests that PIR2 expression may differentially regulate the relative abundance of TAp73 and ΔNp73 proteins. These changes were only seen at the protein level, as PIR2 expression did not change the mRNA level of either p73 isoform (Fig. S4C).
PIR2 Can Bind to p73 Isoforms and Affect Their Stability.
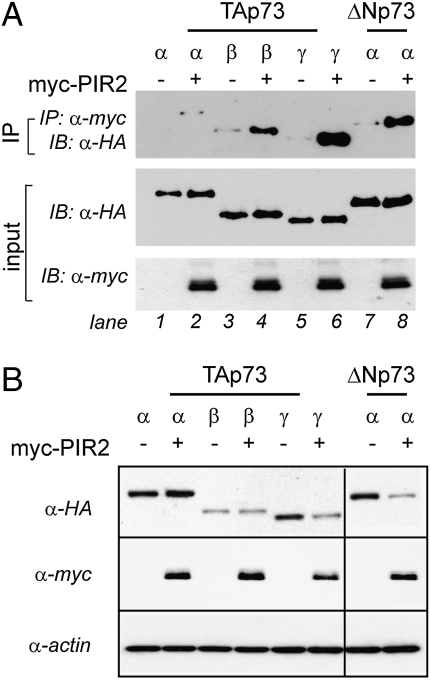
Next, we sought to investigate the mechanism by which PIR2 exerts differential effects on TAp73α and ΔNp73α levels. To this end, we first tested whether PIR2 can bind to p73 isoforms. A coimmunoprecipitation assay, following expression of PIR2 with TAp73α, β, and γ or ΔNp73α in H1299 cells revealed a strong interaction between ΔNp73α and PIR2 (Fig. 3A, lanes 7–8). TAp73 isoforms were also able to bind to PIR2, although TAp73α and β bound with a much lower affinity (Fig. 3A, lanes 1–2). The results were confirmed with a reverse coimmunoprecipitation, where TAp73α and ΔNp73α were pulled-down and the interacting amount of PIR2 was analyzed (Fig. S5).
Fig. 3.
PIR2 binds p73 and affects its stability. (A) PIR2 can bind TAp73γ and ΔNp73α efficiently. HEK293 cells were transiently transfected to express myc-PIR2 alone or in different combinations with HA-tagged TAp73α, TAp73β, TAp73γ, or ΔNp73α. Cell extracts were immunoprecipitated with anti-Myc antibody. The immune complexes were subjected to Western blot analysis with anti-HA antibody. Aliquots of total cell extracts from unprocessed cells were directly subjected to immunoblot analysis with anti-HA antibody or anti-myc antibody (input). (B) PIR2 expresion modulates TAp73γ and ΔNp73α protein levels. Coexpression of PIR2 with TAp73α, TAp73β, TAp73γ, or ΔNp73α resulted in detection of less TAp73γ or ΔNp73α protein. H1299 cells were transfected with indicated plasmids and total protein was used to assess p73 and PIR2 levels.
The binding affinities of the different p73 isoforms to PIR2 correlated well with their stability, as PIR2 expression resulted in reduced amounts of TAp73γ and ΔNp73α isoforms (Fig. 3B).
PIR2 Induces Ubiquitination of ΔNp73.
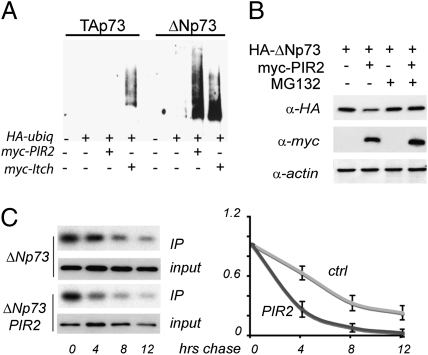
Degradation of most short-lived proteins by the 26S proteasomes is mediated by the ubiquitin system, where ubiquitin is covalently conjugated to the substrate protein in a multistep process (23). We therefore asked if PIR2 targets ΔNp73 for degradation through the ubiquitin-dependent proteasomal pathway. For this purpose extracts of HEK293 cells transfected with plasmids expressing, HA-tagged Ub (Ub-HA), Myc-tagged PIR2 (Myc-PIR2), and Flag-tagged ΔNp73α were subjected to immunoprecipitation with anti-Flag antibodies and detected by Western blotting for HA and Flag. Myc-tagged Itch (Myc-Itch) was used as a positive control. As shown in Fig. 4A, whereas PIR2 was able to ubiquitinate ΔNp73α efficiently; no ubiquitinated TAp73α conjugates were detected upon cotransfection with PIR2. Together these data indicate that PIR2 induces ΔNp73 degradation by promoting its ubiquitination.
Fig. 4.
PIR2 induces ubiquitin mediated degradation of ΔNp73α. (A) PIR2 ubiquitinates ΔNp73. HEK293 cells were transiently cotransfected with expression plasmids for Ub-HA, Flag-TAp73α, Flag-ΔNp73α and Myc-PIR2. Myc-Itch was used as a positive control. Forty-eight hours after transfection, cells were treated with MG132 and then collected. Lysates were subjected to IP using an anti-Flag antibody. Immune complexes were revealed with anti-HA antibody. (B) MG132 blocks PIR2 mediated ΔNp73 degradation. H1299 cells were transfected as in A in the ratio of 1:4. Cells were incubated with 10 μM MG132 for 6 h before harvesting. (C) 35S pulse chase of ΔNp73: effect of PIR2 on protein half-life. H1299 cells were transfected as in B. Forty-eight hours after transfection, cells were labeled for 1 h and chased for the indicated times. Levels of ΔNp73 were evaluated at the indicated time points. Following immunoprecipitation, autoradiography was performed; ΔNp73 bands were quantified and results normalized to both incorporation at time 0 and to total immunoprecipitated HA-p73 protein. The results of three independent experiments (**P < 0.001, *P < 0.05) are represented. A representative example of a 35S pulse-chase experiment showing autoradiographic and Western blotting results for HA-tagged ΔNp73α in the absence and presence of overexpressed myc-tagged PIR2 is shown (Left).
Furthermore, when PIR2 and ΔNp73α were coexpressed in the presence of the proteasome inhibitor MG132, no change in the ΔNp73α level was observed (Fig. 4B). We further confirmed these results by measuring ΔNp73α half-life in the presence or absence of PIR2 by a pulse chase assay using [35S]Met and Cys. Although ΔNp73 levels declined progressively over 12 h in the absence of PIR2, they do so significantly more rapidly when PIR2 is coexpressed (Fig. 4C). Together, these data suggest that PIR2 preferentially targets ΔNp73α for ubiquitination and proteasome-mediated degradation.
PIR2 Expression Modulates the Apoptotic Response Following DNA Damage.
Although TAp73 is an important molecular mediator of apoptosis induced by DNA damage, ΔNp73 confers chemoresistance to cancer cells (6, 7). Therefore, the ratio of the TAp73/ΔNp73 isoforms is an important determinant of the response of a cell to the DNA damaging agents. The opposite effect of PIR2 on TAp73 and ΔNp73 protein stability strongly suggested that the main functional consequence of PIR2 expression might be to alter the TAp73/ΔNp73 ratio and induce sensitivity to DNA damaging agents.
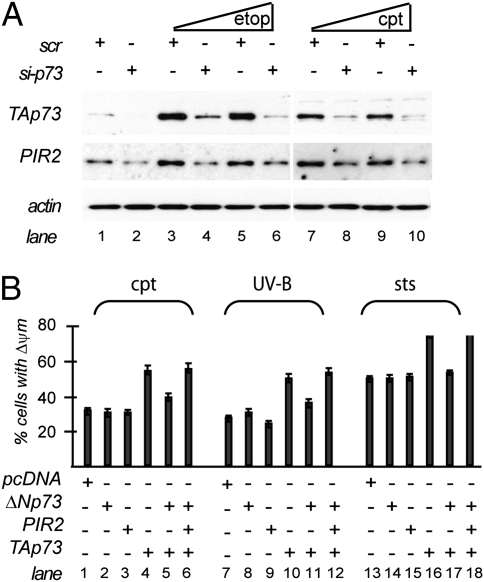
This hypothesis led us first to investigate PIR2 expression following DNA damage. TAp73 accumulates rapidly following DNA damage to induce cell cycle arrest or apoptosis (2, 18, 19). As PIR2 is a target of TAp73, treatment of cell lines with DNA damaging drugs should also result in accumulation of PIR2. To test this, we treated MDA-MB-231 cells with different concentrations of etoposide or cisplatin and compared PIR2 levels by real-time PCR and Western blotting. Both etoposide and cisplatin resulted in induction of PIR2 expression (Fig. 5A, lanes 3, 5, 7, and 9 and Fig. S6A). PIR2 induction also correlated with the increase in TAp73 levels in MDA-MB-231 cells (Fig. S6A). Furthermore, silencing of endogenous TAp73 in MDA-MB-231 cells resulted in reduced PIR2 protein levels (Fig. 5A).
Fig. 5.
PIR2 modulates apoptotic response. (A) Induction of PIR2 following DNA damage in MDA-MB-231 cells is p73 dependent. MDA-MB-231 cells were transfected with scrambled (scr) or short-interfering RNA (siRNA) against p73. Twenty-four hours after transfection, cells were treated with 10 μM or 20 μM etoposide (etop) or cisplatin (cpt) for 24 h. PIR2 and p73 levels were measured by Western blotting. (B) PIR2 reverts ΔNp73 inhibition on TAp73 induced death. HA-TAp73 (1 μg), ΔNp73 (2 μg), and PIR2 (8 μg) expression plasmids were transfected to HeLa cells as indicated. Thirty-six hours after transfection, the medium was changed and cells were treated with 100 μM cisplatin (9 h), 20 mJ/cm2 UV (6 h), or 20 nM staurosporine (16 h). Cells were collected and subjected to flow cytometric analysis to detect mitochondrial membrane depolarization.
To assess the functional consequence of PIR2 expression following DNA damage, we transiently transfected H1299 cells, which do not express detectable levels of PIR2 protein (Fig. S6B), with a plasmid encoding PIR2. Twenty-four hours after transfection, we treated these cells with 100 μM cisplatin, 100 μM etoposide, 100 mJ/cm2 UV-B, or 100 nM staurosporine. H1299 cells transfected with the control vector showed 30, 25, 20, and 40% mitochondrial membrane depolarization 24 h after treatment with cisplatin, etoposide, UV, and staurosporine, respectively (Fig. S6C). Cells that expressed PIR2 showed a very similar mitochondrial membrane depolarization profile, suggesting that PIR2 is not playing a direct role in apoptosis under these conditions. Treatment of H1299 cells with low concentrations of apoptosis inducing drugs resulted in alterations in the cell cycle profile (Fig. S6C). Similarly, PIR2 overexpression did not modify the effect of these drugs on cell cycle profile (Fig. S6C). To further investigate the effect of PIR2 expression on cell cycle or apoptosis, we silenced endogenous PIR2 in MDA-MB-231 cells by short-interfering RNA (siRNA) and treated these cells with staurosporine or cisplatin. Although treatment of MDA-MB-231 cells with these drugs resulted in up-regulation of PIR2 expression, siRNA successfully reduced PIR2 protein level in all treatments (Fig. S6D). The levels of apoptosis and the cell cycle profile of the cells transfected with siRNA were comparable to those of cells transfected with scrambled RNA (scr) (Fig. S6E). In conclusion, PIR2 expression per se has no direct impact on cell cycle or apoptosis under either normal or stress conditions.
The significant up-regulation of PIR2 by TAp73 without a change in the apoptotic potential of the cells or cell cycle profiles, further supported the hypothesis that the biological function of PIR2 may be the fine-tuning of the TAp73/DNp73 ratio in response to stress.
To test this, HeLa cells were transfected with plasmids expressing TAp73, ΔNp73, and PIR2 in different combinations. Forty-eight hours after transfection cells were treated with cisplatin, UV, or staurosporine. Expression of ΔNp73 or PIR2 did not have any synergetic effect on baseline apoptosis caused by the treatment (Fig. 5B). However, expression of TAp73 augmented apoptosis by 60%, 90%, and 40% in the case of cisplatin (lane 4), UV (lane 10), and staurosporine (lane 16), respectively. Expression of ΔNp73 together with TAp73 conferred resistance to TAp73 mediated sensitization to the treatments (lanes 5, 11, and 17) but PIR2 coexpression neutralized this effect by restoring the proapoptotic activity of TAp73 (lanes 6, 12, and 18).
Discussion
Although the p53 protein is mainly inactivated in human cancers through mutations, it is theoretically possible that TA isoforms of p73 could assume its apoptotic function, because TAp73 has been shown to be a genuine tumor suppressor (2). However, transcription of apoptotic genes by TAp73 is inhibited by ΔNp73, which is highly expressed in many cancers (7, 8), thus compromising the potential proapoptotic role of TAp73. This suggests that it is the ratio of TAp73/ΔNp73, which is important in determining the sensitivity of individual cancers to apoptosis induction and thus their chemosensitivity. In cell lines, DNA damage leads to a more rapid decline in ΔNp73 than TAp73 expression, although the underlying molecular mechanisms are not well understood. Here, we demonstrate that the ubiquitin ligase, PIR2, is induced by TAp73 and selectively degrades ΔNp73 and may provide one mechanism whereby the TAp73/ΔNp73 ratio can be modulated.
p73 steady state protein levels are kept low under normal physiological conditions through degradation by the 26S proteasome, at least partly through the HECT-containing E3 ubiquitin ligase Itch (24). This requires the interaction of the WW domain of Itch with the PY region just before the SAM domain of p73, and in particular the Y487 residue of TAp73. The same interaction is conserved in p63 (25). Itch itself is directly regulated by several interactors, including N4BP1 (13), JNK (26), YAP (27). In addition to this major degradation pathway, the E3 ubiquitin ligase FBX045 can also target both TAp73 and ΔNp73 isoforms for proteasomal degradation mediated by the Nedd8/cullin1/SCF complex (25). Another mechanism of p73 degradation involves the ubiquitin-independent 20S proteasomal pathway, regulated by NAD(P)H quinone oxidoreductase 1 together with NADH (26). Although the relative importance of each of these pathways is still under evaluation, none of these mechanisms explains the differential degradation of the TAp73/ΔNp73 isoforms. Under DNA damage, Itch is itself degraded, allowing TAp73 stabilization, whereas ΔNp73 is rapidly degraded in an Itch-independent fashion (13, 24). Thus, selective ΔNp73 degradation remains an important open question.
Here we show that the PIR2 gene is up-regulated by TAp73 and selectively degrades ΔNp73. The sequence and domain structure of PIR2 (also known as RNF144b, IBRDC2, and p53RFP) and alignments with its orthologs are shown in Fig. S1. Although PIR2 has been previously described by two independent groups as a modulator of cell cycle arrest and apoptosis (27, 28), in our experimental model PIR2 was not induced by p53 overexpression and did not induce cell cycle arrest or apoptosis when overexpressed.
We have quantified PIR2 induction by p73 by real-time PCR, defined the p73 responsive elements on the PIR2 promoter and measured PIR2 protein levels following overexpression of p73. Moreover, we have shown that induction of PIR2 after DNA damage in MDA-MB-231 cells was dependent on p73, as silencing of p73 in these cells resulted in a significant reduction in PIR2 levels. We have also shown that PIR2 mRNA levels are lower in TAp73 KO MEFs and PIR2 protein levels are lower in TAp73 KO cortex. Together, these data suggest that TAp73 is an inducer of PIR2 in vitro and in vivo.
Coexpression of PIR2 with TAp73α or ΔNp73α resulted in preferential degradation of ΔNp73α, possibly due to differential binding affinities of PIR2 to the different p73 isoforms. Indeed when PIR2 was expressed with different N and C-terminal truncated isoforms of p73, it showed a high binding affinity to TAp73γ and ΔNp73α only. This suggests that TAp73α-PIR2 binding may be inhibited due to conformational masking of the interaction site resulting from interactions between the TA domain and the long C-terminal sequence.
PIR2 expression did not result in degradation of TAp73α. On the contrary, TAp73α protein was stabilized following PIR2 expression. Although this stabilization effect was less evident on endogenous TAp73α in unstressed cells, at least four times more TAp73α accumulated in PIR2-expressing cells exposed to DNA damage compared with PIR2 negative counterparts. However, the lack of interaction between TAp73α and PIR2 suggests an indirect regulation of TAp73α stability by PIR2 following DNA damage.
These data support the hypothesis that PIR2 may have a key function in regulating p73 function, through fine-tuning the TAp73/ΔNp73 ratio (Fig. S7). In keeping with this, coexpression of PIR2 together with ΔNp73 relieved the inhibitory effect of ΔNp73 on TAp73 mediated apoptosis. These results suggest that due to its ability to differentially regulate of TAp73 and ΔNp73 protein stability, PIR2 may be a potential therapeutic target, especially in tumors exhibiting increased ΔNp73 expression.
Materials and Methods
Cell Culture, Drug Treatments, and Transfections.
All cells were grown as recommended by ATCC. Lipofectamine-2000 reagent (Invitrogen) was used for transfections. PIR2 inducible HCT116 stable cell lines were generated by using the T-Rex system (Invitrogen). Apoptosis and cell cycle analysis was assessed by flow cytometry using TMRE or PI staining, respectively, as described previously (29). Cells were treated with etoposide, cisplatin, UV, or staurosporine (Sigma) as indicated in figure legends. MG132 was from Bio-Mol. Predesigned siRNA targeting PIR2 and p73 were purchased from Ambion Ltd.
Plasmids and Luciferase Assay.
Human PIR2 was cloned in pcDNA4/TO/myc-His vector (Invitrogen). The putative promoter region of PIR2 was identified by using PROSCAN and TSSG software, and the promoter regions were cloned into the pGL3 basic reporter plasmid (Promega). Luciferase assays were performed as described previously (30).
Antibodies and Immunoblot Analysis.
The antibody against PIR2 was generated by using a C-terminal PIR2 peptide. Proteins were denatured, separated on SDS polyacrylamide gels, and then transferred to nitrocellulose membranes. Following protein transfer, blots were incubated in blocking solution and then with a primary antibody for 1 h. Antibodies were used at dilutions: 1:1,000, anti-PIR2; 1:3,000, anti-p73-full length (31); 1:5,000, anti-HA-HRP (Sigma); 1:500 anti-actin (Santa Cruz). Immunoreactive proteins were detected using an enhanced chemiluminescence system (Pierce).
PCR.
RNA extraction and cDNA synthesis were done as described before (32). Equal amount of RNA was used in cDNA synthesis and the quality of cDNA was tested by GAPDH amplification with the primer pair GF 5′ GGCTGAGAACGGGAAGCTTGTCAT and GR 5′ CAGCCTTCTCCATGGTGGTGAAGA. Transcript analysis was done with the following primers SQ-F 5′ ATGGGCTCAGCTGGTAGGC and SQ-R 5′ GTGTCCTGTTGCAGACTGT C. The primers used for real time PCR were PIR2-278F: CCAGGGATTGAGGAGACTGAAG and PIR2-349R: GCGAGATAGTGGAGCCTACCA.
Immunofluorescence.
H1299 were transfected with full-length PIR2-myc construct and plated on coverslips. Immunofluorescence was performed as described previously (33). Slides were analyzed with a confocal laser microscope (ZEISS LSM 510).
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council and grants from the European Union (Blandino; LSHC-CT-2004-503576-Active p53).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911828107/-/DCSupplemental.
References
- 1.Melino G, Lu X, Gasco M, Crook T, Knight RA. Functional regulation of p73 and p63: Development and cancer. Trends Biochem Sci. 2003;28:663–670. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 3.Ishimoto O, et al. Possible oncogenic potential of DeltaNp73: A newly identified isoform of human p73. Cancer Res. 2002;62:636–641. [PubMed] [Google Scholar]
- 4.Stiewe T, Theseling CC, Pützer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: Implications for tumorigenesis. J Biol Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 5.Pozniak CD, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 6.Zaika AI, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergamaschi D, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 8.Vossio S, et al. DN-p73 is activated after DNA damage in a p53-dependent manner to regulate p53-induced cell cycle arrest. Oncogene. 2002;21:3796–3803. doi: 10.1038/sj.onc.1205465. [DOI] [PubMed] [Google Scholar]
- 9.Pützer BM, Tuve S, Tannapfel A, Stiewe T. Increased DeltaN-p73 expression in tumors by upregulation of the E2F1-regulated, TA-promoter-derived DeltaN’-p73 transcript. Cell Death Differ. 2003;10:612–614. doi: 10.1038/sj.cdd.4401205. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa T, et al. Autoinhibitory regulation of p73 by Delta Np73 to modulate cell survival and death through a p73-specific target element within the Delta Np73 promoter. Mol Cell Biol. 2002;22:2575–2585. doi: 10.1128/MCB.22.8.2575-2585.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kartasheva NN, Contente A, Lenz-Stöppler C, Roth J, Dobbelstein M. p53 induces the expression of its antagonist p73 Delta N, establishing an autoregulatory feedback loop. Oncogene. 2002;21:4715–4727. doi: 10.1038/sj.onc.1205584. [DOI] [PubMed] [Google Scholar]
- 12.Grob TJ, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- 13.Oberst A, et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci USA. 2007;104:11280–11285. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mantovani F, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell. 2004;14:625–636. doi: 10.1016/j.molcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo A, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 16.Bernassola F, et al. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaghad M, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 18.Gong JG, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 19.Sayan AE, et al. P73 and caspase-cleaved p73 fragments localize to mitochondria and augment TRAIL-induced apoptosis. Oncogene. 2008;27:4363–4372. doi: 10.1038/onc.2008.64. [DOI] [PubMed] [Google Scholar]
- 20.Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Differ. 2004;11:685–687. doi: 10.1038/sj.cdd.4401376. [DOI] [PubMed] [Google Scholar]
- 21.Müller M, et al. TAp73/Delta Np73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564–1577. doi: 10.1038/sj.cdd.4401774. [DOI] [PubMed] [Google Scholar]
- 22.Melino G, et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 23.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 24.Rossi M, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene. 2009;35:3157–3166. doi: 10.1038/onc.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng CC, Arakawa H, Fukuda S, Kondoh H, Nakamura Y. p53RFP, a p53-inducible RING-finger protein, regulates the stability of p21WAF1. Oncogene. 2003;22:4449–4458. doi: 10.1038/sj.onc.1206586. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, Xu LG, Liu T, Zhai Z, Shu HB. The p53-inducible E3 ubiquitin ligase p53RFP induces p53-dependent apoptosis. FEBS Lett. 2006;580:940–947. doi: 10.1016/j.febslet.2005.09.105. [DOI] [PubMed] [Google Scholar]
- 29.Sayan BS, Sayan AE, Knight RA, Melino G, Cohen GM. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem. 2006;281:13566–13573. doi: 10.1074/jbc.M512467200. [DOI] [PubMed] [Google Scholar]
- 30.Sayan BS, et al. Cleavage of the transactivation-inhibitory domain of p63 by caspases enhances apoptosis. Proc Natl Acad Sci USA. 2007;104:10871–10876. doi: 10.1073/pnas.0700761104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayan AE, et al. New antibodies recognizing p73: Comparison with commercial antibodies. Biochem Biophys Res Commun. 2005;330:186–193. doi: 10.1016/j.bbrc.2005.02.145. [DOI] [PubMed] [Google Scholar]
- 32.Sayan AE, Sayan BS, Findikli N, Ozturk M. Acquired expression of transcriptionally active p73 in hepatocellular carcinoma cells. Oncogene. 2001;20:5111–5117. doi: 10.1038/sj.onc.1204669. [DOI] [PubMed] [Google Scholar]
- 33.Sayan BS, Ince G, Sayan AE, Ozturk M. NAPO as a novel marker for apoptosis. J Cell Biol. 2001;155:719–724. doi: 10.1083/jcb.200106044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.