Abstract
While many functions of the p53 tumor suppressor affect mitochondrial processes, the role of altered mitochondrial physiology in a modulation of p53 response remains unclear. As mitochondrial respiration is affected in many pathologic conditions such as hypoxia and intoxications, the impaired electron transport chain could emit additional p53-inducing signals and thereby contribute to tissue damage. Here we show that a shutdown of mitochondrial respiration per se does not trigger p53 response, because inhibitors acting in the proximal and distal segments of the respiratory chain do not activate p53. However, strong p53 response is induced specifically after an inhibition of the mitochondrial cytochrome bc1 (the electron transport chain complex III). The p53 response is triggered by the deficiency in pyrimidines that is developed due to a suppression of the functionally coupled mitochondrial pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase (DHODH). In epithelial carcinoma cells the activation of p53 in response to mitochondrial electron transport chain complex III inhibitors does not require phosphorylation of p53 at Serine 15 or up-regulation of p14ARF. Instead, our data suggest a contribution of NQO1 and NQO2 in stabilization of p53 in the nuclei. The results establish the deficiency in pyrimidine biosynthesis as the cause of p53 response in the cells with impaired mitochondrial respiration.
Keywords: dihydroorotate dehydrogenase, mitochondrial electron transport chain, NQO1 and NQO2, p53 tumor suppressor, apoptosis
The mitochondrion is the major power station of the cell that generates most of the cell’s supply of ATP. In addition, mitochondria are involved in a range of intracellular processes, such as cell growth and division, differentiation, apoptosis, and intracellular signaling (1). The molecular mechanism of mitochondrial energy transformation involves the electron transport chain (ETC) that consists of electron transfer complexes I-IV embedded in the inner mitochondrial membrane. The ETC converts high energy potential of electrons from NADH and FADH2 into the energy of electrochemical proton gradient across the inner membrane that drives the synthesis of ATP by the ATP-synthase (complex V). Besides, mitochondria participate in the synthesis of many metabolic intermediates, including the de novo biosynthesis of pyrimidines. The latter process is catalyzed by dihydroorotate dehydrogenase (DHODH), an FMN flavoprotein in the inner mitochondrial membrane, which transfers electrons from dihydroorotate to ubiquinone of the ETC for further oxidation (2).
The p53 tumor suppressor mediates important quality control functions by limiting proliferation and survival of abnormal or damaged cells. Mitochondria are critically important for the p53-mediated cell death, because p53 controls transcription of several genes that affect the release of mitochondrial cytochrome c (3). In addition, p53 can induce a transcription-independent apoptosis through the direct interaction with Bcl-2 family proteins (4). On the other hand, p53 also plays homeostatic roles in mitochondria (5) as it controls mtDNA copy number through the p53 regulated M2 subunit of ribonucleotide reductase (6) and stimulates mitochondrial respiration and ATP production through up-regulation of SCO2 and AIF genes (7, 8).
Despite the established significance of p53 in mitochondrial physiology there is little information regarding signals emitted by mitochondria that trigger p53 response. Yet substantial changes in mitochondrial respiration and in the activity of ETC are observed during exposure to hypoxia (9) as the side effects of drugs leading to hepatotoxicity (10) and cardiotoxicity (11) in the inherited succinate dehydrogenase deficiency associated with the development of paragangliomas and pheochromocytomas (12), etc. Activation of p53 in response to an obstruction of mitochondrial ETC may additionally contribute to tissue damage. Mitochondrial ROS were implicated in uncoupling of ETC and in p53 activation in response to hypoxia (13). However, the role of mitochondrial ETC activity in the induction of p53 response remains ambiguous. It was suggested that mitochondrial activity could be required for the stress-induced activation of p53, as inhibitors of complexes I and V mitigate the response to etoposide treatment (14) and inhibitors of complex III interfere with the activation of p53 after treatment with cisplatin (15). On the other hand, it was noticed that certain ETC inhibitors produce a cell senescence phenotype associated with a modest activation of p53, leading to the suggestion that the reduced mitochondrial membrane potential (MMP) could initiate the p53 response (16).
In this study we blocked by specific inhibitors each of the mitochondrial ETC complexes and monitored p53 induction. We conclude that neither the compounds that reduce MMP nor the suppression of ETC activity per se can trigger the p53 response. However, an activation of p53 and an induction of a p53-dependent apoptosis can be elicited specifically by inhibitors of mitochondrial complex III, which cause depletion of pyrimidines through the inhibition of a functionally coupled DHODH. We found that the deficiency in pyrimidines is critical for the induction of p53 in response to ETC complex III inhibitors. The results provide a previously unknown functional link between mitochondrial respiration and the p53 pathway and suggest a contribution of NQO1 and NQO2 in stabilization and nuclear retention of p53 in epithelial cells with exhausted pools of pyrimidine nucleotides.
Results
ETC Complex III Inhibitors Specifically Up-Regulate p53 and Induce a p53-Dependent Apoptosis.
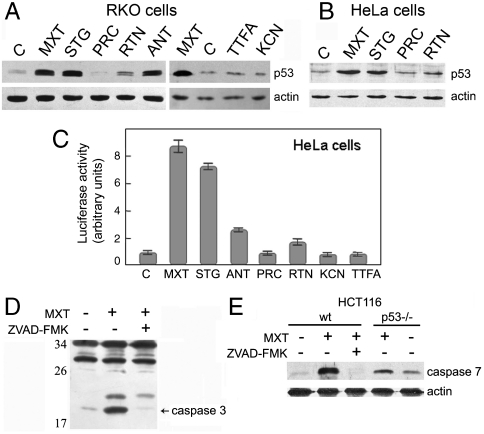
To find whether the deficiency in mitochondrial respiration can elicit p53 response we studied the accumulation of p53 in cells treated with inhibitors of different mitochondrial ETC complexes. In RKO cells the treatment for 16–18 h with complex I inhibitors piericidin and rotenone, complex II inhibitor TTFA, and cytochrome c oxidase (complex IV) inhibitor KCN produced almost no effect on the level of p53. However, a significant accumulation of p53 was observed after the treatment with complex III inhibitors myxothiazol, stigmatellin, and antimycin A (Fig. 1A). Similar effects were observed in A549 and HCT116 cells (Fig. S1 A and B). In HeLa cells there was also a substantial up-regulation of p53 level in response to myxothiazol and stigmatellin but no effect of piericidin and rotenone (Fig. 1B). A p53-dependent reporter was also markedly induced: 7–8-fold after the treatment with myxothiazol and stigmatellin and 2.5-fold after the treatment with antimycin A (Fig. 1C), while there was no effect of the inhibitors in the cells transfected with the control reporter plasmid containing antioxidant response elements (ARE) (Fig. S1D). There was no activation of the p53-dependent reporter after treatment with the inhibitors of complex I (piericidin), complex II (TTFA) and complex IV (KCN or NaN3) (Fig. 1C and Fig. S1C). Similar results were obtained with the RKO reporter cells, although the magnitude of the induction was less explicit due to higher background level (Fig. S1E). We conclude that the induction of p53 response is specifically triggered by the inhibition of ETC at complex III but not by the impairment of the ETC itself.
Fig. 1.
Mitochondrial complex III inhibitors specifically up-regulate p53 and stimulate p53-dependent apoptosis. (A and B) Western analysis of p53 levels in RKO (A) and HeLa (B) cells exposed for 16 h and 18 h, respectively, to the indicated inhibitors: 2 μM myxothiazol (MXT), 1 μM stigmatellin (STG), 2 μM piericidin (PRC), 2 μM rotenone (RTN), 2 μM antimycin A (ANT), 100 μM TTFA (TTFA), 5 mM KCN (KCN), or without drugs (C). (C) Luciferase assay for p53-dependent transcription in HeLa cells exposed for 20 h to the indicated inhibitors, as described in A and B. (D) Western analysis of caspase-3 processing in HCT116 cells exposed to 2 μM myxothiazol (MXT) or MXT + 100 μM ZVAD-FMK. (E) Western analysis of caspase 7 processing in HCT116 wt and p53 -/- cells exposed to 2 μM myxothiazol (MXT) or MXT + 100 μM ZVAD-FMK.
Myxothiazol that demonstrated the most pronounced effect was chosen for further experiments. In RKO cells the treatment with myxothiazol induced a substantial accumulation of p53 in the nuclei, and only trace amounts of p53 were observed in the mitochondrial fraction (Fig. S2). In response to myxothiazol there was a dose-dependent accumulation of p53 (IC50 ∼ 40 nM) reaching maximum at 12–16 h (Fig. S3 A–D and G). In HeLa cells the induction of p53-dependent reporter and accumulation of p53 and the p53-responsive p21 was dose-dependent and started 16–20 h after the addition of the inhibitor (Fig. S3 E–G). In RKO cells we observed an induction of transcripts from the CDKN1A (p21) gene (Fig. S3H) and accumulation of the p21 and Mdm2 proteins (Fig. S3I). Similarly, the induction of p21 was observed in A549 and HCT116 cells.
The myxothiazol treatment induced morphological changes characteristic to apoptotic cell death, which came along with the increase in the fraction of Annexin V-positive cells (Fig. S4) and processing of the effector caspases 3 and 7 (Fig. 1 D and E). The changes were blocked when the myxothiazol treatment was carried out in the presence of the caspase inhibitor ZVAD-FMK (Fig. 1 D and E and Fig. S4) and were substantially suppressed in the p53 knockout HCT116 cells (Fig. 1E and Fig. S4).
The p53 Up-Regulation Induced by Complex III Inhibitors Is Largely ROS- and MMP-Independent.
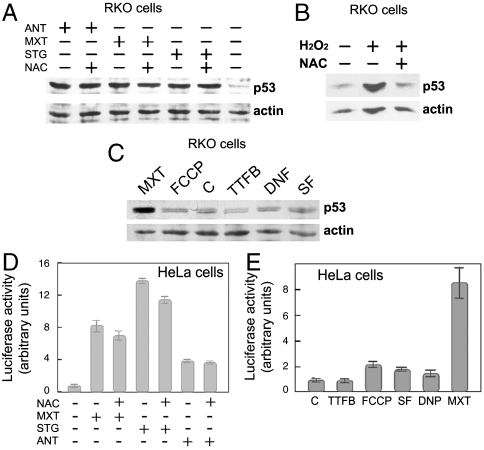
Inhibition of complex III by myxothiazol was previously shown to be associated with increased intracellular levels of ROS (17). Correspondingly, we found a slightly increased level of ROS in the cells treated with complex III inhibitors. The effects were sensitive to the ROS scavenger N-acetylcysteine (NAC) (Fig. S5 A and B). However, pretreatment of RKO and A549 cells with NAC did not prevent the accumulation of p53 in response to complex III inhibitors (Fig. 2A and Fig. S6A), whereas it completely prevented the accumulation of p53 in response to hydrogen peroxide (Fig. 2B). Similarly, in HeLa cells the pretreatment with NAC did not prevent the activation of p53-dependent reporter after treatment with complex III inhibitors (Fig. 2D). Therefore, the observed induction of p53 cannot be explained by a ROS-mediated mechanism.
Fig. 2.
The p53 induction is ROS and MMP-independent. (A) Western analysis of p53 levels in RKO cells exposed to 2 μM antimycin A (ANT), 2 μM myxothiazol (MXT), 1 μM stigmatellin (STG) for 18 h, or without treatment. Where indicated 10 mM NAC was added 1 h before the treatment. (B) Western analysis of p53 levels in RKO cells exposed to 300 μM H2O2 for 20 h. Where indicated 10 mM NAC was added 1 h before the treatment. (C) Western analysis of p53 levels in RKO cells exposed for 18 h to 2 μM myxothiazol (MXT) or uncoupling reagents 0.4 mM dinitrophenol (DNF), 5 μM TTFB, 1 μM FCCP, 1 μM SF or not treated (C). (D) Luciferase assay for p53-dependent transcription in HeLa cells treated for 20 h, as in A. (E) Luciferase assay for p53-dependent transcription in HeLa cells exposed for 20 h to the reagents, as in C.
We also found that treatment with uncoupling reagents SF, TTFB, FCCP, or dinitrophenol fail to induce a notable accumulation of p53 (Fig. 2C and Fig. S6B) or to increase its transcription activity (Fig. 2E). The results argue against the proposed role of mitochondrial membrane depolarization induced by ETC inhibitors in the up-regulation of p53 (16).
Complex III Inhibitors Up-Regulate p53 by Blocking the Dihydroorotate Dehydrogenase Step of Pyrimidine Biosynthesis.
Our results indicate that the p53 response is triggered by the inhibition of ETC at the cytochrome bc1 complex. However, oxidized ubiquinol produced by the cytochrome bc1 complex is essential for the de novo biosynthesis of pyrimidines, namely for the dihydroorotate oxidation by DHODH, an enzyme that is localized at the inner mitochondrial membrane (2). Inhibition of biosynthesis of pyrimidines was observed after blocking complex III with antimycin A (18). We measured changes in the intracellular concentrations of purine and pyrimidine nucleotides following treatment of RKO cells with ETC inhibitors (Table 1). Similar to a specific inhibitor of DHODH leflunomide (19), the treatment with myxothiazol was capable of decreasing the intracellular levels of pyrimidines (approximately fourfold) without affecting purines. Meanwhile, the cytochrome c oxidase (complex IV) inhibitor KCN could affect neither p53 nor the levels of pyrimidines.
Table 1.
Levels of pyrimidine and purine derivatives in the cells treated with myxothiazol, KCN or leflunomide
| Treatment of the cells | Uridine (pmol/106 cells) | Cytidine (pmol/106 cells) | Guanosine (pmol/106 cells) | Adenosine (pmol/106 cells) |
| Control | 4890 ± 222 | 1185 ± 51 | 2580 ± 56 | 8385 ± 225 |
| KCN | 4575 ± 333 | 960 ± 38 | 2835 ± 216 | 7290 ± 312 |
| Leflunomide | 1215 ± 77 | 285 ± 17 | 3540 ± 53 | 9705 ± 266 |
| Myxothiazol | 1260 ± 77 | 270 ± 3 | 3225 ± 141 | 7035 ± 62 |
RKO cells, control or treated with myxothiazol, KCN, or leflunomide were processed as described in Experimental Procedures. The experiments were performed in triplicates. Data are represented as mean ± SEM
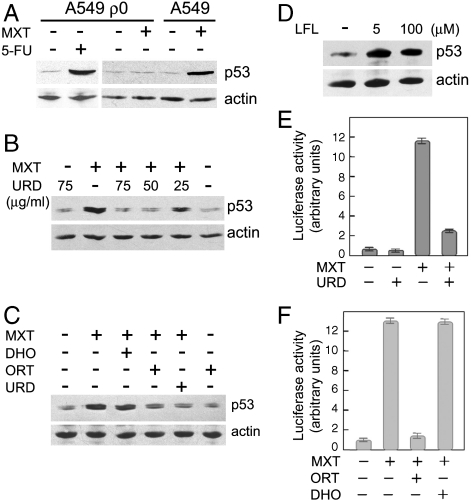
Considering specific role of mitochondrial ETC in p53 up-regulation, we tested an effect of myxothiazol in mtDNA-depleted (ρ0) A549 cells. The ρ0 cells were maintained in a medium supplemented with uridine and pyruvate to complement the metabolic deficiency connected with the defective ETC. Treatment with 5-fluorouracil caused an induction of p53 in the control A549 cells as well as in their ρ0 derivatives. However, myxothiazol was capable of inducing p53 accumulation only in the control cells and failed to activate p53 in the ρ0 A549 cells (Fig. 3A).
Fig. 3.
DHODH deficiency leads to up-regulation of p53 in response to myxothiazol. (A) Myxothiazol fails to activate p53 in the ρ0 cells. Western analysis of p53 levels in A549 ρ0 and A549 wt cells exposed to 2 μM myxothiazol (MXT) or 5-FU (10 μg/ml) for 18 h. (B and C) Uridine and orotate (but not dihydroorotate) suppress myxothiazol-induced accumulation of p53. Western analysis of p53 in RKO cells exposed to 1 μM myxothiazol (MXT) for 16 h in the presence of indicated concentrations of uridine (URD) (B) or in the presence of 50 μg/ml uridine (URD) or 1 mM orotate (ORT) or 1 mM dihydroorotate (DHO) (C). (D) Western analysis of p53 in HeLa cells exposed to 5 or 100 μM DHODH inhibitor leflunomide (LFL) for 48 h. (E and F) Uridine and orotate (but not dihydroorotate) suppress myxothiazol-induced activation of the p53-dependent reporter. Luciferase reporter assay of p53-dependent transcription in HeLa cells exposed to 1 μM myxothiazol (MXT) for 22 h in the presence of 50 μg/ml uridine (URD) (E) or in the presence of 1 mM orotate (ORT) or 1 mM dihydroorotate (DHO) (F).
The result could indicate that the activation of p53 in A549 cells requires an intact mitochondrial function, which is absent in the ρ0 cells. However, as the medium for the ρ0 cells contained uridine, an equally possible explanation was that the uridine supplementation may complement the metabolic deficiency that triggers the p53 response. Indeed, a supplementation with uridine at 50–75 μg/ml prevented the p53 induction in response to myxothiazol (Fig. 3B) and decreased substantially the induction of p53-dependent reporter in response to complex III inhibitors (Fig. 3E and Fig. S7A). Orotate, the product of the reaction catalyzed by DHODH, unlike dihydroorotate, the substrate in the same reaction, prevented the activation of p53 in response to complex III inhibitors (Fig. 3C and F and Fig. S7 A and B). Furthermore, the DHODH inhibitor leflunomide was also capable of inducing p53 accumulation (Fig. 3D). The results indicate that the myxothiazol-induced activation of p53 is driven by the deficiency in pyrimidines rather than by the inhibition of the ETC.
Complex III Inhibitors Activate p53 Through Interfering with Its Degradation in 20S Proteasomes.
In normal cells a p53 protein level is maintained through a controlled degradation in 26S and 20S proteasomes (20, 21). Targeting to the 26S proteasomes is achieved through a polyubiquitinylation by Mdm2 and other E3-ligases (22), and the process is interrupted by stresses, through phosphorylation of p53 at its N termini or through overexpression of the p14ARF inhibitor protein (23). Up-regulation of p53 can be also achieved through binding to NAD(P)H:quinone oxidoreductase 1 (NQO1) and NRH:quinone oxidoreductase 2 (NQO2) that provide protection form ubiquitin-independent degradation of p53 in 20S proteasomes (24, 25).
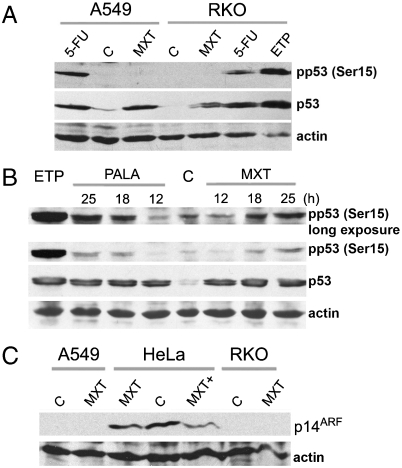
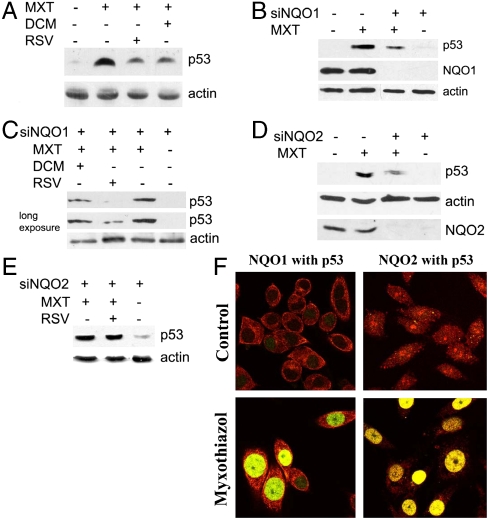
In A549 and RKO cells we found strong stimulation of Ser15 phosphorylation after treatment with 5-fluorouracil or with etoposide but not after treatment with myxothiazol for 12–16 h (Fig. 4 A and B), the time points at which the p53 accumulation reaches maximal levels. Modest accumulation of phosho-Ser15 p53 was observed only by 18–25 h. A similar dynamic of Ser15 phosphorylation was observed in RKO cells after treatment with N-(phosphonacetyl)-L-aspartate (PALA), another inhibitor of de novo pyrimidine nucleotide biosynthesis acting upstream of DHODH (Fig. 4B). Then we compared changes in the p14ARF levels following myxothiazol treatment. In HeLa cells the treatment resulted in a slight decrease in the level of p14ARF (Fig. 4C), while in RKO and A549 cells the levels were undetectable despite the treatment (Fig. 4C). Yet we found that treatment with either NQO1 inhibitor dicoumarol or NQO2 inhibitor resveratrol produced a similar in magnitude suppression of the myxothiazol-induced stabilization of p53 in RKO cells (Fig. 5A) and prevented the stimulation of p53-dependent reporter in HeLa cells (Fig. S8A). Similar inhibition by dicoumarol of the p53 stabilization and reporter activation was observed in RKO and HeLa cells treated with pyrimidine biosynthesis inhibitor PALA (Fig. S8 B and C). Stable inhibition of either NQO1 or NQO2 expression in RKO cells by introduction of lentiviral constructs expressing appropriate shRNAs each resulted in a twofold reduction in myxothiazol-induced p53 stabilization (Fig. 5 B and D). The effect of dicoumarol was absent in the cells with inhibited NQO1, while resveratrol was still potent in reducing the p53 stabilization (Fig. 5C). Similarly, resveratrol did not affect the induction of p53 in RKO cells with knocked-down NQO2 (Fig. 5E). Considering that NQO1 and NQO2 participate in the induction of p53 we tested by confocal microscopy intracellular localization of the proteins following myxothiazol treatment. We found that in untreated RKO cells NQO1 was present exclusively in the cytosol, and NQO2 was distributed throughout the cells at very low level with no apparent nuclear localization. In the cells treated for 12 h with 2 μM myxothiazol both NQO1 and NQO2 produced bright nuclear staining, which perfectly colocalized with the accumulated nuclear p53 (Fig. 5F and Figs. S9 and S10). The result suggests that the accumulation of p53 in response to complex III inhibitors depends on NQO1 and NQO2, which acquire nuclear localization and possibly assist in nuclear retention and protection from the degradation of p53 in 20S proteasomes.
Fig. 4.
Up-regulation of p53 by myxothiazol does not correlate with the p53 phosphorylation at Ser15, or with the accumulation p14ARF. (A) Western analysis of phosphorylated p53 (Ser15) in RKO and A549 cells not treated (C) or exposed to 2 μM myxothiazol (MXT) or DNA-damaging drugs 5-FU (10 μg/ml) and 50 μM etoposide (ETP) for 16 h. (B) Western analysis of phosphorylated p53 (Ser15) in RKO cells exposed to 2 μM myxothiazol (MXT) or 250 μM PALA for 12, 18, or 25 h, or in nontreated cells (C). Treatment with 50 μM etoposide (ETP) for 18 h served as a positive control. (C) Western analysis of p14 ARF in RKO, HeLa, and A549 cells exposed to 2 μM myxothiazol for 18 h (MXT) or for 24 h (MXT+) or in not treated cells (C).
Fig. 5.
p53 induction in response to myxothiazol depends on NQO1 and NQO2. (A) Western analysis of p53 in RKO cells treated with 2 μM myxothiazol (MXT) for 10 h and 300 μM dicoumarol (DCM) for 4 h or 50 μM resveratrol (RSV) for 10 h. (B) Western analysis of p53 level in NQO1 knock-down (siNQO1) and control RKO cells after treatment with 2 μM myxothiazol for 10 h. (C) Western analysis of p53 in NQO1 knock-down RKO cells treated with 2 μM myxothiazol for 10 h, and 300 μM dicoumarol for 4 h or 50 μM resveratrol for 10 h. (D) Western analysis of p53 in NQO2 knock-down (siNQO2) and control RKO cells after treatment with 2 μM myxothiazol for 10 h. (E) Western analysis of p53 in NQO2 knock-down RKO cells treated with 2 μM myxothiazol and 50 μM resveratrol for 10 h. (F) Colocalization of p53 with NQO1 or with NQO2 after myxothiazol treatment. RKO cells were incubated with 2 μM myxothiazol or without (control) for 12 h and stained with antibodies to NQO2 or NQO1 (red) and p53 (green). Then pictures with red and green fluorescence were overlaid. Yellow color depicts colocalization of two proteins.
Discussion
Changes in mitochondrial respiration have been reported in many pathologic conditions (9–12) although not much is known about signals from an impaired electron transport chain that could induce p53 stress response thereby contributing to tissue damage. Previous evidence suggests that mitochondrial ETC could play a role in stress-induced activation of p53 (13–16). However, in the present study we found that the inhibition of mitochondrial ETC does not necessarily generate p53-activating signals. Blocking ETC with complex I inhibitors piericidin and rotenone, or with complex IV inhibitors KCN and NaN3, was not sufficient to induce p53 response in four human carcinoma cell lines expressing wild-type p53. Similarly, no activation of p53 was observed in response to dissipation of MMP by protonophorous uncoupling reagents. The results indicate that simple deficiency in the energy-generating mitochondrial functions is not sufficient to elicit the p53-dependent stress response.
Yet we found that substantial p53 activation can be induced in response to specific inhibitors of complex III. Remarkably, the activation was observed also in HPV-positive HeLa cells, suggesting that the NQO1/2-mediated induction circumvents the E6-mediated degradation of p53. We also found that the inhibitors induce p53 dependent apoptotic cell death. The data suggest that although the inhibition of ETC by itself is not sufficient for p53 activation, a function linked to the mitochondrial complex III could be responsible for triggering the p53 response. Of note, the activation of p53 by complex III inhibitors cannot be abrogated by the ROS scavenger NAC suggesting that it is ROS-independent.
The function of DHODH, an enzyme involved in the de novo biosynthesis of pyrimidines, depends on the activity of complex III. Correspondingly, we found that myxothiazol induced a depletion in pyrimidines without affecting purines. Supplementation with uridine or with orotate, the product of the reaction catalyzed by DHODH, abolished the p53 activation, whereas the DHODH substrate dihydroorotate produced no effect. Finally, similar to complex III inhibitors, the specific inhibitor of DHODH leflunomide induced up-regulation of p53. The results confirm that the impairment of de novo pyrimidine biosynthesis at the DHODH step triggers p53 activation in response to complex III inhibitors.
The smaller N-terminal domain of DHODH forms a tunnel in the inner mitochondrial membrane that provides access of ubiquinone to FMN, which is bound at the active site of the larger catalytic domain [2]. DHODH relies on ubiquinone that is provided by complex III, thereby forming a functional link between mitochondrial ETC and pyrimidine biosynthesis. The inhibition of complex III forces ubiquinone to its completely reduced state, ubiquinol, which is no longer able to accept electrons from dihydroorotate in the reaction catalyzed by DHODH.
Remarkably, complex IV inhibitor KCN, which is known to block oxidation of ubiquinol in isolated mitochondria, did not induce p53 or affect pyrimidine nucleotide levels. Neither did it affect the myxothiazol-induced up-regulation of p53. Probably a subpool of ubiquinone that is controlled by a cyanide-resistant and myxothiazol-sensitive pathway of ubiquinol oxidation is critical for maintaining the DHODH activity.
The depletion of ribonucleotide pools is known to be a strong inducer of p53 response, which in normal fibroblasts results in a p53-dependent arrest at the G1/S phase of the cell cycle (26). Activation of p53 was reported after treatment of normal human fibroblasts with PALA, an inhibitor of aspartate transcarbamylase acting upstream of the DHODH in the de novo pyrimidine nucleotide biosynthesis pathway. In fibroblasts such activation of p53 leads to reversible cell cycle arrest that protects the cells from severe DNA damage and apoptosis (27–29). In the human epithelial carcinoma cell lines used in this study, the activation of p53 response by complex III inhibitors or PALA induced apoptosis. The difference could be attributed to tissue-specific variations, although the question requires additional studies.
Although it was previously reported that in a human fibroblast model the induction of p53 after treatment with PALA proceeds in the absence of DNA damage (29), recently it was found that the response in fact involves a limited accumulation of DNA damage that triggers ATR-CHK1-dependent p53 phosphorylation at Ser15 (30). Yet in human carcinoma models we found that myxothiazol or PALA, unlike etoposide or 5-FU, can up-regulate p53 even without phosphorylation at Ser15. A limited accumulation of phospho-Ser15 p53 was detected at a much later time points when the p53 response reached its maximum. Therefore, in epithelial carcinoma cells, unlike in normal fibroblasts, the DNA damage response pathway is not likely to be a major contributor to the p53 induction under condition of pyrimidine nucleotides deficiency. Neither did we find any myxothiazol-induced changes in the p14ARF levels. Instead, our data suggest that NQO1 and NQO2 appear to contribute to the stabilization and activation of p53. This recently identified pathway involves inhibition of the ubiquitin-independent degradation of p53 in 20S proteasomes through its binding to NQO1 or NQO2 (20, 24, 25). We found that dicoumarol, which is known to interfere with the NQO1 binding to p53, and resveratrol, a highly specific inhibitor of NQO2 (31), can mitigate the stabilization of p53 and the induction of p53-dependent reporter in response to treatment with myxothiazol or PALA. Similar to the drug inhibitors, the p53-inducing effect of myxothiazol was alleviated in the cells with NQO1 or NQO2 downregulated by specific shRNAs. Finally, we observed a substantial intracellular redistribution of NQO1 and NQO2 in response to myxothiazol treatment. The enzymes moved from cytoplasm to nuclei were they colocalized with the induced p53. The results suggest that in response to pyrimidine nucleotides deficiency in epithelial cells both NQO1 and NQO2 participate in the activation of p53, possibly by their association with p53 in the nuclei and protection from its degradation in 20S proteasomes.
Unlike a rather clear understanding of the mechanisms involved in the p53 induction in normal fibroblasts (30), in epithelial cell models the exact pathway of NQO1/2-mediated p53 accumulation in response to exhausted pools of pyrimidine nucleotides is yet to be defined. Previous studies indicate that inhibition of transcription by RNA polymerase II at preelongation step or inhibition of transcription by RNA polymerase I can induce accumulation of p53 even in the absence of DNA damage and without phosphorylation of p53 at Ser15 (32–34). Possibly, the described NQO1- and NQO2-dependent alternative pathway of p53 induction in the cells with inhibited respiration and deficient nucleotide pools is triggered by similar perturbations in RNA biosynthesis. Further studies are needed to identify mechanistic link between the exhausted pools of nucleotides and the p53-stabilizing activity of NQO1 and NQO2.
Experimental Procedures
Cell Lines and Chemicals.
The human cell lines bearing wild-type p53-A549, HeLa, RKO, and HCT116 were grown in DMEM supplemented with 10% fetal calf serum (HyClone). The A549 cells with depleted mtDNA (ρ0 cells) were selected and cultured in the same medium supplemented with 50 μg/ml uridine, 100 μg/ml pyruvate, and 50 ng/ml ethidium bromide, as described (35). Cells were incubated with the chemicals listed below as indicated in the legends to figures. RKO cells with knocked-down expression of NQO1 or NQO2 were obtained by introduction of lentiviral constructs pLKO1 expressing appropriate shRNAs (TRCN0000003769 and TRCN0000036711) obtained from Sigma-Aldrich Inc. and selection in the medium containing 2 μg/ml puromycin for 4 d.
Mitochondrial ETC inhibitors myxothiazol, antimycin A, piericidin, rotenone, and thenoyl trifluoroacetone (TTFA), uncouplers dinitrophenol (DNP), 3,5-di-tert-butyl-4-hydroxybenzylidenemalononitrile (SF), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), 4,5,6,7-tetrachloro-2-trifluoromethylbenzimidazole (TTFB) were from Sigma-Aldrich Inc.; stigmatellin was from Fluka. The inhibitors were added in concentrations necessary to completely block respiration of HeLa cells (myxothiazol, antimycin A, stigmatellin, piericidin, rotenone, KCN) or stimulate respiration suppressed by oligomycin (uncouplers) as described (36).
Antibodies and Western Analysis.
Antibodies to p53 (DO-1), p21 (F-5), NQO2 (H-50) and actin (C-2) were from Santa Cruz Biotechnology Inc.; antibodies to p14ARF (4C6/4) and phospho-p53 (Ser15) (16G8) were from Cell Signaling Technology Inc.; antibodies to NQO1 (A180, ab28947) were from Abcam. Inc.; and antibodies to caspase-3 (4-1-18) and caspase-7 (1-1-10) were a gift of Yuri Lazebnik.
For Western analysis cells were lyzed in reporter lysis buffer (Promega Inc.). For phospho-p53 (Ser15) analysis cells were harvested, washed with PBS, and immediately lyzed by boiling in SDS sample buffer for 5 min. Equivalent amounts of total protein were subjected to 12–18% SDS-polyacrylamide gel electrophoresis and processed as previously described (37).
Reporter Assays.
The luciferase reporter plasmids p53RE-luc, harboring p53 response elements (38), or pARE/luc, harboring antioxidant response elements (39), were transfected into HeLa cells along with β-galactosidase expression plasmid pcDNA4/HisMax/lacZ (Invitrogen) using ExGene 500 transfection reagent (Fermentas). Twenty-four hours after the transfection cells were treated as indicated in legends to figures and incubated routinely for 20 h. Cells were lyzed in reporter lysis buffer (Promega Inc.), and luciferase and β-galactosidase assays were performed with the Luciferase Assay System (Promega Inc.). The reporter luciferase activities were normalized relative to the β-galactosidase activity. All experiments were performed in triplicates. Data are represented as mean ± SEM.
Measuring Concentrations of Purine and Pyrimidine Nucleosides.
50% confluent RKO cells (control or treated with 2 μM myxothiazol or 5 mM KCN for 16 h or 50 μM leflunomide for 40 h) were harvested and washed in PBS. The pellets containing approximately 106 cells were disrupted with 150 μl 10% trichloroacetic acid (TCA) and centrifuged for 2 min at 12,000 rpm. TCA in the supernatants was back-extracted with water-saturated diethyl ether to pH 5.0. To convert the nucleotides (NMPs, NDPs, and NTPs) into the corresponding nucleosides, the extracts were treated with 150 U of bacterial alkaline phosphatase and 1 U of calf intestine alkaline phosphatase (Fermentas) at 37 °C overnight. Concentration of nucleosides in the extracts was analyzed by ACQUITY Ultra Performance LC/MS (Waters).
Confocal Microscopy.
Cells were fixed in 4% formaldehyde/PBS, permeabilized with 4% formaldehyde/0.2% Triton X100/PBS, blocked in 3% BSA/PBS for 1 h at 4 °C, and incubated overnight at 4 °C with anti-p53 monoclonal antibody DO-1 (1∶200), and then for 6 h with anti-mouse-FITC antibodies (1∶200). The cells were again blocked with 3% BSA/PBS, incubated with anti-NQO1 monoclonal antibodies (1∶200), or with rabbit polyclonal anti-NQO2 antibodies (1∶50) overnight at 4 °C, and stained with Rhodamine-conjugated goat anti-mouse or anti-rabbit secondary antibodies (1∶200) for 6 h at 4 °C. Secondary antibodies were from Santa Cruz Biotech, Inc. The cells were washed with PBS and mounted in Tris-glycerol (1∶9, pH 8.0) for microscopy. For negative controls the procedure was the same, except the primary antibodies against NQO1 or NQO2 were omitted from the analysis. Instead, the controls were incubated with PBS overnight at 4 °C. Cells were visualized with a Carl Zeiss LSM 510 confocal laser scanning microscope by using a 100x oil-immersion objective, appropriate filter sets, argon (488 nm), and helium-neon (543 nm) lasers.
Supplementary Material
Acknowledgments.
We thank V.N. Tashlitsky for measuring concentrations of nucleosides by LC/MS and S.V. Melnikov for help with Flow cytometry. Funding support was from National Institutes of Health Grants CA104903 and AG025276 (P.M.C.); from Howard Hughes Medical Institute Grant 55005603 (P.M.C.); from the Russian Foundation for Basic Research Grants 09-04-01246a (A.G.E), 08-04-00686a (P.M.C.), and 07-04-00335a (B.V.C.); from the Russian Academy of Sciences Program on Molecular and Cellular Biology (P.M.C); and from Russian Ministry of Education and Science Program NK-542P, P334 (A.B.V.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910885107/-/DCSupplemental.
References
- 1.McBride HM, Neuspiel M, Wasiak S. Mitochondria: More than just a powerhouse. Curr Biol. 2006;16(14):R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient pathway. J Biol Chem. 2004;279(32):33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH. p53: Death star. Cell. 2000;103(5):691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 4.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17(6):631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Olovnikov IA, Kravchenko JE, Chumakov PM. Homeostatic functions of the p53 tumor suppressor: Regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19(1):32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53- dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404(6773):42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 7.Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: Balancing aerobic respiration and glycolysis. J Bioenerg Biomembr. 2007;39(3):243–246. doi: 10.1007/s10863-007-9083-0. [DOI] [PubMed] [Google Scholar]
- 8.Vahsen N, et al. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23(23):4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405(1):1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 10.Jaeschke H, et al. Mechanisms of hepatotoxicity. Toxicol Sci. 2002;65(2):166–176. doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- 11.Berthiaume JM, Wallace KB. Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol Toxicol. 2007;23(1):15–25. doi: 10.1007/s10565-006-0140-y. [DOI] [PubMed] [Google Scholar]
- 12.Favier J, et al. Hereditary paraganglioma/pheochromocytoma and inherited succinate dehydrogenase deficiency. Horm Res. 2005;63(4):171–179. doi: 10.1159/000084685. [DOI] [PubMed] [Google Scholar]
- 13.Chandel NS, VanderHeiden MG, Thompson CB, Schumacker PT. Redox regulation of p53 during hypoxia. Oncogene. 2000;19(34):3840–3848. doi: 10.1038/sj.onc.1203727. [DOI] [PubMed] [Google Scholar]
- 14.Karawajew L, Rhein P, Czerwony G, Ludwig WD. Stress-induced activation of the p53 tumor suppressor in leukemia cells and normal lymphocytes requires mitochondrial activity and reactive oxygen species. Blood. 2005;105(12):4767–4775. doi: 10.1182/blood-2004-09-3428. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Biju MP, Wang MH, Haase VH, Dong Z. Cytoprotective effects of hypoxia against cisplatin-induced tubular cell apoptosis: Involvement of mitochondrial inhibition and p53 suppression. J Am Soc Nephrol. 2006;17(7):1875–1885. doi: 10.1681/ASN.2005121371. [DOI] [PubMed] [Google Scholar]
- 16.Behrend L, Mohr A, Dick T, Zwacka RM. Manganese superoxide dismutase induces p53-dependent senescence in colorectal cancer cells. Mol Cell Biol. 2005;25(17):7758–7769. doi: 10.1128/MCB.25.17.7758-7769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young TA, Cunningham CC, Bailey SM. Reactive oxygen species production by the mitochondrial respiratory chain in isolated rat hepatocytes and liver mitochondria: Studies using myxothiazol. Arch Biochem Biophys. 2002;405(1):65–72. doi: 10.1016/s0003-9861(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 18.Gattermann N, et al. Severe impairment of nucleotide synthesis through inhibition of mitochondrial respiration. Nucleos Nucleot Nucl. 2004;23(8-9):1275–1279. doi: 10.1081/NCN-200027545. [DOI] [PubMed] [Google Scholar]
- 19.Greene S, Watanabe K, Braatz-Trulson J, Lou L. Inhibition of dihydroorotate dehydrogenase by the immunosuppressive agent leflunomide. Biochem Pharmacol. 1995;50(6):861–867. doi: 10.1016/0006-2952(95)00255-x. [DOI] [PubMed] [Google Scholar]
- 20.Tsvetkov P, Reuven N, Shaul Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010;17(1):103–108. doi: 10.1038/cdd.2009.67. [DOI] [PubMed] [Google Scholar]
- 21.Chumakov PM. Versatile functions of p53 protein in multicellular organisms. Biochemistry-Moscow. 2007;72(13):1399–1421. doi: 10.1134/s0006297907130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21(3):307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10(1):94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 24.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitinindependent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 19(3):316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong X, Kole L, Iskander K, Jaiswal AK. NRH: Quinone oxidoreductase 2 and NAD(P)H:quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 2007;67(11):5380–5388. doi: 10.1158/0008-5472.CAN-07-0323. [DOI] [PubMed] [Google Scholar]
- 26.Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10(8):934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal ML, et al. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc Natl Acad Sci USA. 1998;95(25):14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal MK, et al. Macrophage inhibitory cytokine 1 mediates a p53- dependent protective arrest in S phase in response to starvation for DNA precursors. Proc Natl Acad Sci USA. 2006;103(44):16278–16283. doi: 10.1073/pnas.0607210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidita G, Polizzi D, Costanzo G, Costa S, DiLeonardo A. Differential gene expression in p53-mediated G(1) arrest of human fibroblasts after gammairradiation or N-phosphoacetyl-L-aspartate treatment. Carcinogenesis. 2000;21(12):2203–2210. doi: 10.1093/carcin/21.12.2203. [DOI] [PubMed] [Google Scholar]
- 30.Hastak K, et al. DNA synthesis from unbalanced nucleotide pools causes limited DNA damage that triggers ATR-CHK1-dependent p53 activation. Proc Natl Acad Sci USA. 2008;105(17):6314–6319. doi: 10.1073/pnas.0802080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buryanovskyy L, et al. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry. 2004;43(36):11417–11426. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20(9):3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derheimer FA, et al. RPA and ATR link transcriptional stress to p53. Proc Natl Acad Sci USA. 2007;104(31):12778–12783. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ljungman M. The transcription stress response. Cell Cycle. 2007;6(18):2252–2257. doi: 10.4161/cc.6.18.4751. [DOI] [PubMed] [Google Scholar]
- 35.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 36.Shchepina LA, et al. Oligomycin, inhibitor of the F0 part of H+-ATPsynthase, suppresses the TNF-induced apoptosis. Oncogene. 2002;21(53):8149–8157. doi: 10.1038/sj.onc.1206053. [DOI] [PubMed] [Google Scholar]
- 37.Sukhacheva EA, et al. Sensing prothymosin alpha origin, mutations and conformation with monoclonal antibodies. J Immunol Methods. 2002;266(1-2):185–196. doi: 10.1016/s0022-1759(02)00098-4. [DOI] [PubMed] [Google Scholar]
- 38.Zakharova NI, et al. [Influence of prothymosin alpha and its mutants on activity of the p53 tumor suppressor] Mol Biol (Mosk) 2008;42(4):673–684. [PubMed] [Google Scholar]
- 39.Karapetian RN, et al. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: Implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25(3):1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.