Abstract
It is commonly assumed but not proven that microRNAs (miRNAs) and their targets coevolve. Under this assumption, miRNAs and targets from different species may interact adversely, resulting in reduced fitness. However, the strength of the adverse interactions may not be detectable because even outright deletions of miRNAs often manifest only subtle fitness effects. We tested and measured the strength of heterospecific interactions by carrying out transgenic experiments across Drosophila species by overexpressing the miR310s cluster of Drosophila melanogaster (Dm310s) and Drosophila pseudoobscura (Dp310s) in D. melanogaster. Flies overexpressing the heterospecific Dp310s are only one-third as viable as those overexpressing the conspecific Dm310s. The viability effect is easily detectable in comparison to the effect of the deletion of miR310s. The number of genes significantly misexpressed under the influence of Dp310s is 3–10 times greater than under Dm310s. Importantly, the numbers of predicted targets are similar between them. Expression analysis of the predicted target genes suggests that miRNAs may sometimes function to buffer fluctuations in the transcriptome output. After the buffering function has evolved, heterospecific combinations may cause adverse effects.
Keywords: microRNAs, coevolution, canalization, genetic buffering
Coevolution is a central topic in evolutionary biology. Good examples of phenotypic coevolution abound, but coevolution at the molecular level has been observed only in limited contexts. Genic coevolution can be the molecular basis of the Muller–Dobzhansky (MD) model of hybrid incompatibility (1, 2). In the simplest two-locus MD model, the ancestral A0 and B0 alleles at loci A and B, respectively, evolved to A1B1 in species 1 and A2B2 in species 2. In each lineage, the two genes coevolve such that alleles at the two loci are always compatible. However, alleles A1 and B2 or B1 and A2 may not be compatible because they have never coexisted and coevolved in the same species. Interspecific hybridization brings them together, causing hybrid breakdown. Hidden meiotic drive revealed by interspecific crosses can be viewed in the same framework (3, 4). The MD model has been elaborated extensively in recent years (reviewed in ref. 5).
In this study, we ask whether microRNAs (miRNAs) and their target genes from different species might interact adversely. The answer would be a first step toward elucidating how coevolution between miRNAs and their targets may occur. MiRNAs are small RNAs about 22 nt in length that guide RNA-induced silencing complex to repress translation or to degrade transcripts of target genes (reviewed in 6, 7). In animals, target transcripts are matched to the miRNA sequence in their 3′UTRs but only imperfectly. The seed region (positions 2–8) of each miRNA has a more stringent requirement for matching (8, 9).
Although it is often assumed that miRNAs and their target genes can coevolve, evidence is lacking. The first step should show that orthologous miRNAs from one species do not interact well with the genome of another species. However, there are several hurdles to demonstrate incompatibility between miRNAs and their targets. First, most functionally characterized miRNAs are highly conserved, but the demonstration would require the study of evolving miRNAs. Second, the fitness consequences of evolving miRNAs are likely to be weak. Given that the fitness effects of individual gene knockout are often modest even for conserved miRNAs (10, 11), the substitution of a recently diverged copy for the resident gene should have an even smaller effect. Third, there are no compelling conceptual arguments that miRNAs and their targets should coevolve. Targets could conceivably switch from one miRNA to another during evolution with no fitness consequences. After all, there are often many coexpressed miRNAs performing similar functions. Fourth, if a fitness effect could be detected, the coevolution hypothesis would still require, among other things, demonstration that the effect is mediated through the downstream targets of miRNAs, directly or indirectly.
In Drosophila, many evolving miRNAs have indeed been identified (12–14) but most are evolutionarily transient and may not have been well integrated into the transcriptome (“background miRNAs”) (13). Among Drosophila miRNAs that have been shown to be functional, the miR-310/311/312/313 cluster (or miR310s) are the only ones currently known to be adaptively evolving (12, 15). [A few other possibilities require further rigorous testing (13, 14)]. The miR310s cluster likely originated from duplication of miR92a/b about 40–60 million years ago (12). In this study, we test the hypothesis of the incompatibility of miRNA targets by moving miR310s across species boundaries and analyzing the fitness and transcriptional consequences.
We tested miR310s as a cluster, because members of this cluster are known to have overlapping, or even redundant, functions (15, 16) such that the effects of disrupting an individual member may be compensated for by the activities of other members (15). That is, we treated the cluster as an evolutionary unit. Coevolution between individual members and their respective targets will be amenable for analysis when the collective pattern becomes known.
Results
miRNA Transformation of D. melanogaster.
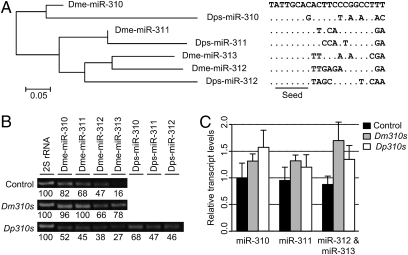
The choice of species for incompatibility studies has to be based on their evolutionary distance, which, in turn, depends on the evolutionary rates of the genes of interest. Fig. 1 shows the sequences of the mature miR310s of Drosophila melanogaster and Drosophila pseudoobscura. The homologous relationship between each member of Dm310s and Dp310s can be seen in their phylogeny (Fig. 1A). The seed region is invariant except for a transition (C→G) at the eighth position in one of the sequences in D. pseudoobscura. For the miR310s cluster, the average number of nucleotide differences between miRs within the same species of the melanogaster group (D. melanogastor, 6.00; Drosophila sechellia, 6.33; Drosophila simulans, 6.33; Drosophila yakuba, 6.33; Drosophila erecta, 6.17; Drosophila ananassae, 7.33) ranges from 6 to 7.33, whereas the average difference between species ranges from 4.75 to 5.69, which is consistently smaller than the divergence within species. The average difference is 7.33 between miRs from D. melanogaster and D. pseudoobscura, and the average difference within D. pseudoobscura is 9.33, which is unusually large for paralogs of the same species. At the level of the comparison between D. melanogaster and D. pseudoobscura, the divergence between species became larger than the divergence within D. melanogaster (between paralogous genes). This would be the level at which we study the cross-species interactions between miRNAs and targets.
Fig. 1.
Sequences and overexpression of miR310s. (A) Mature sequences of the miR310s cluster in D. melanogaster (Dm310s) and D. pseudoobscura (Dp310s) and their phylogeny. The seed is highly conserved except for a C→G change at the eighth position of one Dps310s sequence. Nucleotides that differ from the Dme-miR-310 sequence are indicated. Phylogenetic reconstruction among the mature sequences of miR310s was done using the neighbor-joining method with Kimura's two-parameter distances. (B) RT-PCR of Dm310s and Dp310s. RT-PCR was performed for each member of miR310s in the progeny L3 larvae of the cross of NP5941 and w1118 (control; Top), Dm310s (Middle), and Dp310s (Bottom) with the 2S rRNA as a control. Band intensities were normalized to 2S rRNA, and levels relative to 2S rRNA are indicated. (C) Relative transcript levels of miR310s in the control, Dm310s, and Dp310s lines were determined by semiquantitative RT-PCR. Band intensities were normalized to 2S rRNA as in Fig. 1B and then relative to the mean value of Dme-miR-310 in controls. In the Dp310s line, the values of homologous members of Dm310s and Dp310s were added. Bars indicate SEs (n = 3).
The miR-310/311/312/313 cluster from D. melanogaster (referred to as Dm310s) and the homolog from D. pseudoobscura (Dp310s) were used to transform D. melanogaster. The transgenic constructs were under the control of the upstream activation sequence (UAS) promoter, which can be regulated by tissue-specific GAL4s (17). In total, we obtained 8 Dm310s and 18 Dp310s transformant lines. The expression of the transgenic miR310s was driven in 3 GAL4 lines: (i) Act5C (ubiquitous expression), (ii) DJ715 (brain and salivary gland expression) (ref. 18 and Fig. S1 A–E), and (iii) NP5941 (brain, salivary gland, and imaginal disk expression) (SI Text and Fig. S1 F–O).
The enhancer trap line, NP5941-GAL4, is the main one used in this study. The GAL4 construct is inserted 173 bp upstream of miR310s, and hence is suspected to mimic the native miR310s in expression. In our analysis, the reporter (UAS-GFP, driven by the GAL4 in the NP5941 line) is expressed in larval salivary glands, imaginal disks, brain, and diverse tissues of the adult body and head (SI Text and Fig. S1 F–O). The tissue distribution of the reporter expression closely matches the expression profiles of miR310s obtained by deep-sequencing studies (19, 20). Furthermore, we wished to know the expression level of UAS-miR310s under the control of the NP5941-GAL4. We chose to study third instar larvae, which showed substantial differences in viability as a function of the expression of miR310s (SI Text and Fig. S2).
The expression of miR310s in the Dm310s and Dp310s transgenic lines and in the WT control was assayed by RT-PCR. The primers used were listed in Table S1. One of the replicates is shown in Fig. 1B, and the overall results across replicates are given in Fig. 1C. The total level of miR310s expression in the Dm310s line is 54% higher than in the WT control, whereas the level in the Dp310s line is 46% greater than that in the control (Fig. 1C). The key comparison is between the two transgenic lines because they share the same genetic background in which various biological effects were measured. It is also interesting to note that in the transgenic lines, roughly half of the transcripts came from the transgenes and the rest came from the native miR310s. The relative contributions can be measured in the Dp310s line, as shown in Fig. 1B. Finally, a knockout line that carries a deletion of 1,416 bp spanning the entire miR310s cluster was also created. This knockout line, referred to as Dm310s(−), was used mainly to provide supplementary information.
Fitness Consequence of the Transgenic miRNAs.
We first compared the viability effect of the conspecific and heterospecific transgenic miRNAs. Crosses were made such that half of the progeny would have the GAL4-[UAS-Dm310s] genotype and the other half would have the balancer chromosome in place of the [UAS-Dm310s]-bearing chromosome. An identical procedure was used for the Dp310s transgene. Thus, the viabilities of both Dp310s- and Dm310s-bearing flies were measured against a common standard bearing the balancer chromosome (SI Materials and Methods).
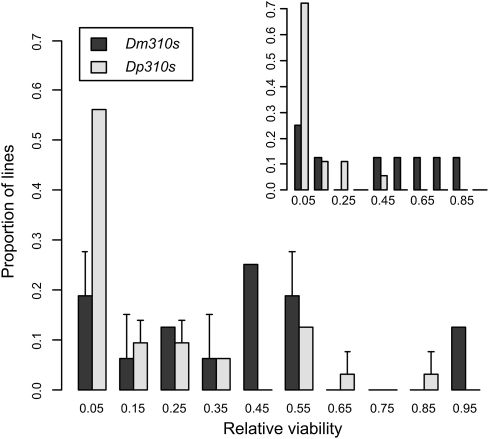
When the ubiquitously expressed Act5C-GAL4 was used to drive UAS-miR310s expression, lethality in the progeny was complete for both Dm310s and Dp310s transformants. This expression pattern was thus not pursued further. The viabilities of the transgenic lines with DJ715-GAL4 or NP5941-GAL4 are shown in Fig. 2. As expected, the viabilities of all lines carrying either the Dm310s or Dp310s transgene were reduced relative to the balancer-chromosome control.
Fig. 2.
Viabilities of the 8 Dm310s and 18 Dp310s transformant lines. Viabilities of Dm310s- and Dp310s-bearing lines were measured against the same balancer-bearing standard (SI Materials and Methods). All the transgenic lines carrying either Dm310s or Dp310s showed reduced viabilities relative to the control. The reduction is more severe in Dp310s lines than in Dm310s lines. The pattern is similar when UAS-miR310s were driven by the NP5941-GAL4 or DJ715-GAL4 (Inset) line. The viability assay using the NP5941-GAL4 line was repeated twice independently, and bars indicate SEs.
A salient observation of Fig. 2 is the low viabilities of the lines bearing Dp310s relative to those bearing Dm310s. The average viability across transgenic lines driven by NP5941-GAL4 was only 13.7% (±4.5%) for Dp310s-bearing lines but three times as high at 40.9% (±10%) for Dm310s-bearing lines. The difference in the average viability under the DJ715-GAL4 driver was even more striking: 5.0% (±2.3%) vs. 45.3% (±12%) for Dp310s and Dm310s, respectively (Fig. 2, Inset). The differences are significant by the Mann–Whitney U test (P < 0.05; Materials and Methods). Therefore, relative to the native Dm310s, Dp310s are poorly compatible with the transcriptome of D. melanogaster. The difference in viability between the Dm310s and Dp310s transgenic lines is, in fact, larger than that between the Dm310s(−) line and the control. In the latter comparison, there is no detectable difference in viability (although there is a substantial difference in female fecundity).
Although these lines all appear normal in regular cultural conditions, there was substantial variation in lethality when driven by GAL4, even among lines with the same transgene. A possible explanation is position effect of the insertion site, with each site possibly differing somewhat in accessibility to the UAS sequences. We therefore expected the line effect on lethality under one GAL4 driver to be correlated with that under a different GAL4 driver. For Dm310s, the correlation coefficient in viability under the control of the two drivers shown in Fig. 2 is 0.755 (Spearman's ρ, P < 0.05). For Dp310s, it is 0.677 (Spearman's ρ, P < 0.01). There is indeed a significant position effect.
In the remainder of this study, we analyzed gene expression using lines that were closest in viability to the median of Fig. 2 for both Dm310s and Dp310s transgenes. As a check, we examined the expression of miR310s in these “median” lines. Because many larvae died in late third instar (L3) and others failed to develop in pupation, the assay was done on the L3 larvae. Both Dp310s and Dm310s were expressed in L3 larvae when driven by NP5941-GAL4 (SI Text and Fig. S2). As shown in Fig. 1 B–C, Dp310s were expressed at a comparable level as (or at a slightly lower level than) Dm310s, suggesting that the two sets of transgenic lines have a similar median in their expression of miR310s. Despite the similarity in median expression, the median viability of the Dm310s lines is 10 times higher than that of the Dp310s lines.
Effects of the Transgenic miRNAs on Total Expression.
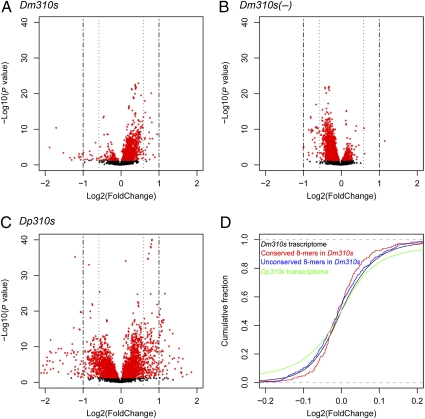
We next compared whole-genome expression (WGE) of the Dp310s and Dm310s transgenic lines in L3 larvae. Total RNAs were extracted from whole larvae and processed for microarray analysis. Fig. 3 shows the “volcano plots” of transcript expression profiles assayed with an Affymetrix tiling array. For comparison, the knockout line, Dm310s(−), was also included. Misregulation in each case was determined by comparison with the WT of the identical genetic background. In the Dm310s lines, overexpression was more common than underexpression among misregulated genes (Fig. 3A), although miRNAs are expected to repress target gene expression. (Overcompensation of expression is plausible if cells read the levels of protein to decide on the level of mRNA output.) The pattern appeared to be reversed in the Dm310s(−) line (Fig. 3B). Thus, overexpressing and underexpressing Dm310s have opposite effects on WGE.
Fig. 3.
Impact of miR310s misexpression on WGE in D. melanogaster. The negative log10-transformed P values of a paired t test (Materials and Methods) are plotted against the log 2-fold change for a total of 19,878 transcripts. (A) Dm310s line. (B) Dm310s(−) line. (C) Dp310s line. Fold change is the ratio of the expression in the experimental line over that in the control. (D) Empirical cumulative distributions of the plots in A (black line) and C (green line). Cumulative distributions of the plots in Fig. 4A (red and blue lines for conserved and unconserved targets, respectively) are also included. Note that the range on the x axis of D is much smaller than that in A–C for the purpose of showing the trend near X = 0. In that small interval, the large number of points in the scattered plot of A–C obscures the pattern.
It is most noteworthy that the extent of misregulation was much greater in the Dp310s line (Fig. 3C) than in the Dm310s line (Fig. 3A). The Kolmogorov–Smirnov test on the cumulative plot shows that the differences are highly significant (P < 2.2 × 10−16 ; Fig. 3D, black vs. green curves). Transcript WGE was also assayed on a cDNA array. The numbers of misregulated genes on each platform are shown in Table 1. There are 3–10 times as many misregulated genes in the Dp310s line as in the Dm310s line. Most misregulated genes in the Dm310s line are also misregulated in the Dp310s line (overlap is shown in Table 1).
Table 1.
Summary of microarray analyses in Dm310s, Dp310s, and Dm310s(−) lines
| Expression difference | Dm310s | Dp310s | Overlap* | Dm310s(−) |
| Tiling array (n = 19,878) | ||||
| 1.2-fold | 838 | 2,011 | 478 | 815 |
| 1.5-fold | 61 | 508 | 37 | 35 |
| 2-fold | 20 | 143 | 13 | 1 |
| cDNA array (n = 13,506) | ||||
| 1.5-fold | 59 | 677 | 48 | NA |
| 2-fold | 59 | 520 | 46 | NA |
The number of genes that are significantly misexpressed relative to the WT control at a false discovery rate of 5% is shown. Note that the number of misregulated genes is much larger in the Dp310s line than in the Dm310s line. NA, data not available.
*Genes that are misregulated in Dp310s and either of the two Dm310s lines were considered to overlap.
The results from the Dp310s transformants may represent the transcriptome's response to “unfamiliar” miRNAs. Table S2 tallies the misregulated genes by gene ontology. Among the 13 genes that are underexpressed in both transgenic lines (see the overlap set at the 2-fold cutoff), 8 are glue proteins or secreted proteins of the salivary gland, including six Sgs genes (Sgs1, CG3047; Sgs3, CG11720; Sgs4, CG12181; Sgs5, CG7596; Sgs7, CG18087; Sgs8, CG6132), Eig71Ee (CG7604), and ng3 (CG10788). The expression timing is thus consistent with the observed L3 lethality. Cuticle proteins constitute another category of genes that are preferentially misregulated mainly in the Dp310s line. The difference may account for the stronger lethality associated with Dp310s.
Effects of the Transgenic miRNAs on the Expression of Predicted Target Genes.
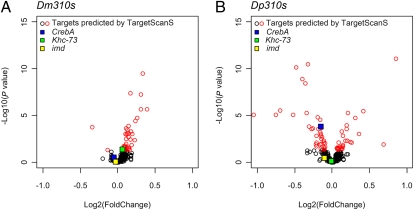
Ultimately, miRNAs act on the transcriptome through their direct targets. The expression patterns of the target genes inferred by TargetScanS (21) deviated much more strongly from the control in the Dp310s line than in the Dm310s line (Fig. 4 A vs. B). More predicted target genes were significantly misregulated by Dp310s than by Dm310s, and the absolute fold changes of targets are significantly smaller in the Dm310s line (P = 5.30 × 10−5, one-sided Wilcoxon paired-sample test). Again, the Kolmogorov–Smirnov test on the cumulative plot shows that the difference is highly significant (P < 0.001; Fig. S3A). Because target prediction remains imprecise, we attempted two other methods, Pictar (22) and Probability of Interaction by Target Accessibility (PITA) (23), and reached the same conclusion (Fig. S3 B and C).
Fig. 4.
Impact of Dm310s and Dp310s on the expression of the predicted targets in D. melanogaster. (A) Dm310s. (B) Dp310s. The axes are the same as in Fig. 3. Putative conserved targets were downloaded from TargetScanFly version 5.1 (http://www.targetscan.org/fly_12/), and 346 target genes were plotted. The same set of targets was used for both Dm310s and Dp310s lines (SI Text). Significantly misexpressed genes are labeled in red. Three genes that have previously been experimentally verified as targets are indicated by colored squares: CrebA (red), Khc-73 (green), and imd (orange).
The main reason for the difference between Fig. 4 A and B is that many of the predicted target genes were not significantly underexpressed in the Dm310s line (Fig. 4A). Three target genes that have been experimentally verified (24, 25), which are color-marked in Fig. 4, are such examples. To see this point more clearly, we may compare the expression profiles of the target genes (Fig. 4A) and the whole transcriptome (Fig. 3A). Indeed, the cumulative distributions of the two profiles suggest that target genes are no more underexpressed than the transcriptome as a whole (Fig. 3D).
In Fig. 3D, we further separate the putative target genes into two groups: the evolutionarily conserved vs. unconserved targets. To simplify the comparison, target genes are defined as genes for which the 3′UTR contains sequences with a perfect match to the 8-mer miRNA seed (SI Materials and Methods). Such genes are known to be enriched for miRNA targets, and the enrichment is greater for genes with evolutionarily conserved 8-mer sites (26, 27). Fig. 3D shows that genes bearing evolutionarily unconserved 8-mer sites have an expression profile very similar to the transcriptome as a whole. In contrast, those bearing evolutionarily conserved sites have proportionately fewer misexpressed (both overexpressed and underexpressed) genes than the rest of the transcriptome.
In summary, overexpressing the conspecific Dm310s in D. melanogaster appears to have a relatively small effect on the abundance of the inferred target transcripts. The effect on the expression of the direct targets appears to be even smaller than the effect on the transcriptome in general. In contrast, overexpressing the heterospecific Dp310s has a strong effect on their putative targets in D. melanogaster.
Discussion
The much reduced viability and massive misregulation in the Dp310s lines suggest negative fitness interactions between miRNAs and targets from different species. During the divergence between D. pseudoobscura and D. melanogaster, Dp310s and the transcriptome of D. melanogaster have become, to some extent, incompatible in the MD framework. The nature of this incompatibility lies in the interactions between miRs and their direct targets. The overexpression of Dp310s destabilizes the transcriptome of D. melanogaster, whereas Dm310s affect it to a much smaller extent. This is true for the expression of both direct and indirect targets.
There may be several potential sources of error. First, the assays of WGE might not be reliable. However, WGE was measured on two different platforms, and the results were consistent in terms of the proportion of misregulated genes. (Slightly more than half of the misregulated genes are shared between the two platforms.) Second, target prediction might not be sufficiently accurate [although the criteria used generally yield false-positive results at a rate lower than 30% (e.g., 21, 22, 28)]. With such false-positive rates, a trend of repression by the overexpressed miR310s should still have been observable. Curiously, in the case of the conserved targets of Dm310s, the trend is, in fact, opposite to the predicted repression. In other words, target prediction has to be worse than random selection to account for the observations. We consider this possibility unlikely. In summary, although there may be many caveats against experimental errors, the parallel experiments comparing Dm310s and Dp310s serve as an internal control. Potential errors in, for example, WGE measurement, GAL4 expression, and target prediction should not have given rise to very different patterns induced by Dm310s and Dp310s.
The greater extent of WGE misregulation in the Dp310s line may not be surprising, because “wrong” genes were targeted by Dp310s, leading to a cascade of downstream misexpression. However, the low level of misregulation of the direct targets, relative to the misregulation of the entire transcriptome in the Dm310s line (Fig. 3D), is quite unexpected. Under the simple repression model in which miRs directly repress the expression of these targets with little feedback (29–31), the directed targets are expected to be most severely affected by the overexpression of miRs. The effects then cascade down to WGE, which should be, at most, as variable as the expression of the direct targets themselves. In this simple model, it should not matter whether the miRs were Dp310s or Dm310s. Indeed, the effects on direct targets and WGE are both large and nearly distinguishable in the Dp310s line. This makes the effect of Dm310s on their direct targets surprisingly small in both absolute and relative terms (against WGE).
Because there is a 1-bp change in the seed of Dps-miR-310, its effect on WGE was examined in (SI Text). Furthermore, the strength of miRNA-target interactions might be different for Dm310s and Dp310s. This possibility is also addressed in SI Text. Neither factor could account for the contrasting patterns of gene expression between Dm310 and Dp310s lines. Details can be found in SI Text.
The results of Fig. 4 therefore suggest buffering of target expression when conspecific miR310s were disturbed. Buffering requires (i) the existence of a feedback loop onto the miRNAs themselves [or a feedforward loop (e.g., 32, 33)] and (ii) sufficient time for the feedback to take effect. Many previous studies assayed transient responses of the transcriptome soon after miRNAs were misexpressed. Buffering was hence not expected to be observed. Neither do we expect buffering in those studies that deleted the miRNAs as feedback loops were severed. Indeed, we observed patterns closer to simple repression in the Dm310s(−) knockout line than in the Dm310s line (Fig. S3D).
An interesting issue is how often miRNAs play a functional role in reducing expression variances (33–37) vs. how often they repress their target expression with little feedback from downstream events. An example of simple target repression may be CrebA (CG7450, blue box in Fig. 4), which is a transcription factor in the salivary gland, regulating its secretory activity. It is also known to play a role in the development of larval cuticle (38). The expression of CrebA is consistently in the expected direction of simple repression: underexpression in Dp310s and Dm310s lines and overexpression in the Dm310s(−) lines. We suspect that the targeting of CrebA by Dp310s (and, to a lesser extent, Dm310s) results in the extensive misexpression of downstream salivary gland-expressed genes and L3 lethality, as reported above.
Simple repression, however, may not be the most common circuitry. Otherwise, the putative direct targets should have been misregulated at least as much as WGE in Dm310s lines. The weak repression observed in Fig. 4A suggests that putative targets are often buffered via some mechanisms in Dm310s lines. These mechanisms may include negative feedback or an incoherent feedforward loop (37). In Fig. S4, we hypothesize a feedback mechanism regulating Ras85D (CG9375), which is ubiquitously expressed in Drosophila and occupies a key position in the Ras pathway (39). Ras85D in Dp310s lines is strongly repressed, but in the Dm310s line, it is not repressed at all. Among the 113 interactor genes of Ras85D in our study (BioGRID database, http://www.thebiogrid.org;), 31 are significantly misregulated in the Dp310s line.
We hypothesize that genes whose expression needs to be stabilized may often be wired to miRNAs for genetic buffering. Previous studies have shown that miRNAs predominantly target positive regulator motifs, highly connected scaffolds, and downstream network components (40). Wired in such a manner, miRNAs may help to maintain cellular homeostasis and facilitate robust signal transduction. A good example is miR-7, which stabilizes E(spl)-mediated Atonal expression against environmental fluctuation during sense organ precursor determination in Drosophila (32). Many motifs are indeed known to buffer gene expression (41). According to this hypothesis, direct targets of miRNAs would appear to be more stably expressed than the transcriptome as a whole. The observation in Fig. 3D that genes with conserved targets appear to be more strongly buffered than those with unconserved ones is consistent with this hypothesis. It has also been reported that cross-species variation in gene expression is significantly lower for miRNA targets than for non-miRNA targets (35).
Finally, stabilizing gene expression might be an (evolutionarily) early function of newly evolved miRNAs like miR310s. As recently suggested (37), recently emerged miRNAs may be more easily integrated into the genome if they mainly help to stabilize WGE. It is harder to see how recently emerged miRNAs could evolve if they immediately assume a repression role by turning down the expression of many target genes. In this view, simple repressive functions might evolve gradually later on.
Materials and Methods
The generation of Dm310s or Dp310s transgenic lines and Dm310s deletion lines is described in SI Materials and Methods. Balanced female transformant lines were crossed to GAL4 homozygote male lines, and the viability of the UAS-GAL4 progeny was assessed relative to the balancer-bearing control. The overexpression of Dm310s and Dp310s was detected using Northern blot analysis and semiquantitative RT-PCR assay. The WGE profiling was performed using DGRC-1 Amplicon Transcriptome Microarrays (Drosophila Genomics Resource Center, Indiana University, Bloomington, IN) and Drosophila Tiling 1.0F Arrays (Affymetrix) (SI Materials and Methods). Target predictions from TargetScan release 5.1 (21, 42), PicTar (single miRNA target predictions with setting S1) (22), and the PITA “TOP” category with 3/15 flank version 6 (23) were obtained from their respective Web sites. Testing for overrepresentation of functional categories was carried out using Database for Annotation, Visualization, and Integrated Discovery (DAVID) tools (43).
Supplementary Material
Acknowledgments
We thank Richard W. Carthew and reviewers for invaluable comments and advice. We thank Jaejung Kim for execution of the tiling array experiments, Haijun Wen for help with semiquantitative RT-PCR, and Xu Zhang for instructive discussions in tiling array analysis. This work was supported by National Institutes of Health Grants GM058686 and GM30998 (to C.-I.W. and W.-H.L.); National Science and Technology Major Project of China Grant 2009ZX08010-017B (to S. S. and C.-I.W.); and 973 Program Grant 2007CB815701, National Science Foundation of China Grants 30730008 and 30970208, and State Key Laboratory of Biocontrol of China Grant 09A06 (to S.S.). T.T. was supported by Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry. T.T. and Y.S. were supported by the Junior Faculty Development Program, SunYat-sen University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (SuperSeries accession no. GSE15863).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007591107/-/DCSupplemental.
References
- 1.Dobzhansky T. Further data on the variation of the Y chromosome in Drosophila Pseudoobscura. Genetics. 1937;22:340–346. doi: 10.1093/genetics/22.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller HJ. Isolating mechanisms, evolution and temperature. Biological Symposium. 1942;6:71–125. [Google Scholar]
- 3.Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biol. 2007;5:e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. PLoS Biol. 2007;5:e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 10.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 11.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, et al. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol. 2008;25:929–938. doi: 10.1093/molbev/msn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, et al. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leaman D, et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Zhao T, et al. A Drosophila gain-of-function screen for candidate genes involved in steroid-dependent neuroendocrine cell remodeling. Genetics. 2008;178:883–901. doi: 10.1534/genetics.107.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Seroude L, Brummel T, Kapahi P, Benzer S. Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging Cell. 2002;1:47–56. doi: 10.1046/j.1474-9728.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 19.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLOS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 24.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 28.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 29.Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding—A new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 35.Cui Q, Yu Z, Purisima EO, Wang E. MicroRNA regulation and interspecific variation of gene expression. Trends Genet. 2007;23:372–375. doi: 10.1016/j.tig.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Martinez NJ, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CI, Shen Y, Tang T. Evolution under canalization and the dual roles of microRNAs: A hypothesis. Genome Res. 2009;19:734–743. doi: 10.1101/gr.084640.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrew DJ, Henderson KD, Seshaiah P. Salivary gland development in Drosophila melanogaster. Mech Dev. 2000;92:5–17. doi: 10.1016/s0925-4773(99)00321-4. [DOI] [PubMed] [Google Scholar]
- 39.Schnorr JD, Holdcraft R, Chevalier B, Berg CA. Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics. 2001;159:609–622. doi: 10.1093/genetics/159.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006 doi: 10.1038/msb4100089. 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 42.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.