Abstract
The multisubunit Golgi-associated retrograde protein (GARP) complex is required for tethering and fusion of endosome-derived transport vesicles to the trans-Golgi network. Mutation of leucine-967 to glutamine in the Vps54 subunit of GARP is responsible for spinal muscular atrophy in the wobbler mouse, an animal model of amyotrophic lateral sclerosis. The crystal structure at 1.7 Å resolution of the mouse Vps54 C-terminal fragment harboring leucine-967, in conjunction with comparative sequence analysis, reveals that Vps54 has a continuous α-helical bundle organization similar to that of other multisubunit tethering complexes. The structure shows that leucine-967 is buried within the α-helical bundle through predominantly hydrophobic interactions that are critical for domain stability and folding in vitro. Mutation of this residue to glutamine does not prevent integration of Vps54 into the GARP complex but greatly reduces the half-life and levels of the protein in vivo. Severely reduced levels of mutant Vps54 and, consequently, of the whole GARP complex underlie the phenotype of the wobbler mouse.
Keywords: Golgi apparatus, vesicle trafficking, protein stability, X-ray crystallography
Continuous transfer of material between organelles of the endomembrane system is required to maintain the overall metabolic balance of the cell. This transfer is mediated by transport vesicles that bud from a donor compartment and fuse with an acceptor compartment. The first physical link between a transport vesicle and its target compartment is established by tethering factors, which also promote pairing of fusogenic SNARE proteins, while small GTPases of the Rab, Rho, and Arl subfamilies orchestrate the overall process (1, 2). To date, eight multisubunit tethering complexes (MTCs) have been shown to act at different steps of secretory and endosomal/lysosomal pathways. The transport protein particle I (TRAPI), transport protein particle II (TRAPII), Dsl1p, and the conserved oligomeric Golgi (COG) complexes participate in endoplasmic reticulum (ER)-Golgi and intra-Golgi traffic (1, 3), whereas the exocyst complex facilitates fusion with the plasma membrane (4, 5). The class C core vacuole/endosome tethering (CORVET) and the homotypic fusion and vacuole protein sorting (HOPS) complexes, on the other hand, function in the endosomal/lysosomal system, while the Golgi-associated retrograde protein (GARP) complex participates at the trans-Golgi network (TGN) (1, 6). Sequence analysis of MTC subunits has revealed that Dsl1, COG, GARP, and exocyst, denoted as the DCGE subfamily (7), share a distant relationship among themselves but are unrelated to TRAPI, TRAPII, CORVET and HOPS (8–10). Furthermore, the crystal structures of the full-length Tip20p subunit of the Dsl1 complex and the Exo70p subunit of the exocyst complex reveal a similar domain organization consisting of a tandem arrangement of α-helical bundles (11–13).
GARP, also referred to as VFT (Vps fifty three), is an evolutionarily conserved heterotetrameric complex composed of Vps51, Vps52, Vps53 and Vps54 subunits, which is involved in retrograde transport from endosomes to the TGN (14–17). RNAi of mammalian GARP subunits blocks retrieval of recycling proteins such as the cation-independent mannose 6-phosphate receptor (CI-MPR) and TGN46, as well as retrograde transport of the Shiga toxin B subunit from endosomes to the TGN (18). The block in CI-MPR retrieval results in missorting of its cargo, acid hydrolases, into the extracellular space. As a consequence, lysosomes become enlarged due to accumulation of undegraded materials within their lumen (18).
A spontaneous missense mutation (Leu967 to Gln) near the C terminus of Vps54 is responsible for the wobbler mouse phenotype (19) characterized by spinal muscular atrophy and defective spermiogenesis (20). The neurological phenotype of wobbler mice is similar to that of humans with spontaneous spinal neurodegeneration, including ALS (also known as Lou Gehrig’s disease). To date, there is no evidence for mutations of Vps54 in patients with ALS (21). However, similarities in motor neuron degeneration in the wobbler model and ALS patients raise the possibility of common pathways to cell death, perhaps including impaired vesicle traffic.
To elucidate the structural basis for the wobbler phenotype, we have determined the crystal structure at 1.7 Å resolution of the C-terminal 145 amino acid residues (836–977) of mouse Vps54. The structure reveals an α-helical bundle that is strikingly similar to domains found in other MTCs such as Dsl1, COG, and exocyst. Leu967 is completely buried against the α-helical bundle through predominantly hydrophobic interactions that are critical for domain stability and folding in vitro. The L967Q Vps54 mutant assembles with other GARP subunits but has a shortened half-life, resulting in lower levels of the mutant relative to the wild-type protein in vivo. The phenotype of wobbler mice thus results from destabilization of Vps54 and consequently reduced levels of this protein and of the GARP complex.
Results
Structure of the Vps54 C-Terminal Domain.
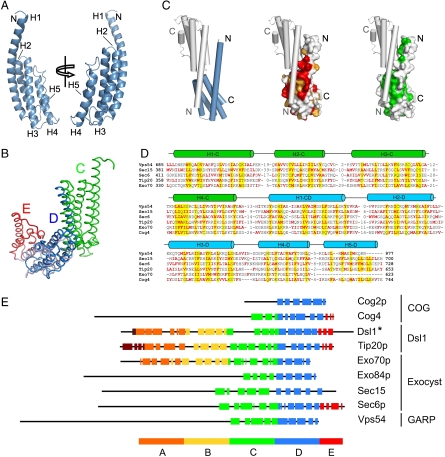
The structure of the Vps54 C-terminal domain (Vps54-CT, residues 836–977) corresponds to a five-stranded antiparallel coiled-coil bundle (Fig. 1A) of 65-Å length and 22-Å width. Pairwise alignment using the Dali server (22) revealed a notable resemblance to the D domain of other MTC subunits, namely, Sec6 (23) (Fig. 1B), Sec15 (24), and Exo70p (25) from the exocyst, Tip20p (13) from the Dsl1 complex, and Cog2 (26) and Cog4 (27) from the COG complex. Therefore, we refer to this bundle (residues 848–977) as the D domain.
Fig. 1.
Structure of the Vps54 C-terminal region (residues 836–974), crystal packing interactions, and comparative sequence analysis. (A) Ribbon backbone trace of Vps54 C-terminal domain (Vps54-CT) colored in blue. (B) Backbone trace of Vps54-CT structure (dark blue) superimposed on the D domain of Sec6 (light blue) illustrating that both domains share the same α-helical bundle with an rmsd of 3.2 Å. (C) Vps54-CT showing the surface properties of the contact region with the lattice-related molecule (gray) in tube drawing representation. The molecular surface is colored by residue conservation (red and orange denote strictly and highly conserved residues, respectively) or hydrophobicity (green). The packing highlights the potential presence of a C domain in full-length Vps54. (D) Multiple sequence alignment of Vps54 (residues 656–977) and domains C and D from structures of representative tethering complex subunits Sec15 (2a2f), Sec6 (2fji), Tip20 (3fhn), Exo70 (2pfv), and Cog4 (3hr0). Columns of residues that are likely to form the hydrophobic core of the structure have yellow background. Apolar residues are indicated in red. Cylinders show α-helical regions. Insertion regions between the aligned blocks are replaced by the numbers of residues. (E) Graphical representation of the α-helical bundle domain organization in N- to C-terminal order of Vps54 (836–974) with known structures of COG, exocyst, and Dsl1 subunits. Each α-helix is represented by a color-coded box along the sequence. Shown are Cog2p (PDB ID 2JQQ, residues 109–262 out of 262), Cog4 (3HRO, residues 536–785 out of 785), Dsl1* (combined model of Dsl1Clactis, PDB 3K8P, residues 333–684 and Dsl1p, PDB 3ETU, residues 37–355 out of 754), Tip20p (3FHN, residues 4–701 out of 701), Exo70p (2PFV, residues 67–623 out of 623), Exo84p (2D2S, residues 525–753 out of 753), Sec15 (2A2F, residues 382–699 out of 766), and Sec6p (2FJI, residues 411–805 out of 805).
The first helix (H1), the longest in the domain structure, is noticeably bent, with its N-terminal portion packed against the hydrophobic groove formed by helices H1 and H2 from the symmetrically related monomer, forming a pseudo-three-helix bundle. This interaction is mediated by highly conserved hydrophobic residues and stabilizes one of the major symmetry contacts in the crystal lattice (Fig. 1C and Fig. S1). Despite this intermolecular crystal contact, Vps54-CT behaves in solution as a monomer by size exclusion chromatography coupled to multiangle laser light scattering analysis. It is commonly assumed that crystal packing represents an energetically optimal arrangement where favorable contacts are preserved. When crystals are grown in the absence of the normal interaction partner, those contacts sometimes mimic native interactions. Therefore, it is likely that the intermolecular contacts between H1 from one monomer and H1-H2 from the symmetrically related monomer may be replaced by intramolecular contacts in the full-length Vps54. This packing behavior could involve a predicted C domain (Fig. 1B), in a similar manner as C and D domains interact in other MTC subunits. This notion is supported by the consensus positions at which nonpolar core residues are distributed within C and D domains in other MTC subunits with similar fold (Fig. 1D).
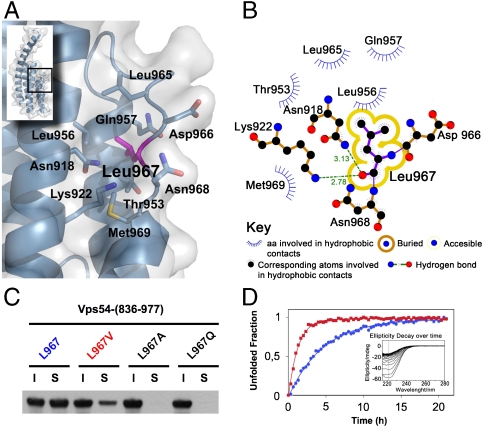
Loops that connect α-helices are relatively short and well defined in the structure. The last loop (L4), spanning residues 959–968, has a meander shape with conserved hydrophobic contacts such as Leu962 and Leu967 lying along the interhelical groove between H3 and H4 (Fig. 2A). Notably, the conserved Leu967 is completely buried into the hydrophobic interior (Fig. 2A) suggesting that its mutation to glutamine imposes different steric and hydrophobic constraints.
Fig. 2.
Leu967 interactions contribute to D-domain stability. (A) Stick model shown under translucent surface of the main hydrophobic pocket in the vicinity of L967 highlighted in magenta. (B) Diagram showing hydrophobic contacts, hydrogen bonds and solvent accessibility of L967. (C) Soluble (S) and insoluble (I) fractions of wild-type and mutant (L967V, L967A, and wobbler) GST-Vps54-CT after overexpression in E. coli. Proteins were detected by immunoblotting with antibody to GST conjugated to HRP. (D) Unfolding rate of wild-type (blue circles) and L967V mutant (red squares) Vps54-CT measured by ellipticity decay at 222 nm over time at 30 °C (inset shows raw data for the L967V mutant). Normalized ellipticity signal was used to calculate the decay rate constant from nonlinear-curve fitting.
In Vitro Destabilization of the Vps54 C-Terminal Domain by the Wobbler Mutation.
Protein stability upon single amino acid mutation strongly depends on the physical-chemical properties, location, and solvent accessibility of the mutated residue. Particularly in buried mutations, the amino acid hydrophobicity strongly correlates with stability (28). In the Vps54-CT crystal structure, the Leu967 side chain is packed among the methyl group of Thr953, Leu956, and the α and β carbons of Gln957 from H4, indicating predominant hydrophobic interactions (Fig. 2 A and B). The wobbler mutation (L967Q) barely reduces (by 7 Å3) the van der Waals volume of the residue, but the side-chain intrinsic hydrophobicity is considerably reduced from 73% to 6% (29), prompting us to examine changes in the stability and/or conformation caused by the L967Q mutation within D domain.
Not surprisingly, the L967Q mutant overexpressed in Escherichia coli was completely insoluble, in contrast to the native crystallized construct (Fig. 2C). We further investigated the importance of Leu967 burial by decreasing the length of the aliphatic side chain. We observed that mutation of Leu967 to Val967 or Ala967 drastically reduced the solubility of the D domain and only L967V remained partially soluble as compared to wild type (Fig. 2C). This result indicated that preserving the hydrogen bond between the carbonyl oxygen of the backbone and the amide proton of Asn918 or Lys922 is insufficient for correct folding (Fig. 2 B and C).
We examined the thermal denaturation of Vps54-CT by circular dichroism (CD); however, this process proved highly irreversible, precluding estimation of thermodynamic parameters (Fig. S2). Therefore, we used CD spectroscopy to further analyze the stability of the D domain by characterizing the isothermal ellipticity decay at 222 nm over time (Fig. 2D). Vps54-CT exhibited an exponential ellipticity decay with an apparent kinetic rate (kapp) of spontaneous unfolding that is concentration independent in the explored range (5–15 μM). The L967Q mutant could not be analyzed using this method because of its complete insolubility. The decay rate of the partially soluble L967V mutant, however, was 3.5-fold faster than that of the wild-type protein. These results indicate that the Leu967 side chain establishes critical hydrophobic contacts within the cavity volume, implying that the L967Q mutation affects the stability and/or correct folding of Vps54.
Assembly and Function of Vps54 Alleles in HeLa Cells.
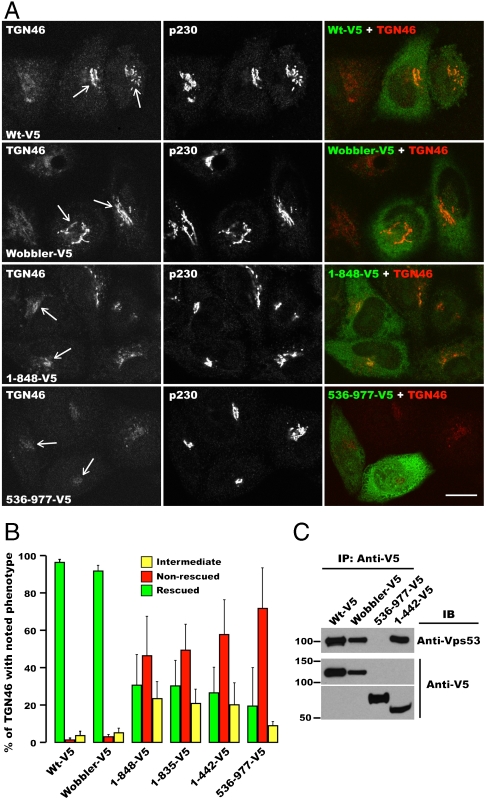
To determine the importance of the D domain and the impact of the wobbler mutation on GARP complex function, we knocked-down (KD) Vps54 in HeLa cells using a specific siRNA and rescued the cells by transfection with siRNA-resistant constructs encoding wild-type or mutant mouse Vps54 proteins tagged with a C-terminal V5 epitope (Fig. 3 and Fig. S3). As shown before (18), Vps54 KD greatly decreased staining for TGN46 at the TGN (Fig. S3A), reflecting inhibition of retrograde transport from endosomes. Transfection with different Vps54 constructs resulted in various extents of rescue, as exemplified in Fig. 3A and Fig. S3B, and quantified in Fig. 3B. We observed that both wild-type and wobbler Vps54 rescued the localization of TGN46 to the TGN (Fig. 3A) in the majority of transfected cells (Fig. 3B). In contrast, N-terminal (1–848, 1–835, and 1–442; all lacking a complete D domain) or C-terminal (536–977; containing the D domain) fragments of Vps54 largely failed to rescue TGN46 localization (Fig. 3 A and B and Fig. S3; mostly nonrescued or intermediate rescue phenotypes). Furthermore, coprecipitation analyses showed that both wild-type and wobbler Vps54 were incorporated into the GARP complex, as determined by coisolation with endogenous Vps53 (Fig. 3C). An N-terminal Vps54 construct (residues 1–442) was also incorporated into GARP (Fig. 3C), whereas a C-terminal Vps54 construct (residues 536–977) was not (Fig. 3C). Taken together, these experiments indicated that the N-terminal region of Vps54 is required for assembly into the GARP complex. The C-terminal region of Vps54 (including the D domain) is not involved in assembly but is required for function in TGN46 retrieval to the TGN. The fact that the wobbler mutation does not affect either function or assembly of the complex indicates a more subtle defect that is not functionally evident when the protein is overexpressed, as is the case in the RNAi-rescue assay used in these experiments.
Fig. 3.
Function and assembly of different Vps54 mutants. (A) HeLa cells depleted of Vps54 were transfected with the indicated siRNA-resistant Vps54 alleles containing a C-terminal V5 tag, fixed, and triple-labeled with antibodies to TGN46, V5, and p230. Arrows indicate the TGN in cells expressing the different Vps54 alleles. (Scale bar, 10 μm.) (B) Quantification of the percentage of transfected cells (n = 3; > 100 cells each) showing the indicated TGN46 phenotype for each Vps54 mutant allele tested, as described in Materials and Methods and in Fig. S3. (C) Assembly of Vps54 with endogenous GARP subunits depends on its N-terminal region. The indicated Vps54 alleles were expressed by transfection in HeLa cells, and protein lysates were prepared and immunoprecipitated with anti-V5 antibody. Endogenous Vps53 and transfected Vps54-V5 in the immunoprecipitates were analyzed by SDS-PAGE and immunoblotting. Molecular mass markers are shown on the left.
Reduced Stability of the Wobbler Vps54 Mutant Expressed in HeLa Cells.
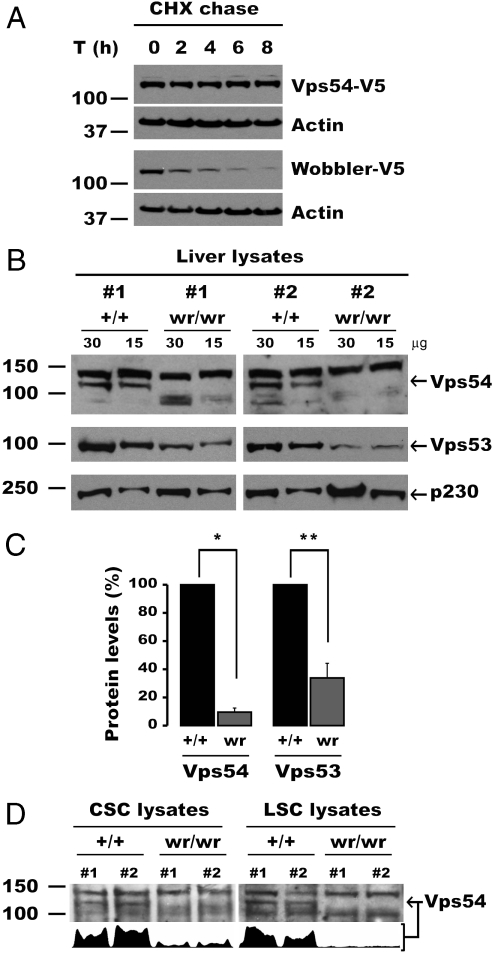
The experiments described above indicated that the wobbler mutation reduced the solubility and stability of the Vps54 D domain in vitro (Fig. 2), but did not affect the activity of the whole protein when it was overexpressed in vivo (Fig. 3). We noticed, however, that wobbler Vps54 was expressed at lower levels relative to wild-type Vps54 in HeLa cells (Fig. 3C). This observation prompted us to compare the stability of these Vps54 constructs by cycloheximide chase experiments (Fig. 4A). Indeed, we observed that the wobbler Vps54 mutant was rapidly degraded, whereas wild-type Vps54 was stable over 8 h. From these observations, we concluded that the unfolding and aggregation propensity of the mutant D domain in vitro (Fig. 2) was manifested as a faster degradation rate of the full-length protein in vivo (Fig. 4A).
Fig. 4.
The L967Q mutation destabilizes Vps54, resulting in reduced levels of GARP in the wobbler mouse. (A) Stability of Vps54 alleles analyzed by cycloheximide (CHX) chase. HeLa cells were transiently transfected with the indicated Vps54 constructs for 16 h, after which CHX was added to the medium to stop protein synthesis, and aliquots of cells were taken at different time points. Ten micrograms of lysate protein per lane were resolved by SDS-PAGE and immunoblotted with antibody to the V5 tag. A nonspecific reactive band of around 57 kDa was used as an internal loading control. One representative experiment out of three with similar results is shown. (B–D) Reduced levels of Vps54 and GARP in the wobbler mouse. (B) Thirty or 15 μg of extract proteins from the livers of two wobbler (wr/wr) mouse and two control Vps54 +/+ (+/+) littermates were resolved in contiguous lanes by SDS-PAGE. The levels of endogenous Vps54 (Top) and Vps53 (Bottom) in both groups of mice were compared by immunoblotting. The trans-Golgi p230 protein was used as loading control. (C) Quantification of Vps54 and Vps53 levels in control (+/+) and wobbler (wr) mice. Mean ± SD from three pairs of mice are shown. *, p < 0.001; **, p < 0.01. (D) Vps54 levels measured by immunoblotting (15 μg/lane) in relevant tissues of wild-type and wobbler mice. (Lower) Integrated intensity profile corresponding to Vps54 band. CSC, cervical spinal cord; LSC, lumbar spinal cord. Molecular mass markers are shown on the left. Arrows indicate Vps54.
Reduced Vps54 Levels in Tissues from Wobbler Mice.
The above data showed that, despite being rapidly degraded (Fig. 4A), the wobbler Vps54 allele retains functionality upon overexpression in a cultured cell system (Fig. 3 A and B). The instability of this mutant Vps54, however, could compromise the assembly and function of the GARP complex when expressed at endogenous levels. To test this hypothesis, we determined the levels of Vps54 in liver from wild-type (+/+) Vps54 mice and homozygous wobbler (wr/wr) littermates, killed at 8 weeks after birth, when motor defects are clearly discernible. Immunoblotting with an antibody raised to the whole D domain showed that Vps54 was present at ∼10-fold lower levels in the liver of wobbler mice as compared to that of control littermates [Fig. 4 B (arrow) and C]. In addition, total Vps53 levels were decreased ∼3-fold (Fig. 4 B and C), indicative of destabilization of the whole GARP complex.
The wobbler phenotype is characterized by degeneration of motor neurons, more prominently in cortical spinal cord than in lumbar spinal cord. Because the wobbler mutation affects the intrinsic stability of Vps54, however, we expected decreased Vps54 levels independently of the tissue analyzed and its pathology. Indeed, this was the case, as cervical spinal cord, lumbar spinal cord (Fig. 4D), brain cortex, and cerebellum all showed reduced Vps54 levels. From these experiments we concluded that the wobbler phenotype arises from greatly decreased levels of Vps54—and consequently of the GARP complex—in all tissues of wobbler mice. The residual GARP complex containing mutant Vps54 appears sufficient to support the viability of wobbler mice but not the health of motor neurons.
Discussion
The resolution of the crystal structure of the C-terminal region of mouse Vps54 (Vps54-CT) reported here demonstrates that this GARP subunit is structurally similar to other subunits of the DCGE subfamily of tethering complexes and that the similarity extends to the C-terminal regions of the proteins. Vps54-CT comprises a five-α-helix bundle similar to those found in the Tip20p subunit of Dsl1 (13), the Cog2p and Cog4 subunits of COG (26, 27), and the Sec6p, Sec15, Exo70p, and Exo84p subunits of the exocyst (11, 23–25). All of these proteins consist of an elongated, tandem array of α-helical-bundle modules designated A, B, C, D, and E, in N- to C-terminal order (11, 13) (Fig. 1E). The Vps54-CT structure most strongly resembles the D domain, a designation that we have adopted for this fragment. The fact that this domain occurs at the very C terminus of Vps54 indicates that this protein lacks an E domain. The nonphysiological stacking of monomers within the crystal hints at the continuation of the α-helical-bundle structure toward the N terminus of the protein, similar to that found in other MTC subunits. Interestingly, this crystal packing behavior with C-D-domain interactions has been previously observed for Cog4 (27). Multiple sequence alignments of Vps54-CT, and the corresponding regions of Sec15, Sec6p, Tip20p, Exo70p, and Cog4 shows the presence of nonpolar buried residues at consensus positions along the C and D domains, consistent with the existence of a C domain N-terminal to the D domain of Vps54 (Fig. 1D). These findings provide structural evidence that a GARP subunit shares a common ancestral relationship with subunits of Dsl1, COG, and exocyst, and suggest that all of these complexes might function in a similar manner at different stages of the endomembrane system.
The N-terminal region of both yeast (15, 30) and mammalian Vps54 (31) is involved in assembly with the other subunits of the GARP complex, whereas the C-terminal region including the D domain is dispensable for assembly. The requirement of the D domain to rescue TGN46 recycling to the TGN, however, indicates that this domain is essential for Vps54 function. Likewise, the 296 C-terminal residues from yeast Vps54p are required for retrieval of the v-SNARE, Snc1, from endosomes to the TGN (30). Moreover, the C-terminal region of mammalian Vps53 participates in tethering through interactions with SNAREs (31). The free C-terminal domains might thus project away from the assembled N-terminal core, allowing for long-range interactions with other components of the vesicular transport machinery. The identity of such interactions partners remains to be established.
The choice of the C-terminal part of Vps54 for our initial structural analysis of GARP was prompted by the occurrence of the spontaneous L967Q mutation in the wobbler mouse, an animal model of motor neuron degeneration similar to that in ALS. The crystal structure of Vps54-CT shows that Leu967 is completely buried within a groove formed by helices H3 and H4 through predominantly hydrophobic interactions. Both the hydrophobicity and size of the Leu967 side chain contribute to folding and stabilization of the D domain in vitro. Substitution of this side chain by the hydrophilic glutamine or smaller valine side chains causes insolubility and conformational instability of the D domain. This instability is reflected as a higher rate of degradation and lower levels of the full-length Vps54 L967Q mutant as compared to wild-type Vps54 upon expression in HeLa cells. The Vps54 L967Q mutant is nonetheless capable of rescuing TGN46 recycling to the TGN in HeLa cells depleted of endogenous Vps54. Hence, this mutant protein is intrinsically active, unlike the D-domain deletion mutants, which are largely inactive. The ability of the Vps54 L967Q mutant to correct the TGN46 retrieval defect despite its reduced instability is likely due to overexpression of the mutant protein in the RNAi-rescue protocol, a common property of hypomorphic mutants.
The hypomorphic nature of the Vps54 L967Q mutant is also evident from the fact that wobbler mice are viable despite exhibiting neurological and male reproductive defects, which is in contrast to Vps54 null mice, which die early during embryonic development (19). Therefore, the Vps54 L967Q mutant also appears to retain some activity in the mice. However, the structural instability and lower levels of this protein when expressed at endogenous levels in the mice render it limiting for assembly of the GARP complex. Indeed, various tissues from wobbler mice show greatly reduced levels of not only Vps54 but also Vps53. This reduction is likely a consequence of degradation of the other GARP subunits in the absence of Vps54, as previously demonstrated in yeast (14). Thus, the wobbler phenotype likely results from reduced levels of GARP complex. The fact that defects are most dramatically manifested in some cell types (i.e., motor neurons and sperm cells) is a common occurrence in genetic defects of ubiquitous proteins, and reflects greater susceptibility of those cell types to reduced GARP levels. Motor neurons, in particular, have long axons (up to 1 m) and their health might therefore be more dependent on the integrity of intracellular traffic. It remains to be determined how reduced levels of GARP lead to the wobbler phenotype. The impaired sorting of acid hydrolases to lysosomes in GARP-depleted cells results in accumulation of undegraded materials and swelling of lysosomes (18), a phenotype similar to that of lysosomal storage disorders (32). The intense vacuolization and morpho-functional alterations observed in wobbler motor neurons are reminiscent of such a phenotype (20). Indeed, animal models of acid hydrolase deficiency such as CatD-deficient mice and sheep manifest lysosomal dysfunction, impaired autophagy, and massive neurodegeneration (33, 34). Defective autophagy has also been proposed to contribute to motor neuron death in wobbler mice (35–37) and some ALS patients (38). These findings highlight the importance of endosomal/lysosomal traffic for maintenance of motor neuron integrity and warrant further studies of the role of GARP in this process.
Materials and Methods
Cloning, Protein Expression, Purification and Crystallization.
See SI Materials and Methods for details.
Structure Determination.
Native and selenomethionyl proteins crystallized within the same monoclinic (C2) lattice with two molecules per asymmetric unit. The structure was determined at 2.4 Å by combining the two wavelength anomalous diffraction dataset from the selenomethionyl crystal and the native dataset (2WMADN) using the BP3 program from the CCP4 suite (39) (Fig. S4 and Table S1). Subsequent solvent flattening, phase improvement, and model building was carried out using COOT (40) and the PHENIX suite (41).
The structure of native Vps54-CT crystallized in P212121 space group was solved by molecular replacement from data collected to 1.7 Å using the Vps54-CT structure crystallized in C2 space group as a search model in PHENIX.automr (41). The final model has a good geometry with no residues in disallowed regions of the Ramachandran plot. Together, the two crystal forms present images of four copies of Vps54-CT that adopt very similar conformations (Fig. S5). Crystallographic statistics are summarized in Table S1.
Vps54 KD and Rescue in HeLa Cells.
To deplete endogenous Vps54, HeLa cells were transfected twice at 48 h intervals with 50 nM of an siRNA oligonucleotide (UCACGAUGUUUGCAGUUAAUU, J-021174-07) from Dharmacon, Inc. targeting human Vps54 but not the murine gene. The extent of Vps54 depletion was monitored by immunofluorescence microscopy for TGN46, which depends on GARP to recycle from endosomes to the TGN (18). Penetrance of the interference phenotype ranged from 70% to 93% of treated cells. For functional analysis of the different Vps54 mutant alleles, depleted cells were processed as described in ref. 18. Fluorescently labeled cells were examined using an inverted confocal laser scanning microscope (model LSM 510; Carl Zeiss MicroImaging, Inc.) equipped with Ar, HeNe, and Kr lasers, and a 63 × 1.4 N.A. objective. Identical settings were used for all images taken in each panel. The staining pattern for TGN46 in each transfected cell was scored visually, distinguishing three phenotypic groups: (i) rescued cells, having TGN46 in the TGN area with high intensity (similar to nondepleted cells); (ii) nonrescued cells, in which TGN46 remained mislocalized and not concentrated at the TGN (similar to depleted cells); or (iii) intermediate, with TGN46 delineating the TGN but without reaching the normal intensity at this organelle. Examples for the three phenotypes are given in Fig. S3B. This experiment was performed three independent times, scoring at least 100 transfected cells per condition in each experiment.
Supplementary Material
Acknowledgments.
We thank the staff of the European Synchrotron Radiation Facility (Grenoble, France) for assistance with X-ray data collection under the Block Allocation Group MX-924. We are very grateful to Adriana L. Rojas for discussion and comments on the manuscript. This research was supported by the Carlos III Health Institute Grant PI081739 (to A.H.) and the intramural program of the National Institute of Child Health and Human Development (to J.S.B.). G.A.-P. has been supported by fellowship from the Basque Government (BFI08.53). A.H. is the recipient of a “Ramón y Cajal” contract from the Spanish Ministry of Science and Innovation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3N1B and 3N1E).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004756107/-/DCSupplemental.
References
- 1.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12(5):671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Pfeffer SR. Transport-vesicle targeting: Tethers before SNAREs. Nat Cell Biol. 1999;1(1):E17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 3.Lupashin V, Sztul E. Golgi tethering factors. Biochim Biophys Acta. 2005;1744(3):325–339. doi: 10.1016/j.bbamcr.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am J Physiol—Cell Physiol. 2006;290(1):C11–26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 5.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13(7):577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 6.Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: Gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21(4):543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sztul E, Lupashin V. Role of vesicle tethering factors in the ER-Golgi membrane traffic. FEBS Lett. 2009;583(23):3770–3783. doi: 10.1016/j.febslet.2009.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1(4):527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 9.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei J, Ma C, Rizo J, Grishin NV. Remote homology between Munc13 MUN domain and vesicle tethering complexes. J Mol Biol. 2009;391(3):509–517. doi: 10.1016/j.jmb.2009.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12(12):1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 12.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139(6):1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16(2):114–123. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11(1):305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277(50):48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- 16.Conibear E, Cleck JN, Stevens TH. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell. 2003;14(4):1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J Biol Chem. 2003;278(7):5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Victoria FJ, Mardones GA, Bonifacino JS. Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell. 2008;19(6):2350–2362. doi: 10.1091/mbc.E07-11-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt-John T, et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet. 2005;37(11):1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- 20.Boillee S, Peschanski M, Junier MP. The wobbler mouse: A neurodegeneration jigsaw puzzle. Mol Neurobiol. 2003;28(1):65–106. doi: 10.1385/MN:28:1:65. [DOI] [PubMed] [Google Scholar]
- 21.Meisler MH, et al. Evaluation of the Golgi trafficking protein VPS54 (wobbler) as a candidate for ALS. Amyotroph Lateral Sc. 2008;9(3):141–148. doi: 10.1080/17482960801934403. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13(6):555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 24.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12(10):879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 25.Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J Mol Biol. 2007;371(2):410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavanaugh LF, et al. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem. 2007;282(32):23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- 27.Richardson BC, et al. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci USA. 2009;106(32):13329–13334. doi: 10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlassi M, Cesareni G, Kokkinidis M. A correlation between the loss of hydrophobic core packing interactions and protein stability. J Mol Biol. 1999;285(2):817–827. doi: 10.1006/jmbi.1998.2342. [DOI] [PubMed] [Google Scholar]
- 29.Mant CT, Kovacs JM, Kim HM, Pollock DD, Hodges RS. Intrinsic amino acid side-chain hydrophilicity/hydrophobicity coefficients determined by reversed-phase high-performance liquid chromatography of model peptides: Comparison with other hydrophilicity/hydrophobicity scales. Biopolymers. 2009;92(6):573–595. doi: 10.1002/bip.21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quenneville NR, Chao TY, McCaffery JM, Conibear E. Domains within the GARP subunit Vps54 confer separate functions in complex assembly and early endosome recognition. Mol Biol Cell. 2006;17(4):1859–1870. doi: 10.1091/mbc.E05-11-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol Cell Biol. 2009;29(19):5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settembre C, Fraldi A, Rubinsztein DC, Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy. 2008;4(1):113–114. doi: 10.4161/auto.5227. [DOI] [PubMed] [Google Scholar]
- 33.Koike M, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20(18):6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyynela J, et al. A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J. 2000;19(12):2786–2792. doi: 10.1093/emboj/19.12.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popper P, Farber DB, Micevych PE, Minoofar K, Bronstein JM. TRPM-2 expression and tunel staining in neurodegenerative diseases: Studies in wobbler and rd mice. Exp Neurol. 1997;143(2):246–254. doi: 10.1006/exnr.1996.6364. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez Deniselle MC, et al. Progesterone neuroprotection in the Wobbler mouse, a genetic model of spinal cord motor neuron disease. Neurobiol Dis. 2002;11(3):457–468. doi: 10.1006/nbdi.2002.0564. [DOI] [PubMed] [Google Scholar]
- 37.Bigini P, et al. Lack of caspase-dependent apoptosis in spinal motor neurons of the wobbler mouse. Neurosci Lett. 2007;426(2):106–110. doi: 10.1016/j.neulet.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 38.Fornai F, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2008;105(6):2052–2057. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anonymous. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60(Pt 12, Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.