Abstract
Areas and layers of the cerebral cortex are specified by genetic programs that are initiated in progenitor cells and then, implemented in postmitotic neurons. Here, we report that Tbr1, a transcription factor expressed in postmitotic projection neurons, exerts positive and negative control over both regional (areal) and laminar identity. Tbr1 null mice exhibited profound defects of frontal cortex and layer 6 differentiation, as indicated by down-regulation of gene-expression markers such as Bcl6 and Cdh9. Conversely, genes that implement caudal cortex and layer 5 identity, such as Bhlhb5 and Fezf2, were up-regulated in Tbr1 mutants. Tbr1 implements frontal identity in part by direct promoter binding and activation of Auts2, a frontal cortex gene implicated in autism. Tbr1 regulates laminar identity in part by downstream activation or maintenance of Sox5, an important transcription factor controlling neuronal migration and corticofugal axon projections. Similar to Sox5 mutants, Tbr1 mutants exhibit ectopic axon projections to the hypothalamus and cerebral peduncle. Together, our findings show that Tbr1 coordinately regulates regional and laminar identity of postmitotic cortical neurons.
Keywords: arealization, Auts2, microarray
The mammalian cerebral neocortex has a conserved modular organization comprised of areas parcellating the cortical surface and layers stratifying the cortical thickness (1–4). The development of neocortical areas and layers is coordinated by specialized programs of neurogenesis and neuronal subtype specification (5–7). Areal and laminar identity are initially specified in cortical progenitor cells and then, implemented through subsequent processes of fate acquisition in postmitotic neurons (3, 4, 8). Some transcription factors (TFs) have been implicated in the regulation of both areal and laminar identity. For example, Pax6, a TF expressed in progenitor cells, promotes both rostral identity (9) and upper layer neurogenesis (10). Similarly, Bhlhb5, a TF expressed in postmitotic neurons, is required for the acquisition of caudal motor and sensory areal identity and layer 5 corticospinal motor neuron (CSMN) identity (8).
In the present study, we investigated whether Tbr1, a T-box TF expressed in postmitotic projection neurons (11), might regulate areal and laminar identity of cortical neurons. Previously, we have shown that Tbr1 is necessary for the differentiation of preplate and layer 6 neurons (11). Whereas Tbr1 is expressed at the highest levels in developing frontal cortex (12–14), we hypothesized that Tbr1 might implement frontal identity. This hypothesis accords with evidence that Tbr1 is expressed downstream of Pax6 through a TF cascade from Pax6+ radial progenitors to Tbr1+ postmitotic projection neurons (15–18). In addition, we examined whether Tbr1 might suppress alternative regional and laminar fates.
To test our hypothesis, we profiled changes of regional and laminar identity in Tbr1 null mutant neocortex (11, 19) using gene-expression markers. We found that markers of frontal and layer 6 differentiation were markedly down-regulated, whereas markers of caudal cortex and layer 5 were significantly increased in Tbr1 mutants. Furthermore, the expression of caudal markers was shifted rostrally, and early-born neurons shifted from layer 6 to layer 5 identity. Finally, we found that Tbr1 implements frontal identity in part by transcriptional activation of Auts2, a frontal marker gene (20) linked to autism (21) and mental retardation (22).
Our findings show that Tbr1 modulates the balance of cortical areas and layers by regulating gene expression in postmitotic neurons. Perturbations of regional and laminar identity may be important factors in neurodevelopmental diseases.
Results
Previous studies have shown that Tbr1 exhibits high rostral and low caudal expression in developing neocortex (12–14). This suggests that Tbr1 may contribute to the implementation of frontal cortex identity. Alternatively, Tbr1 expression could simply reflect the rostrocaudal gradient of neurogenesis (23).
To further characterize Tbr1 expression along the rostrocaudal axis, we compared Tbr1 with Bhlhb5, a caudal marker (8) in developing mouse cortex (Fig. S1). These experiments revealed opposing gradients of Tbr1 (high rostral) and Bhlhb5 (high caudal), first discernible in the cortical plate (CP) on embryonic days (E) 13.5–14.5 (Fig. S1 A–C). Tbr1 and Bhlhb5 also exhibited complementary laminar patterns, most obvious from E16.5 to postnatal day (P) 0.5 (Fig. S1 F–U). In frontal cortex, Tbr1 was highly expressed in all layers, whereas Bhlhb5 was virtually absent. In more caudal regions, Tbr1 was highly expressed in layer 6, subplate (SP), and Cajal-Retzius (C-R) neurons, whereas Bhlhb5 was highly expressed in layers 2–5 as noted previously (8). Tbr1 was not absent from layers 2–5 but was expressed at much lower levels than in early-born neurons.
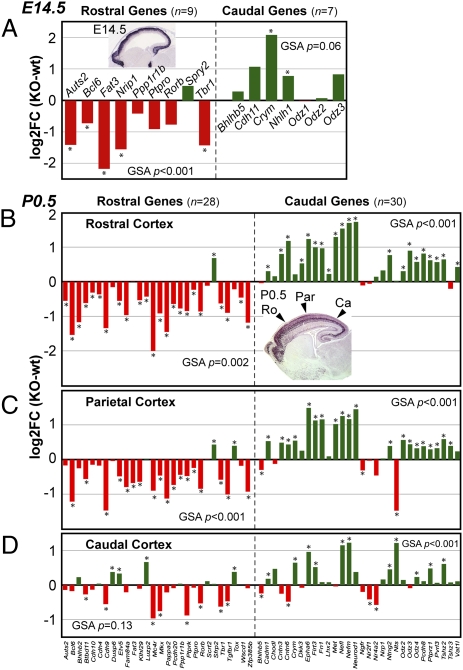
To determine whether Tbr1 is required for the acquisition of frontal identity, we studied cortical regionalization in Tbr1 null mice, which die shortly after birth (11, 19). Because anatomical landmarks of cortical areas (e.g., somatosensory barrels) are not developed on P0.5, we assessed regional identity using molecular expression patterns (12, 13). Previous studies have described several regional markers in embryonic and neonatal cortex, such as Bcl6 rostrally and Odz3 caudally (14, 24, 25). To find additional regional markers, we mined online databases (26–28), previous publications, and our microarray data (Dataset S1 and Dataset S2). This approach yielded 20 rostral and 12 caudal markers in E14.5 cortex and 28 rostral and 30 caudal markers in P0.5 cortex (Tables S1 and S2). Regional markers were assembled into panels for gene-set analysis (GSA), a statistical method for testing the significance of coordinate changes in the expression of multiple genes (29, 30).
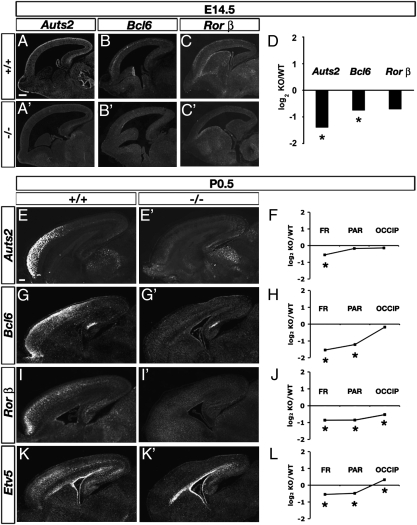
Regional marker analysis revealed profound defects of frontal differentiation in Tbr1 mutant neocortex (Fig. 1). Early in CP development (E14.5), frontal markers Auts2, Bcl6, and Rorb all had reduced expression, shown anatomically by in situ hybridization (ISH) and quantitatively by microarray profiling (Fig. 1 A–D). Among eight markers of rostral identity (excluding Tbr1) in E14.5 CP and intermediate zone (IZ), seven had decreased expression in Tbr1 null cortex by microarray (Fig. 2A). The reduction of rostral markers in Tbr1 null E14.5 CP/IZ was highly significant (GSA P < 0.001). Notably, these defects of frontal gene expression preceded the onset of increased apoptosis in frontal cortex (Discussion). Interestingly, the only rostral marker with increased expression in Tbr1 null E14.5 CP/IZ was Spry2 (Fig. 2A). Because Spry2 is regulated by FGF signaling (31, 32), this could suggest that FGF signaling was increased in Tbr1 mutant cortex. This interpretation was supported by expression data on some other FGF signaling-related molecules, including Fgf15, Fgf17, Spry1, and Etv1 (Fig. S2, Dataset S1, and Dataset S2). These results suggest that Tbr1 may activate genes that suppress FGF signaling, among other possibilities (Discussion).
Fig. 1.
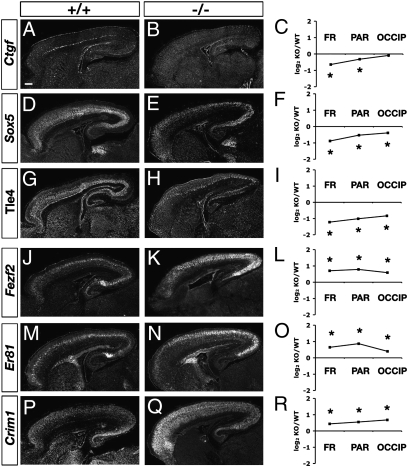
Down-regulation of rostral marker genes in Tbr1 mutant cortex. (A–D) E14.5. Auts2 (A and A′), Bcl6 (B and B′), and Rorb (C and C′) mRNA were reduced in Tbr1 null rostral cortex (A′–C′) compared with controls (A–C). Microarray results (D) confirmed down-regulation of these genes, although only Auts2 and Bcl6 reached statistical significance. *P < 0.05. (E–L) P0.5. Auts2 (E and E′), Bcl6 (G and G′), Rorb (I and I′), and Etv5 (K and K′) mRNA were reduced in Tbr1 null frontal cortex (E′, G′, I′, and K′) compared with controls (E, G, I, and K). Microarray profiling (F, H, J, and L) showed significant reductions of these genes mainly in frontal (FR) and parietal (PAR) regions of Tbr1 mutant cortex. Microarray results are expressed as log2 of the ratio between knockout (KO) and wild-type (WT) normalized mRNA levels. OCCIP, occipital cortex. [Scale bars (A–K′): 200 μm.]
Fig. 2.
Microarray analysis of rostral and caudal marker genes in Tbr1 null cortex. (A) E14.5. Most rostral markers decreased (red) and caudal markers increased (green) in Tbr1 null cortex. GSA P values indicate the probability that sets of genes (rostral or caudal) increased or decreased by chance. (Inset) Tbr1 ISH, E14.5 (28). (B–D) P0.5. Rostral and caudal markers were analyzed in rostral (B), parietal (C), and caudal (D) regions of Tbr1 KO cortex versus controls. (B Inset) Tbr1 immunohistochemistry, E18.5. n, number of genes in the set.
Later in CP development (P0.5), frontal differentiation remained severely defective in Tbr1 mutants. Auts2, Bcl6, Rorb, and Etv5 all showed decreased expression on P0.5 (Fig. 1 E–L). Among 28 markers of rostral identity in P0.5 CP and IZ, 27 showed decreased expression in Tbr1 null frontal cortex by microarray (Fig. 2B). Frontal markers were reduced in all layers of cortex, including Bhlhb2 in layers 2–3, Rorb in layer 4, Etv5 in layers 4–6, and Wscd1 in layer 6 and SP. Many frontal markers were reduced in parietal and occipital cortex as well, consistent with graded expression (Fig. 2 C and D). The decrease of rostral markers was highly significant in Tbr1 null frontal (GSA P < 0.002) and parietal (GSA P = 0.001) cortex. The only rostral marker to increase in P0.5 Tbr1 null frontal cortex was Sfrp2 (Fig. 2B), possibly regulated through the Fgf15–Pax6–Sfrp2 pathway (33, 34).
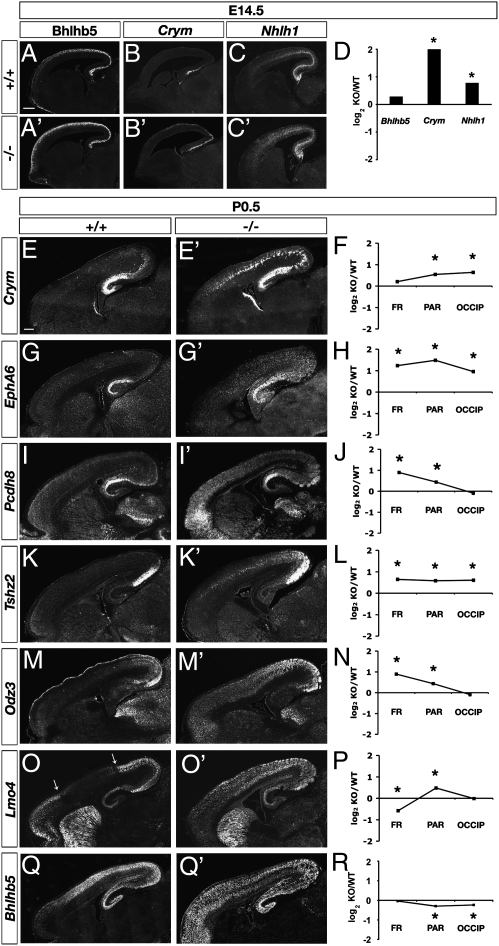
To determine if Tbr1 regulates caudal identity, we studied caudal markers in Tbr1 mutants. On E14.5, caudal markers Bhlhb5, Crym, and Nhlh1 all showed up-regulation (Fig. 3 A–D). Significantly, the expression boundaries of these caudal genes were shifted rostrally in Tbr1 null cortex, indicating that they were expressed ectopically. The rostral shift of Bhlhb5 expression in E14.5 cortex (Fig. 3A) was especially significant, because Bhlhb5 mediates the postmitotic acquisition of caudal identity (8). In a panel of seven caudal CP/IZ markers, six had increased expression by microarray (Fig. 2A). The overall increase of E14.5 caudal gene expression almost reached statistical significance (GSA P = 0.06). The only caudal marker that exhibited a (slight) decrease of expression in E14.5 Tbr1 null cortex, Odz1, is expressed mainly in SP (25), and SP differentiation is severely defective in Tbr1 mutants (11). This observation illustrates one way in which regional and laminar identity may interact.
Fig. 3.
Up-regulation of caudal marker genes in Tbr1 mutant cortex. (A–D) E14.5. Immunofluorescence for Bhlhb5 (A and A′) and ISH for Crym (B and B′) and Nhlh1 (C and C′) showed increased rostral expression of these genes in Tbr1 null cortex (A′–C′) compared with controls (A–C). (D) Microarray results showed that Crym and Nhlh1 expression changes were statistically significant. (E–R) P0.5. Crym (E and E′), Epha6 (G and G′), Pcdh8 (I and I′), Tshz2 (K and K′), Odz3 (M and M′), Lmo4 (O and O′), and Bhlhb5 (Q and Q′) mRNA were increased in Tbr1 null cortex. The Lmo4 low-expression domain (O, between arrows) was shifted rostrally in Tbr1 null cortex (O′). Bhlhb5 expression was disorganized and increased rostrally in the malformed Tbr1 null cortex (Q′) compared with control (Q). Microarray results are shown in F, H, J, L, N, P, and R. [Scale bars (A–Q′): 200 μm.]
Caudal CP/IZ markers remained elevated in P0.5 Tbr1 mutant cortex. Crym, Epha6, Pcdh8, Tshz2, and Odz3 all showed increased expression with rostral shifts (Fig. 3 E–N). Lmo4, which normally has expression domains in caudal and rostral but not parietal cortex (Fig. 3O), likewise showed a marked rostral shift (Fig. 3 O′ and P). In a panel of 30 caudal markers in P0.5 CP/IZ, 26 showed increased expression in Tbr1 null frontal cortex (Fig. 2B). The up-regulation of caudal markers was highly significant (GSA P < 0.001). Many caudal markers were also increased in parietal and occipital regions (Fig. 2 C and D). Bhlhb5 was increased in P0.5 Tbr1 mutant prefrontal cortex but did not seem elevated in other regions (Fig. 3 Q and Q′). Bhlhb5 expression normally declines in neonatal mice and becomes restricted to primary sensory-input areas (8). This could reflect dependence on thalamic innervation, which is defective in Tbr1 mutants (11).
The decline of rostral identity and rise of caudal identity in Tbr1 null cortex could have several explanations. First, Tbr1 could regulate regional identity autonomously in postmitotic neurons; our data seem consistent with this idea. Second, Tbr1 might affect regional identity nonautonomously (e.g., by feedback to progenitor cells). However, regional TF gradients in the ventricular zone (VZ) and subventricular zone (SVZ) were not significantly altered in Tbr1 null cortex (Fig. S3). Third, Tbr1 might regulate the genesis or survival of neurons in different cortical regions. Neuronal production seems intact in Tbr1 mutants (11). To investigate whether apoptosis is increased in Tbr1 mutants, we tested for expression of activated caspase-3 protein, a sensitive marker of apoptosis (35). Apoptotic cells were rare in normal cortex but were markedly increased in Tbr1 mutant rostral cortex beginning on E16.5 (Fig. S4). The death of frontal neurons undoubtedly accounted for some changes in regional marker expression on P0.5, but because apoptosis was not increased on E14.5, cell death could not explain defects of rostral identity at this age. Additionally, apoptosis could not explain the rostral shift of caudal markers (Fig. 3). Together, our results suggest that Tbr1 is necessary for the direct implementation of rostral identity in postmitotic neurons.
We next analyzed laminar identity in Tbr1 mutants using panels of layer-specific markers in E14.5 and P0.5 neocortex (Tables S3 and S4). On E14.5, we assayed the differentiation of early-born neuron types, including C-R, SP, and CP neurons. Microarray indicated that C-R neuron markers Reln (Reelin) and Calb2 (calretinin) were significantly decreased in E14.5 Tbr1 null neocortex, confirming previous ISH results (11). Also, the E14.5 microarray data showed decreased expression of SP markers Kitl, Odz1, and Mef2c and CP markers Cdh13, Cnr1, Bcl11b (Ctip2), Sox5, and Zeb2 (SIP1). Reductions of Bcl11b, Mef2c, and Sox5 were further validated by ISH (Fig. S5). These findings supported the conclusion that Tbr1 is required for the differentiation of preplate and layer 6 identity (11).
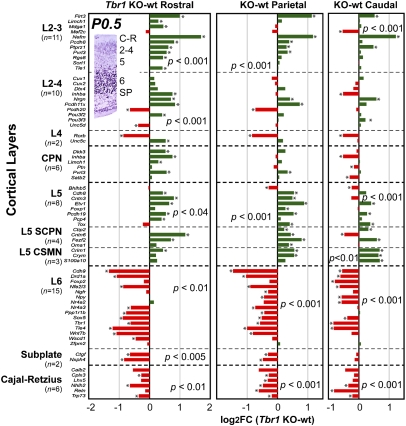
In P0.5 cortex, we analyzed markers for all layers. Microarray indicated severe defects of C-R, SP, and layer 6 differentiation in Tbr1 null P0.5 cortex (Fig. 4). For example, Wnt7b, a layer 6/SP marker, and Reln, a C-R marker, were down-regulated on microarray, supporting previous ISH results (11). Additional ISH experiments validated the down-regulation of SP marker Ctgf and layer 6 markers Sox5 and Tle4 (Fig. 5 A–I). Reduction of Sox5 was noteworthy, because Sox5 suppresses aspects of layer 5 identity, including Fezf2 expression (36, 37). Interestingly, layer 6/SP markers restricted to caudal cortex, such as Nr4a2 (Nurr1) and Ngfr, did not decrease as much as other layer 6/SP markers (Fig. 4). The relative sparing of caudal layer 6/SP markers may reflect an interaction of regional and laminar fate determination, balancing up-regulation of caudal identity with down-regulation of layer 6 and SP identity.
Fig. 4.
Microarray profiling of laminar marker genes in P0.5 Tbr1 mutant mice. Layer 6, SP, and C-R cell markers were down-regulated in all regions of Tbr1 null cortex. Layer 5 markers were up-regulated in all regions, including markers of subcerebral projection neurons (SCPNs) and corticospinal motor neurons (CSMNs). Markers of layers 2 and 3 were also mostly up-regulated in Tbr1 null cortex. Callosal projection neurons (CPN) were not significantly elevated overall, as determined by GSA. (Inset) Tbr1 immunohistochemistry (P0.5) showing laminar expression in parietal cortex.
Fig. 5.
Expression of subplate, layer 6, and layer 5 marker genes in Tbr1 null cortex. (A–I) SP and layer 6 markers Ctgf (A–C), Sox5 (D–F), and Tle4 (G–I) had decreased expression in Tbr1 null cortex by ISH and microarray. (J–R) Layer 5 markers Fezf2 (J–L), Er81 (M–O), and Crim1 (P–R) had increased expression. [Scale bars (A–Q): 200 μm.]
Microarray also showed increased expression of some layer 2–5 markers in P0.5 Tbr1 null cortex (Fig. 4). Layer 5 markers were up-regulated most consistently, including subcerebral projection neuron (SCPN) and corticospinal motor neuron (CSMN) markers (Fig. 4). For example, Fezf2, a determinant of SCPN identity (38–40), and Etv1 (Er81), an established layer 5 marker (Tables S3 and S4), were significantly increased in all regions (Fig. 4). Up-regulation of Fezf2, Er81, and Crim1 (a CSMN marker) was confirmed by ISH (Fig. 5 J–R). Interesting exceptions to the generally increased expression of upper-layer genes included Rorb, a layer 4 marker, and Pcdh20, a layer 2–4 marker (Fig. 4). Because these genes are also P0.5 rostral markers (Tables S1 and S2), their down-regulation was consistent with impaired acquisition of frontal identity in Tbr1 mutant cortex.
Increased expression of layer 5 markers, together with decreased expression of Sox5, suggested that some early-born cells switched from layer 6 to layer 5 identity in Tbr1 null mutants. To more definitively test for changes in laminar fate, we used two approaches: BrdU birthdating followed by analysis of laminar fate markers and axon tracing of corticofugal fibers (Fig. S6). Cells born on E12.5 (BrdU+) had decreased layer 6 fates (Tle4+) and increased layer 5 fates (Er81+, Ctip2+) in all regions (Fig. S6 A–L), including occipital cortex where apoptosis was not a factor (Fig. 4). The Sox5+ fate index did not change, probably because Sox5 is expressed not only in layer 6 but also at lower levels in layer 5 (36), and our cell counts did not distinguish between high and low Sox5 immunoreactivity.
To examine whether cortical axon projections shifted from layer 6 corticothalamic to layer 5 subcerebral or ectopic pathways in Tbr1 mutants, we studied projections from frontal cortex. We focused on frontal cortex, because parietal and occipital regions have severe generalized defects of axon growth (11). These experiments revealed exuberant subcerebral projections in Tbr1 mutants, whereas projections to thalamus were reduced but not absent (Fig. S6 M and N). These findings fit well with the increased Ctip2 fate index in Tbr1 null cortex (Fig. S6L), because Ctip2 promotes subcerebral axon projections (41). Ectopic projections to hypothalamus were also noted (Fig. S6N). Together, the exuberant subcerebral projections and increased layer 5 markers suggested that some early-born neurons switched from layer 6 to layer 5 identity in Tbr1 mutants.
Most T-box transcription factors function as transcriptional activators (42). To test whether Tbr1 might directly activate layer 6 or frontal marker genes, we used Tbr1 overexpression and ChIP assays. Tbr1, along with GFP reporter in the same cells, was overexpressed by plasmid electroporation from the lateral ventricles in E13.5 cortex followed by cortical slice culture for 1 d (Fig. S7). Electroporated cells were probed for up-regulation of candidate Tbr1 target genes, including Auts2 (frontal cortex), Sox5 (layer 6), Tle4 (layer 6), and FOG2 (layer 6). As controls, we tested for induction of genes predicted not to be targets of Tbr1, including Bhlhb5 (caudal cortex/layers 2–5) and Ctip2 (layer 5). Tbr1 overexpression consistently induced ectopic Auts2 (Fig. 6 A–H) but did not induce Sox5, Tle4, FOG2, Bhlhb5, or Ctip2. These data suggested that Tbr1 might bind and activate Auts2 in vivo.
Fig. 6.
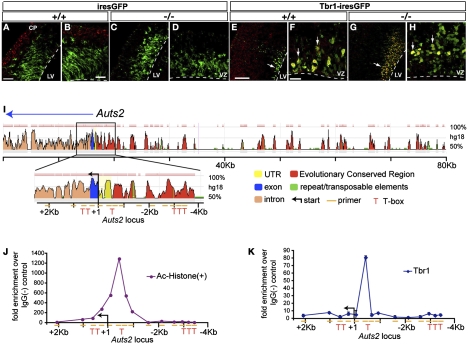
Tbr1 binds and activates the Auts2 gene in neocortex in vivo. (A–H) Tbr1 expression plasmid (Tbr1–ires GFP), but not GFP only (ires–GFP), drove ectopic expression of Auts2 (red) in electroporated GFP+ cells (green) in control (A, B, E, and F) and Tbr1 null (C, D, G, and H) VZ. The electroporated region from A, C, E, and G is shown at higher magnification in B, D, F, and H. [Scale bars (A and C): 50; (B and D): 30 μm; (E and G): 100 μm; (F and H): 25 μm.] (I) Sequence analysis of the Auts2 promoter and proximal coding region identified six candidate Tbr1 binding sites (red T). (J) ChIP of acetylated histones from E14.5 forebrain showed that the Auts2 transcriptional start site was highly enriched in open chromatin. (K) Tbr1 ChIP showed Tbr1 binding to a candidate site in the region of open chromatin (J) adjacent to the Auts2 transcriptional start site.
To determine if Tbr1 binds the Auts2 promoter, we used Tbr1 antibodies for ChIP of E14.5 cortex. Sequence analysis identified six potential Tbr1 binding sites near the Auts2 transcriptional start site (Fig. 6I). One of these candidate Tbr1 binding sites was highly enriched (>50-fold) in chromatin from Tbr1 ChIP (Fig. 6K). This Tbr1 binding site was located in a region of transcriptionally active open chromatin adjacent to the Auts2 transcriptional start site, as shown by acetylated histone ChIP (Fig. 6J). Whereas Tbr1 is required for cortical Auts2 expression (Figs. 1 and 2), induces Auts2 expression ectopically in cortex (Fig. 6 A–H), and binds the Auts2 promoter (Fig. 6K), we conclude that Auts2 is a direct transcriptional target of Tbr1 in developing neocortex.
Discussion
Our findings indicate that Tbr1 is required for the implementation of regional and laminar identity in postmitotic neurons. Unlike Bhlhb5, which mediates the acquisition of caudal and layer 5 fates (8), Tbr1 also suppresses alternative identities—specifically, caudal and layer 5 fates (Figs. 2–5). Thus, Tbr1 probably functions upstream of Bhlhb5 in a transcriptional network to implement cortical-neuron subtype identity.
In the last decade, much has been learned about factors that promote frontal identity of progenitor cells, such as FGF8 and Pax6 (3, 5, 13). Tbr1 is a TF found to promote frontal identity in postmitotic neurons, presumably implementing fate that is initially specified in radial progenitor cells. Interestingly, Tbr1+ neurons are produced both directly from Pax6+ radial progenitors and indirectly from intermediate neuronal progenitors (INPs) that express neither Pax6 nor Tbr1 (15). Therefore, we predict that INPs must express other TFs that maintain regional identity. One candidate could be Tbr2 (Eomes), a T-box factor related to Tbr1 that is specifically expressed in INPs (15) and moreover, is regulated by Pax6 (16, 17).
Our data revealed an unexpected interaction between Tbr1 and FGF signaling. Increased FGF signaling was suggested by elevated levels of Fgf17, Spry1, Spry2, and other molecular reporters of FGF signaling (Fig. S2, Dataset S1, and Dataset S2). Different FGFs have diverse effects on telencephalic development (13, 34, 43–45), and the impact of these FGF signaling changes is unclear. Considering that FGFs regulate mainly progenitor cells and Tbr1 controls postmitotic neurons, our data could suggest that Tbr1 activates a feedback loop to dampen FGF signaling. Further studies will be necessary to investigate this and other possibilities.
Laminar identity, like regional identity, is specified in progenitors and transmitted into postmitotic neurons (4, 8). Among TFs known to implement laminar fate in postmitotic neurons, Tbr1 and Sox5 are important for layer 6 and SP (36, 37). Our present results suggest that they are part of the same transcriptional network (Fig. S6O). Sox5 expression was decreased in Tbr1 null cortex on E14.5 (Fig. S5) and P0.5 (Figs. 4 and 5 D–F). Phenotypic similarities between Sox5- and Tbr1-deficient mice also support this link. Inactivation of Tbr1, like Sox5 (36, 37), led to increased Fezf2 expression (Figs. 4 and 5) along with ectopic corticofugal projections from frontal cortex into the hypothalamus and cerebral peduncle (Fig. S6). Outside frontal cortex, axon growth and guidance are so defective in Tbr1 mutants that most cortical efferents do not grow beyond the internal capsule (11). Whereas layer 5 SCPN markers Fezf2 and Ctip2 were up-regulated in P0.5 Tbr1 mutant cortex, callosal projection neuron (CPN) markers such as Satb2 (4) increased little or not at all (Fig. 4). This indicated that early-born Tbr1 null neurons switched to layer 5 SCPN, not CPN identity, perhaps explaining callosal agenesis in Tbr1 mutants (11). Implementation of laminar identity, like regional identity, may also require expression of specific TFs in INPs.
The present study and others (1, 5, 8) show that regional and laminar fates are regulated coordinately by overlapping transcriptional programs involving some of the same TFs. For individual TFs such as Tbr1, such dual roles may be difficult to reconcile. For example, although Tbr1 promotes frontal cortex and layer 6 identity, how does Tbr1 regulate layer 6 identity in caudal cortex? Some insights come from studying markers with distinct expression patterns, such as Ngfr, a marker of caudal layer 6. Ngfr was down-regulated in Tbr1 mutant occipital cortex (Fig. 4) in contrast to the up-regulation of most other caudal markers (Fig. 2). This suggests that Ngfr was more sensitive to the effects of Tbr1 on laminar than on regional identity. Other informative examples include Rorb (rostral layer 4) and Pcdh20 (rostral layers 2/3). These genes were down-regulated in Tbr1 mutant cortex (Fig. 2), although most other upper layer markers were up-regulated (Fig. 4). Thus, Rorb and Pcdh20 were more sensitive to Tbr1 regulation of regional than laminar identity. Because Tbr1 does not induce rostral identity in caudal layer 6 or layer 6 identity in rostral upper layers, additional regulators must be postulated. These could include combinatorial interactions with other TFs, epigenetic regulation of chromatin, or posttranslational modifications of Tbr1.
Finally, the present study identified Auts2, a frontal cortex marker gene (20) linked to autism (21) and mental retardation (22), as a direct target of Tbr1 binding and activation (Fig. 6). Other potential targets of Tbr1 activation proposed in previous studies include Reln, Grin1, and Grin2b (46–48). Our previous (11) and current results (Fig. 4) support Reln as a direct target of Tbr1. However, by microarray, Grin1 and Grin2b were not reduced in Tbr1 mutant cortex on E14.5 or P0.5, except for a modest reduction of Grin2b (log2FC = −0.17) in P0.5 frontal but not somatosensory or occipital cortex. In future studies, we hope to evaluate these and other candidate targets of Tbr1 through additional ChIP and Tbr1 overexpression assays.
Methods
Detailed Methods.
Additional procedural details are available in SI Methods.
Animals and Tissue Processing.
Tbr1 null mice (11, 19) were maintained on a CD1 background and genotyped by PCR. The plug date was designated E0.5. ICC and ISH were done as described (15, 49) and repeated in at least three brains. Immunocytochemistry antibodies and ISH probes are listed in SI Methods.
Axon Tracing with DiI.
Axon tracing was done as described (11) using P0.5 brains (n = 3 Tbr1 mutant; n = 3 littermate controls) fixed by perfusion with 4% paraformaldehyde. DiI crystals were injected into frontal cortex. After storage in fixative for 8 wk, brains were sectioned coronally at 100 μm, counterstained with DAPI, mounted for fluorescence microscopy, and photographed digitally.
RNA Isolation and Microarrays.
E14.5 and P0.5 brains were removed, and neocortexes were immediately dissected. RNA samples from control (n ≥ 3) and Tbr1 null (n ≥ 3) embryos were hybridized on Affymetrix mouse gene arrays (U430 2.0 or ST 1.0) in the Microarray Core of the University of Washington Center for Ecogenetics and Environmental Health. Data were analyzed statistically by GSA as described in SI Methods.
Identification of Regional and Laminar Markers.
Regional and laminar markers were assembled by mining previous studies, online databases (26–28), and results in the present study. Genes were organized in two age ranges corresponding to embryonic CP differentiation (E13.5–E15.5) and perinatal CP differentiation (E18.5–P4).
Ex Utero Electroporation.
E13.5 embryos were harvested into cold (4 °C) buffer. Plasmids encoding GFP only (ires–GFP) or Tbr1 and GFP (Tbr1–ires–GFP) were injected into the ventricles and electroporated with paddle electrodes across the cerebrum. The brain was sliced coronally (400 μm), cultured for 24 h, and fixed with cold buffered 4% paraformaldehyde for cryosectioning and ICC. Colocalization of GFP and TFs was assessed by confocal microscopy.
ChIP.
Chromatin was precipitated from E14.5 forebrain and further processed as described in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01NS050248 (to R.F.H.). Microarray experiments were supported in part by University of Washington Center for Ecogenetics and Environmental Health National Institute of Environmental Health Services Grant P30ES07033. This research was facilitated by resources provided by National Institute of Child Health and Human Development Grant P30 HD02274. F.B. was supported by a grant from Universita degli Studi di Milano. R.D.H. was supported by fellowships from the Heart and Stroke Foundation of Canada and the American Heart Association.
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray datasets have been deposited in the NCBI GEO database under accession number GSE22371.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002285107/-/DCSupplemental.
References
- 1.Rakic P, Ayoub AE, Breunig JJ, Dominguez MH. Decision by division: Making cortical maps. Trends Neurosci. 2009;32:291–301. doi: 10.1016/j.tins.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 3.O'Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rash BG, Grove EA. Area and layer patterning in the developing cerebral cortex. Curr Opin Neurobiol. 2006;16:25–34. doi: 10.1016/j.conb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Polleux F, Dehay C, Kennedy H. The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. J Comp Neurol. 1997a;385:95–116. [PubMed] [Google Scholar]
- 7.Polleux F, Dehay C, Moraillon B, Kennedy H. Regulation of neuroblast cell-cycle kinetics plays a crucial role in the generation of unique features of neocortical areas. J Neurosci. 1997b;17:7763–7783. doi: 10.1523/JNEUROSCI.17-20-07763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi PS, et al. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop KM, Goudreau G, O'Leary DD. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- 10.Schuurmans C, et al. Sequential phases of cortical specification involve neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:309–311. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 12.Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, Rubenstein JL. Early neocortical regionalization in the absence of thalamic innervation. Science. 1999;285:906–909. doi: 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- 13.Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 14.Leamey CA, et al. Differential gene expression between sensory neocortical areas: Potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex. 2008;18:53–66. doi: 10.1093/cercor/bhm031. [DOI] [PubMed] [Google Scholar]
- 15.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm PC, et al. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Sansom SN, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hevner RF. From radial glia to pyramidal-projection neuron: Transcription factor cascades in cerebral cortex development. Mol Neurobiol. 2006;33:33–50. doi: 10.1385/MN:33:1:033. [DOI] [PubMed] [Google Scholar]
- 19.Bulfone A, et al. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21:1273–1282. doi: 10.1016/s0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- 20.Bedogni F, et al. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns. 2010;10:9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sultana R, et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 22.Kalscheuer VM, et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet. 2007;121:501–509. doi: 10.1007/s00439-006-0284-0. [DOI] [PubMed] [Google Scholar]
- 23.Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: A general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 24.Funatsu N, Inoue T, Nakamura S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: Identification of several region- and layer-specific genes. Cereb Cortex. 2004;14:1031–1044. doi: 10.1093/cercor/bhh063. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Bishop KM, O'Leary DD. Potential target genes of EMX2 include Odz/Ten-M and other gene families with implications for cortical patterning. Mol Cell Neurosci. 2006;33:136–149. doi: 10.1016/j.mcn.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 27.Magdaleno S, et al. BGEM: An in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Bolado G, Eichele G, et al. Analysing the developing brain transcriptome with the GenePaint platform. J Physiol. 2006;575:347–352. doi: 10.1113/jphysiol.2006.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: A developing story. Nat Rev Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 32.Minowada G, et al. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- 33.Kim AS, Anderson SA, Rubenstein JL, Lowenstein DH, Pleasure SJ. Pax-6 regulates expression of SFRP-2 and Wnt-7b in the developing CNS. J Neurosci. 2001;21:RC132. doi: 10.1523/JNEUROSCI.21-05-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borello U, et al. FGF15 promotes neurogenesis and opposes FGF8 function during neocortical development. Neural Develop. 2008;3(17):1–18. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narasimhaiah R, Tuchman A, Lin SL, Naegele JR. Oxidative damage and defective DNA repair is linked to apoptosis of migrating neurons and progenitors during cerebral cortex development in Ku70-deficient mice. Cereb Cortex. 2005;15:696–707. doi: 10.1093/cercor/bhh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwan KY, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai T, et al. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 38.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 41.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 43.Sansom SN, et al. Genomic characterisation of a Fgf-regulated gradient-based neocortical protomap. Development. 2005;132:3947–3961. doi: 10.1242/dev.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwata T, Hevner RF. Fibroblast growth factor signaling in development of the cerebral cortex. Dev Growth Differ. 2009;51:299–323. doi: 10.1111/j.1440-169X.2009.01104.x. [DOI] [PubMed] [Google Scholar]
- 45.Sahara S, O'Leary DD. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsueh YP, Wang TF, Yang FC, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 47.Wang GS, et al. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron. 2004;42:113–128. doi: 10.1016/s0896-6273(04)00139-4. [DOI] [PubMed] [Google Scholar]
- 48.Wang TF, et al. Identification of Tbr-1/CASK complex target genes in neurons. J Neurochem. 2004;91:1483–1492. doi: 10.1111/j.1471-4159.2004.02845.x. [DOI] [PubMed] [Google Scholar]
- 49.Fink AJ, et al. Development of the deep cerebellar nuclei: Transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.