Abstract
Gene regulatory networks are based on simple building blocks such as promoters, transcription factors (TFs) and their binding sites on DNA. But how diverse are the functions that can be obtained by different arrangements of promoters and TF binding sites? In this work we constructed synthetic regulatory regions using promoter elements and binding sites of two noninteracting TFs, each sensing a single environmental input signal. We show that simply by combining these three kinds of elements, we can obtain 11 of the 16 Boolean logic gates that integrate two environmental signals in vivo. Further, we demonstrate how combination of logic gates can result in new logic functions. Our results suggest that simple elements of transcription regulation form a highly flexible toolbox that can generate diverse functions under natural selection.
Keywords: evolution, gene regulation, regulatory logic, signal integration
Processes in free-living cells are modulated to fit the environmental conditions. The possible programs a given cell can execute are defined by the cell's genome, and the optimal program is selected based on the level of environmental signals sensed by the cell. Regulation of gene expression is a main component in the selection of the optimal program (1). A major class of regulators of gene expression consists of transcription factors (TFs) that sense the level of small molecule signals and bind specific sites in the gene's cis-regulatory region. The logic of TF-mediated regulation depends on whether the TF is an activator (positive control) or a repressor (negative control) and whether the apo or the holo form of the TF is active. However, expression of many genes depends on more than one signal. In these cases the combined effect of the incoming signals often results in a regulatory logic that is different from the sum of the logic observed in case of the individual signals. In the simplest case, where only two signals are integrated, the combined effect of two regulators, responding to different signal molecules, can be qualitatively described using Boolean functions, e.g., AND or OR logic gates (2–8). Such genetic logic gates are essential building blocks of next-generation synthetic gene networks (9). Colonization of new environments can be facilitated by reprogramming regulatory networks to obtain new functions. The degree to which networks can be reprogrammed by mutations may be termed “genetic flexibility.”
Previous studies suggested that gene regulatory sequences and interactions between TFs play a major role in network flexibility (3, 5). The pattern of gene regulation—and therefore network function—can be easily changed merely by repositioning of repressor and activator binding sites in gene regulatory sequences (10, 11). Analysis of the lac regulatory region demonstrated that even point mutations in TF binding sites can significantly change the logic of signal integration (5). Theoretical studies suggested that regulatory elements (promoter sequences, TFs and their binding sequences, etc.) constitute a “molecular Lego” set that can be used to assemble a gene regulatory sequence with a desired function (3).
In this work we aimed to explore the potentials and limitations in the function of synthetic regulatory regions constructed from the elements of a natural genetic regulatory system.
Results
Model System.
We chose the galactose network of Escherichia coli as a model system for our study (6). Sugar utilization systems are preferred models to study signal integration because they are usually two-input systems (5, 6, 12–15). One input is the presence of a specific sugar (d-galactose in our system), sensed by a sugar-specific transcription factor (GalR in our case). The other input is the level of cAMP, corresponding to the level of glucose shortage (16). cAMP is synthesized intracellularly and sensed by the cAMP Receptor Protein (CRP). Another advantage of using a sugar utilization system is the versatility in the control mechanisms (17) and in the function of the regulators involved. TFs involved in these systems are often dual-function regulators, acting as repressors or activators depending on the context of their binding sites in the cis-regulatory regions (6, 18–20). There are two major differences between sugar utilization systems and the model system used in the theoretical studies (3). In the sugar utilization systems, levels of the active TFs are primarily dependent on signal molecule concentrations, and there is no glue-like interaction between the two different TFs.
Experimental Setup.
We used E. coli CH1200 cells in our experiments, which are cyaA mutant derivatives of MG1655 (21). These cells are unable to synthesize cAMP, therefore intracellular cAMP and galactose levels can be controlled by the addition of these molecules to the growth medium. In the CH1200 cells we replaced the natural regulatory region of the uidABC (gus) operon (encoding a β-d-glucuronidase and a β-d-glucuronide transport system) with synthetic regulatory sequences constructed from the elements of the gal regulatory system (Fig. 1). The most important elements of the gal “Lego set” used for the construction are different promoter elements of the gal regulon (e.g., −10, extended −10, and −35 boxes), cAMP-CRP binding sites, and GalR binding sites. The strength of specific GalR-DNA and cAMP-CRP-DNA interactions can be tuned by mutations in the binding sites (15, 22–24). A crucial feature of the system is that GalR can bind distant operator sites on a DNA molecule forming a DNA loop, leading to a strong repression of the promoters located between the operator sites (25, 26). This feature is not unique in sugar regulatory systems (19, 20, 27, 28).
Fig. 1.
(A) Representation of a promoter as a logic gate. Small molecule signals (here cAMP and D-gal) interact with specific transcription factors (here GalR and CRP) that bind a specific sequence in the regulatory region and influence transcription of the reporter gene (here uidA). The logic of signal integration, i.e., the activity of the promoter at different combinations of signals (B), depends on the activity and on the combined action of regulatory proteins. In this case, (Lower Right) (D-gal) represents the basal promoter activity (none of the regulators bind to DNA), whereas (Lower Left) (no signals) represents promoter activity when GalR is bound. In the presence of cAMP (Upper Left) both GalR and cAMP-CRP can bind DNA, whereas in the presence of both signals (Upper Right) only cAMP-CRP can regulate the promoter. (C) Schematic drawing of the reporter construct used for testing signal integration at the engineered promoters. Red arrows represent ORFs. Regulatory sequences are inserted between the EcoRI and PstI sites.
Construction of Synthetic Logic Gates.
In a previous work we found that regulatory regions with similar arrangements of TF binding sites and promoter elements can perform different signal integration logics (6). Signal integration depends on several factors that are not easily predictable based on sequence information. Synthetic regulatory regions, aimed to perform different logics of signal integration, were designed based on the available knowledge on signal integration at the natural regulatory regions of the gal regulon. First, sequences were synthesized by PCR and inserted in a cloning plasmid between a terminator region and the 5′ part of the uidA coding sequence. We often created pools of plasmid clones differing in the TF binding sequences or in the promoter elements by having randomized positions in the oligonucleotides used in the PCR. Then the constructed regulatory regions, together with the upstream terminator region, were inserted upstream of the uidA ORF in the E. coli CH1200 chromosome by recombineering (29). Signal integration in the constructed cells was evaluated on a population level, by plating cells on four different media, covering all four possible combinations of signals (cAMP, galactose, both, none) (Fig. 2), and a chromogenic substrate of β-d-glucuronidase (X-gluc). The DNA sequence of regulatory regions in cells performing Boolean-type signal integration was determined. The criterion of Boolean-type integration is that the high and low states of reporter gene expression (blue or white color of the colony on screening plates) can be clearly distinguished. We note here that the high levels present at different signal combinations are not necessarily the same. We tested 40 different constructs made by rearranging TF binding sites and modifying the basal activity of promoters. By this approach we managed to create 11 different Boolean-type signal integration patterns of the 16 possible (Fig. 2).
Fig. 2.
All 16 possibilities for the Boolean-type integration of two input signals. For the 12 gates successfully created the schematic structures of regulatory regions are shown (Left) of the logic gates (boxes). The figure is not drawn to scale. Red boxes represent predicted GalR binding sites, yellow boxes represent predicted cAMP-CRP binding sites, black arrows represent promoters. Numbers in the boxes show the distance of the operator site (Center) from the transcription start site of the promoter. The activity of the reporter gene (uidA) was monitored in the four possible combinations of D-gal and cAMP. The presence of signals is indicated in (A), and the same arrangement was used in B–L. Blue color results from successful conversion of the chromogenic substrate (X-gluc) by the UidA protein, indicating that the reporter gene is expressed (ON) in a given combination of input signals. The lack of blue colors indicates that expression is OFF. Examples of reporter enzyme activities in logarithmic phase cells are provided in the Supporting Information. GenBank accession numbers are shown for constructs longer than 50 bp.
The TRUE and the FALSE gates were constructed so that they do not respond to any of the input signals. The TRUE construct is based on the regulatory region of the galETKM operon, in which the P2galE promoter is inactivated by the G-19T point mutation (30), the −35 box of P1galE is enhanced (CACTTT to TAGTGT) and the GalR and cAMP-CRP binding sites are deleted. The FALSE construct contains a sequence with no promoter activity.
Four of the constructed gates respond only to one of the signals (D-gal, ND-gal, cAMP, NcAMP).
The D-gal logic was obtained in several variants of the galETKM regulatory region containing the natural gal operator sites (OE and OI). The construct shown in Fig. 2C contains the G-14A mutation that reduces the activity of the P1galE promoter (30). In this case the DNA loop inhibits transcription when d-galactose is absent. In the presence of d-galactose DNA looping is inhibited, allowing transcription at the basal level of the promoter, which can be varied by mutations.
The wild-type mglBAC regulatory region, was previously shown to perform cAMP logic (6). The PmglB promoter is not active in the absence of cAMP-CRP. Although a GalR binding site is present, GalR binding only partially inhibits the cAMP-CRP mediated activation of the PmglB promoter (Fig. 2E). This construct also contains the sequence encoding the 5′ untranslated region (5′ UTR) of the mglBAC mRNA.
The ND-gal gate was obtained unexpectedly when the GalR binding site was deleted from the mglBAC regulatory region used for the cAMP gate. Although a preliminary analysis shows that the sequence between +39 to +149 contains information that is necessary for performing ND-gal logic, further studies are needed to elucidate the mechanism of regulation.
The NcAMP gate is based on the P2galE promoter, which is inhibited by the cAMP-CRP complex. In this construct the P1galE promoter is inactivated by several mutations (G-14A, C-13A, ∇13T, Δ-2T, A+1C) (30, 31). Because only partial repression was obtained using the wild-type or the consensus cAMP-CRP binding sequence at the natural position (resulting in TRUE-like logic), we used the consensus sequence in the construct shown in Fig. 2F, together with a second cAMP-CRP binding site inserted at +5. Because the P2galE promoter has a basal activity it allows transcription in the absence of cAMP-CRP. The requirement for two binding sites for sufficient repression can be explained by the fact that transient occupancy of the binding site can result in strong activation; however, for strong repression the TF has to occupy the regulatory site most of the time.
The rest of the gates where the two signals are actually integrated fall into two groups, one group contains a promoter with basal activity (OR, XOR, NAND, cAMP implies D-gal, D-gal nimplies cAMP, colored in the bottom right box), and the other group has a promoter with no basal activity (AND, NOR, EQUAL, D-gal implies cAMP, cAMP nimplies D-gal, white in the bottom right box).
From the first group, three gates were constructed (OR, cAMP implies D-gal, D-gal nimplies cAMP). These are the most complex in structure among the gates created. The OR construct is based on the galETKM regulatory region and contains overlapping GalR and cAMP-CRP binding sites at the positions of the natural OE and OI operator sites. Transcription is inhibited by the GalR mediated DNA loop in the absence of signals. Formation of the DNA loop is inactivated in the presence of galactose and also in the presence of cAMP because cAMP-CRP binding inhibits GalR binding to the operator(s) (Fig. 2G).
The cAMP implies D-gal gate is based on the galETKM regulatory region in which the P2galE promoter is partially inhibited by the cAMP-CRP complex. In this construct the P1galE promoter is inactivated by several mutations to eliminate its activity (G-14A, C-13A, ∇13T, Δ-2T, A+1C, A+3T) (30, 31). The OI operator is replaced by a weaker binding site (GTGAAAGCGATGTCAC), therefore GalR mediated repression is also incomplete. However, in the presence of cAMP and absence of d-galactose the two independent mechanisms, partial hindrance of the promoter by cAMP-CRP and partial repression by DNA looping add up and result in stronger repression.
The D-gal nimplies cAMP gate is also based on the galETKM regulatory region. In this construct the −10 element of the P2galE promoter is replaced by the consensus sequence (TATAAT) and the P1galE promoter is inactivated. The promoter is strongly repressed by both GalR mediated DNA looping and cAMP-CRP (similar to the NcAMP gate), therefore transcription occurs only in the presence of d-galactose (DNA looping is inhibited) and absence of cAMP (cAMP-CRP binding is not allowed).
From the second group the cAMP nimplies D-gal gate (Fig. 2J) was constructed from the ND-gal construct (Fig. 2D) by inserting a cAMP-CRP binding site downstream of the PmglB promoter.
For the AND construct we used the PgalP promoter, which was previously shown to perform AND logic (6, 15). This promoter is inactive in the absence of cAMP-CRP. Because GalR binding inhibits cAMP-CRP mediated activation, both signals are required for transcription (Fig. 2H).
Because the second group needs active recruitment of RNAP for transcription, gates belonging to this group cannot be constructed using only repressors. However, all gates can be obtained by using at least one activator. Activators are more versatile than repressors because they are typically dual-function regulators. Activator binding in the core promoter region can interfere with RNAP binding and therefore can result in decreased level of transcription. RNAP binding to promoters is often regulated by activators and repressors that influence each other's activity, therefore our finding that combination of activation and repression mechanisms can result in the widest range of regulatory logic is not surprising.
Combination of Logic Gates.
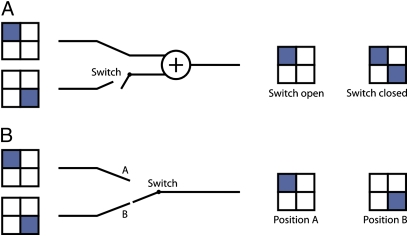
Combinations of logic gates can result in new logic of signal integration (3). Genetic logic gates can be combined in two different ways, which we term parallel and serial connections (Fig. 3). In the parallel setup, both logic gates are fused to the reporter gene separately, which mimics gene duplication followed by expression divergence due to mutations in the regulatory regions of the gene duplicates (32). Because expression of one gene duplicate is independent of the expression of the other, in this case the overall logic of regulation is the sum of the two separate logics (e.g., combination of a “cAMP” and a “D-gal” gate results in “OR” logic). In the serial setup the two logic gates (regulatory sequences) are fused to the same reporter gene, analogous to genes transcribed from more than one promoter regulated by the same TFs. We tested how serial connection of logic gates can result in new logic functions. We tried to obtain the “XOR” (“D-gal nimplies cAMP” + “cAMP nimplies D-gal”), “NAND” (“D-gal nimplies cAMP”+“ND-gal”), and “D-gal implies cAMP” (“AND”+“ND-gal”) gates. However, from the six constructs made only one gave the expected sum function, which resembles Boolean “XOR” logic (Fig. 2L). In the other combinations we did not obtain Boolean-type integration or one of the gates dominated.
Fig. 3.
Strategies for combining logic gates. In the serial connection (A) the regulatory regions (gate 1 and gate 2) are placed sequentially upstream of the reporter gene (red arrows). In the parallel combination (B) the two regulatory regions are fused to the reporter gene separately.
Discussion
Flexibility of Gene Expression Logic.
In this work we constructed small synthetic regulatory networks that integrate two environmental signals. We used the simplest elements and interactions of gene regulation: (i) RNA polymerase-promoter, (ii) TF-DNA binding site, and (iii) TF-small signal molecule. Signal integration in the constructed circuits was characterized by the output in the four extreme situations: (i) in the absence of signals, (ii) in the presence of one signal at high concentration, (iii) in the presence of the other signal at high concentration, and (iv) in the presence of both signals at high concentrations. This characterization resembles Boolean logic where inputs and outputs are 0 (absent) or 1 (present). Although the proper response in natural systems often depends on the integration of signals present at intermediate concentrations, our qualitative analysis is suitable to demonstrate that regulatory networks in bacteria are highly flexible regarding signal integration logic and therefore computational power. We have created and determined the DNA sequence of 12 regulatory regions performing different logic of signal integration. In principle the four other gates can also be constructed using our existing modules. For example, the “NOR” gate could be constructed by cAMP-CRP mediated repression of the “ND-gal” construct and then all of the other three logics can be obtained by proper parallel combinations (EQUAL = NOR+AND, D-gal IMPLIES cAMP = NOR+cAMP, NAND = ND-gal+NcAMP). Although 12 different logics were obtained by our approach, the relation between regulatory structure and function is unclear in some cases. The most interesting is the “ND-gal” construct (Fig. 2D), where we could not identify a GalR binding site by sequence analysis and did not observe cAMP dependence despite the presence of the cAMP-CRP binding site. We find that transcription activation, i.e., recruitment of RNAP to promoter sequences is a crucial determinant of the flexibility of regulatory logic. There is only a limited set of logics that can be performed using two repressors. However, in theory, all possible logics can be performed by activators. The function of an activator can be easily tuned between recruiting RNAP and inhibiting RNAP binding by repositioning its binding site(s). Although generally activators bind DNA weaker than repressors, stronger binding required for steric hindrance of the promoter can be achieved by mutations or by using multiple binding sites.
Apart from the regulatory regions, the regulator genes encoding the TFs can also contribute to the genetic flexibility of networks. The regulatory logic can be significantly altered even by single missense mutations. For instance, analysis of the Lac repressor showed that point mutations can affect DNA binding strength, the ability to form a DNA loop, binding of the small molecule, and even the induction profile of the repressor can be switched, which can completely revert the regulatory logic (in this case TF binding to the operator DNA depends on the presence of the inducer, whereas in the wild-type system the inducer inhibits operator binding) (33, 34). In the galactose system, a GalR mutant having a reverse induction profile could convert a “ND-gal” gate to a “D-gal” gate, or a “NOR” gate to a “D-gal nimplies cAMP” gate.
Combination of Logic Gates.
There are examples for both parallel and serial connections of logic gates in natural regulatory networks (35, 36). The parallel setup can be obtained by gene duplication followed by divergence of the regulatory sequences. Evolution by gene duplication can resolve adaptive conflicts in the regulatory region. For instance, certain regulatory logics may not be feasible by mutations or rearrangements of the regulatory elements but can be obtained by the parallel combination of two simpler logic gates, resulting from divergent mutations in the regulatory elements of the gene duplicates. In this case, the subfunctionalized regulatory elements complement each other's function, thereby facilitating the retention of duplicate gene copies. Indeed, genome analyses of yeast and higher eukaryotes suggest that duplicated genes are more likely to have divergent expression profiles than single-copy genes both within and between genomes (37). However, evolution of regulatory logic by gene duplication in bacteria is strongly limited because of the high rate of recombination between homologous DNA sequences (38). In cells where homologous recombination is highly active, the serial connection of logic gates is more feasible. In fact, several E. coli genes are transcribed from multiple promoters, regulated by the same TFs (36). Our results show that serial connection of logic gates can indeed result in new regulatory logic. However, the combined logic obtained from serial connection is less predictable than in the case of parallel connections. There are several factors that can influence the combined logic, including:
(i) Emergence of a new promoter. New promoters can be obtained by using a strong activator, which is used to repress a promoter in one of the regulatory sequences. In this case a new promoter can emerge downstream of the repressed promoter. In some cases, the fusion of two regulatory sequences can also result in a DNA sequence with significant promoter activity. Such new promoters can change the regulatory logic by either transcribing the reporter gene or by promoter interference (39).
(ii) Transcriptional road block. In this case strong binding of a TF can inhibit transcription of the coding sequence by a promoter located upstream of the TF binding site (40). This mechanism can explain how a downstream logic gate can dominate in the overall function of two serially connected gates.
(iii) Communication at a distance. Many TFs can interact with RNAP or other TFs bound to distant sites on DNA (20, 41). Therefore a TF intended to regulate the upstream promoter may also influence transcription of the downstream promoter in a serial setup.
In this work we showed an example where serial connection of two logic gates, “D-gal nimplies cAMP” and “cAMP nimplies D-gal,” results in an overall “XOR” logic. However, because of the above mentioned factors, the order of the two sequences can strongly influence the overall logic. In this case the GalR-DNA complex in the “D-gal nimplies cAMP” gate might inhibit transcription initiated at an upstream promoter (42). Therefore the “D-gal nimplies cAMP” gate can potentially dominate the overall function of a serial setup when it is placed downstream.
One advantage of using connected logic gates instead of a complex single gate to perform a given logic operation is that the connected setup allows differential regulation of the separate units. Therefore, the overall logic of integration of the two input signals can be selected by using a third signal, similar to multiplexing in electronics. In Fig. 4, we show examples how the choice can be made between the different logic operations performed by the individual gates used, or between the combined logic and the logic performed by one of the gates, depending on the regulatory circuit sensing the third signal. In summary we conclude that the logic of regulatory networks can be highly flexible. Network logic can be easily engineered by mutations in regulatory sequences and/or by combinations of regulatory regions with a specific logic, although construction of a regulatory region with a predefined logic, using building blocks of a specific system, is not always straightforward. However, a large pool of variations can occur in natural populations (especially in “r strategists”), which are tested by natural selection. Indeed, a recent study demonstrated that bacteria can tolerate radical changes in their regulatory circuitry (43). The genetic flexibility of networks may explain why living organisms are so successful in colonizing of new environments.
Fig. 4.
Logic selector circuits. Schematic for showing how logic gates can be combined to get more complex behavior. Depending on the position of the switch, the output responds to the two inputs with different logic. In cells such schemes could be used to adapt the logic gates to environmental conditions. For example, scheme (A) could be implemented by a transcription factor that regulates the lower gate. A change in the environmental condition (e.g., presence of a small molecule) can activate or inactivate the transcription factor. In this case contribution of the lower gate to the output depends on the activity of the TF. In scheme (B) the output logic depends on which of the two gates is active.
Materials and Methods
Strain Construction.
Synthetic regulatory regions were created by PCR and inserted between the EcoRI and PstI sites in plasmid pSEM2027. Plasmid pSEM2027 was created from pSEM2008 (44) in two steps. The pEM7/zeo DNA (Invitrogen) was digested by EcoRI and XhoI, the ends were made blunt using DNA Polymerase I Klenow fragment (Fermentas) and the zeocin cassette was inserted at the previously filled in Acc65I site of plasmid pSEM2008. From the resulting plasmids we selected one in which the orientation of the zeocin cassette was such that the 5′ region of the gene was proximal to the terminator region of pSEM2008, to reduce the background level of reporter gene activity. Next, the uidA ORF together with the Shine-Dalgarno sequence was amplified by PCR using the “GusUP” (5′-AGAGACTGCAGAGGAGGCCCTTATGTTACGTCCTGT AGAAAC-3′) and “GusDN” (5′-CACAAAGCTTAGATCTTCATCATTGTTTGCCTCCCTGCTGCG GT-3′) primers, digested with PstI and BamHI, and the 523-bp fragment containing the 5′ region was inserted between the PstI and BamHI of the selected plasmid.
pSEM2027 derivatives containing the synthetic regulatory regions were digested by BamHI and used as a template for PCR amplification of the region containing the zeocin cassette, the rrnBT1T2 terminators, the cloned promoter region and part of the gusA ORF. This region was amplified using the “uidRZdn” (5′-ACCCGG ATCCTCAATGCTGCCAGAGAGATTTTTTCAGAAAATGGATTTCACGGAATTCTCAGTCCTGCTCCTCGGCCAC -3′) and Gusseqdn (5′-TTCTTGTAACGCGCTTTCCCACCAAC-3′) primers. The ends of the resulting PCR fragment contained sequences of the uid region of the E. coli chromosome (50/128 bp), allowing efficient insertion using recombineering. The PCR product was purified from an agarose gel. Recombineering was performed according the protocol described by Datsenko and Wanner (45). As a result, the region upstream of the uidA ORF, between chromosomal positions 1694107 and 1694987 (GenBank U00096) was replaced by the synthetic construct. Recombinants were selected on LB plates containing 80 μg/mL zeocin.
The construct performing “FALSE” logic (not containing any promoter) contained the sequence “GAGCTCGGTACCCGGGGATCGATCCTCTAGAGTCGAC” between the EcoRI and PstI sites of pSEM2027.
Constructs with serially connected gates were created in a similar way, except that a BglII site was inserted upstream of the EcoRI site in pSEM2027. The upstream regulatory construct was inserted between the BglII and EcoRI sites, whereas the downstream regulatory construct was inserted between the EcoRI and PstI sites.
Screening the Logic of Gene Regulation.
To monitor gene expression at different combinations of d-galactose and cAMP, we prepared four LB plates containing 80 μg/mL zeocin, 50 μg/mL X-gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, Fermentas), and (i) no D-gal, no cAMP, (ii) 8 mM D-gal, (iii) 0.16 mM cAMP, and (iv) 8 mM D-gal and 0.16 mM cAMP. We spotted 2 μL of cell suspension on each LB agar plate. In case of the “ND-gal” construct (Fig. 2D), 16 mM d-galactose was used to obtain full repression.
Verifying the DNA Sequence of the Synthetic Regulatory Regions in Strains.
Cells were grown overnight and total genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega). The regulatory region upstream of the uidABC operon was amplified by PCR (Platinum HiFi supermix; Invitrogen) using the “KpnT1T2” (5′-ATATATGGTACCAAGCTTCTGTTTTGGCGGATGAGA-3′) and “Gusseqdn” primers. The DNA sequence was determined by the Biomi Ltd. DNA sequences were deposited in GenBank (see Fig. 2 for accession numbers).
Supplementary Material
Acknowledgments
We thank our colleagues in the laboratory for various inputs, in particular Kim Sneppen, Sankar Adhya, László Orosz, Sine Svenningsen, and András Holczinger for useful discussions and Takácsné Botond Judit for excellent technical assistance. This research was supported by the Hungarian Scientific Research Fund Grant PD75496 (to S.S.), and the Danish National Research Foundation. S.S. is grateful for the János Bolyai Fellowship of the Hungarian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ872202, GQ872203, GQ872204, GQ872205, GQ872206, GQ872207, and GQ872208).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0915003107/-/DCSupplemental.
References
- 1.Covert MW, et al. Metabolic modeling of microbial strains in silico. Trends Biochem Sci. 2001;26:179–186. doi: 10.1016/s0968-0004(00)01754-0. [DOI] [PubMed] [Google Scholar]
- 2.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 3.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kauffman S. Homeostasis and differentiation in random genetic control networks. Nature. 1969;224:177–178. doi: 10.1038/224177a0. [DOI] [PubMed] [Google Scholar]
- 5.Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 2006;4:e45. doi: 10.1371/journal.pbio.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semsey S, Krishna S, Sneppen K, Adhya S. Signal integration in the galactose network of Escherichia coli. Mol Microbiol. 2007;65:465–476. doi: 10.1111/j.1365-2958.2007.05798.x. [DOI] [PubMed] [Google Scholar]
- 7.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 8.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ptashne M. Repressors. Curr Biol. 2007;17:R740–R741. doi: 10.1016/j.cub.2007.06.053. [DOI] [PubMed] [Google Scholar]
- 11.Murphy KF, Balázsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan S, Bren A, Dekel E, Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol Syst Biol. 2008;4:203. doi: 10.1038/msb.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan S, Bren A, Zaslaver A, Dekel E, Alon U. Diverse two-dimensional input functions control bacterial sugar genes. Mol Cell. 2008;29:786–792. doi: 10.1016/j.molcel.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlman T, Zhang Z, Saier MH, Jr, Hwa T. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6043–6048. doi: 10.1073/pnas.0606717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna S, Orosz L, Sneppen K, Adhya S, Semsey S. Relation of intracellular signal levels and promoter activities in the gal regulon of Escherichia coli. J Mol Biol. 2009;391:671–678. doi: 10.1016/j.jmb.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastan I, Perlman R. Cyclic adenosine monophosphate in bacteria. Science. 1970;169:339–344. doi: 10.1126/science.169.3943.339. [DOI] [PubMed] [Google Scholar]
- 17.Adhya S, Geanacopoulos M, Lewis DE, Roy S, Aki T. Transcription regulation by repressosome and by RNA polymerase contact. Cold Spring Harb Symp Quant Biol. 1998;63:1–9. doi: 10.1101/sqb.1998.63.1. [DOI] [PubMed] [Google Scholar]
- 18.Roy S, Semsey S, Liu M, Gussin GN, Adhya S. GalR represses galP1 by inhibiting the rate-determining open complex formation through RNA polymerase contact: A GalR negative control mutant. J Mol Biol. 2004;344:609–618. doi: 10.1016/j.jmb.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 19.Schleif R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 2000;16:559–565. doi: 10.1016/s0168-9525(00)02153-3. [DOI] [PubMed] [Google Scholar]
- 20.Semsey S, Virnik K, Adhya S. A gamut of loops: Meandering DNA. Trends Biochem Sci. 2005;30:334–341. doi: 10.1016/j.tibs.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Geanacopoulos M, et al. GalR mutants defective in repressosome formation. Genes Dev. 1999;13:1251–1262. doi: 10.1101/gad.13.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebright RH, Ebright YW, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semsey S, Tolstorukov MY, Virnik K, Zhurkin VB, Adhya S. DNA trajectory in the Gal repressosome. Genes Dev. 2004;18:1898–1907. doi: 10.1101/gad.1209404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XP, Ebright RH. Substitution of 2 base pairs (1 base pair per DNA half-site) within the Escherichia coli lac promoter DNA site for catabolite gene activator protein places the lac promoter in the FNR regulon. J Biol Chem. 1990;265:12400–12403. [PubMed] [Google Scholar]
- 25.Aki T, Choy HE, Adhya S. Histone-like protein HU as a specific transcriptional regulator: Co-factor role in repression of gal transcription by GAL repressor. Genes Cells. 1996;1:179–188. doi: 10.1046/j.1365-2443.1996.d01-236.x. [DOI] [PubMed] [Google Scholar]
- 26.Choy HE, Adhya S. Control of gal transcription through DNA looping: Inhibition of the initial transcribing complex. Proc Natl Acad Sci USA. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adhya S. Multipartite genetic control elements: Communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 28.Müller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 29.Thomason L, et al. Recombineering: Genetic engineering in bacteria using homologous recombination. In: Frederick M. Ausubel., editor. Current Protocols in Molecular Biology. Hoboken, NJ: John Wiley & Sons Inc; 2007. pp. 11–24. [DOI] [PubMed] [Google Scholar]
- 30.Bingham AH, Ponnambalam S, Chan B, Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41:67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- 31.Lewis DE, Adhya S. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol Microbiol. 2004;54:692–701. doi: 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- 32.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 33.Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 34.Suckow J, et al. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J Mol Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 35.Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- 36.Keseler IM, et al. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database issue):D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence JG, Hendrix RW, Casjens S. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 2001;9:535–540. doi: 10.1016/s0966-842x(01)02198-9. [DOI] [PubMed] [Google Scholar]
- 39.Shearwin KE, Callen BP, Egan JB. Transcriptional interference—a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: The role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Courey AJ, Jia S. Transcriptional repression: The long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- 42.Lewis DE. Identification of promoters of Escherichia coli and phage in transcription section plasmid pSA850. Methods Enzymol. 2003;370:618–645. doi: 10.1016/s0076-6879(03)70052-4. [DOI] [PubMed] [Google Scholar]
- 43.Isalan M, et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitarai N, et al. Dynamic features of gene expression control by small regulatory RNAs. Proc Natl Acad Sci USA. 2009;106:10655–10659. doi: 10.1073/pnas.0901466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.