Abstract
Retrotransposons including endogenous retroviruses and their solitary long terminal repeats (LTRs) compose >40% of the human genome. Many of them are located in intergenic regions far from genes. Whether these intergenic retrotransposons serve beneficial host functions is not known. Here we show that an LTR retrotransposon of ERV-9 human endogenous retrovirus located 40–70 kb upstream of the human fetal γ- and adult β-globin genes serves a long-range, host function. The ERV-9 LTR contains multiple CCAAT and GATA motifs and competitively recruits a high concentration of NF-Y and GATA-2 present in low abundance in adult erythroid cells to assemble an LTR/RNA polymerase II complex. The LTR complex transcribes intergenic RNAs unidirectionally through the intervening DNA to loop with and modulate transcription factor occupancies at the far downstream globin promoters, thereby modulating globin gene switching by a competitive mechanism.
Keywords: BAC transgenic mice, NF-Y bound at CCAAT motif, transcriptional regulation, globin gene switching
Retrotransposons including long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), endogenous retroviruses (ERVs), and the solitary long terminal repeats (LTRs) of ERVs are repetitive DNA elements composing >40% of the human genome (1, 2). They have been considered selfish DNAs serving no useful host functions, as retrotransposon insertions disrupt gene expression and cause diseases (3, 4). Hence, host cells have been shown to develop mechanisms to silence retrotransposon expression (5). Yet, retrotransposons contain promoters and enhancers (1) and can serve relevant host function in promoting mRNA synthesis of the immediately downstream host genes (6, 7). The majority of retrotransposons, however, are located in the intergenic or intronic regions up to hundreds of kilobases from the promoters of linked genes (2, 8). Whether these distant retrotransposons serve a beneficial host function is not known.
The human endogenous retroviruses (HERVs) and their solitary LTRs compose ∼10% of the human genome (1, 2). They were inserted into the germ cells of primates millions of years ago and many are stably inherited during primate evolution (9). There are ∼50 copies of the ERV-9 endogenous retroviruses and ∼4,000 copies of solitary ERV-9 LTRs in the human genome (10, 11). The solitary LTRs contain the U3 region spanning the retroviral enhancer and promoter; the R region, whose 5′ end marks the initiation site of retroviral RNA synthesis; and the transcribed U5 region but no retroviral gag, pol, and env genes (12). Compared with the LTRs of other families of HERVs, the ERV-9 LTRs exhibit an unusual sequence feature: The U3 enhancer region spans 5–17 tandem repeats of 40 bases that contain recurrent CCAAT and GATA motifs (13–15).
The ERV-9 LTRs are associated with a number of hematopoietic and disease gene loci, including the β-like globin genes (14), the major histocompatibility (MHC) genes (16), the axin gene, located near the α-globin gene cluster (15), the selectin genes (AL022146), and the BRCA1 gene (L78833). In the β-globin gene locus, an ERV-9 LTR retrotransposon is stably integrated near the 5′ end of the locus control region (LCR), at 40–70 kb upstream of γ- and β-globin genes in higher primates from orangutans to humans (15). The ERV-9 LTRs in the human and chimpanzee globin gene loci, in reporter gene assays, exhibit prominent enhancer and promoter activities in embryonic and hematopoietic progenitor cells (14, 15, 17). To investigate whether this intergenic LTR retrotransposon serves a host function in regulating transcription of the far downstream globin genes, we generated transgenic (Tg) mice carrying the entire 100-kb human globin gene locus with or without the ERV-9 LTR. Locus-wide analyses of transcription factor occupancies, transcriptome status, and in vivo chromatin conformation provided unique experimental evidence that an intergenic retrotransposon serves a long-range, beneficial host function.
Results
Generation of Transgenic Mice Carrying the Human Globin Gene Locus with or Without the ERV-9 LTR by cre-loxP-Mediated in Situ Recombination.
To investigate whether the intergenic ERV-9 LTR regulates transcription of the far downstream globin genes, we generated Tg mice carrying a 100-kb BAC DNA spanning the entire human globin gene locus with a floxed ERV-9 LTR (Fig. 1A). We then used Cre-loxP-mediated in situ recombination by crossing the BAC Tg mice with cre Tg mice to delete the ERV-9 LTR in the zygotes to generate the ΔLTR Tg mice (Fig. 1B and Fig. S1). This strategy ensured that the parental BAC lines and the derived ΔLTR lines contained the human globin gene locus integrated into identical host sites to eliminate the unpredictable effects of different host sites on transgene expression. Thus, any changes in globin gene expression observed in the deletion mutants cannot be due to position-of-integration effects and must be due solely to deletion of the ERV-9 LTR.
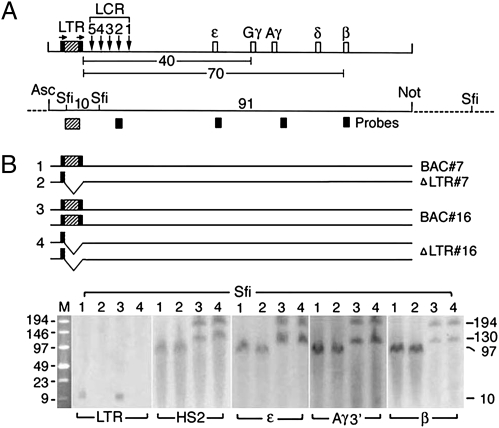
Fig. 1.
Structural analysis of the human β-globin gene locus in Tg mice. (A) (Upper) Map of BAC DNA spanning the 100-kb human globin gene locus. Hatched box flanked by black bars shows the ERV-9 LTR flanked by loxP sites; arrows designate direction of loxP sites, inserted into the BAC DNA by Chi-recBCD-mediated homologous recombination in E. coli (18) (SI Materials and Methods). Vertical arrows, DNase I hypersensitive sites HS 5–1 underlying the cores of the LCR; open boxes, embryonic ε-, fetal Gγ- and Aγ-, and adult δ- and β-globin genes; numbers, distances in kilobases between the LTR and the globin genes. (Lower) Map of restriction enzyme sites in the integrated BAC DNA (solid horizontal line) and the flanking mouse DNA (dotted lines) used for excision of the BAC DNA (AscI and NotI sites) and Southern blots (SfiI sites); numbers, sizes in kilobases of DNA fragments generated by SfiI and NotI digestions; hatched and solid boxes, the LTR, HS2, and the globin gene probes used in Southern blots. (B) (Upper) Horizontal lines grouped under 1, 2, 3, and 4 depict the sizes and copy numbers of the BAC DNA in the parental BAC and the derived ΔLTR Tg mice of lines 7 and 16. (Lower) Southern blots following SfiI digestion and pulsed field gel electrophoresis (PFGE). Lanes 1 and 2 and lanes 3 and 4 show bands generated by BAC and ΔLTR Tg mice of lines 7 and 16, respectively. M, size markers in kilobases; numbers in the right margin, sizes in kilobases of the Southern bands.
The integration sites of the human globin gene locus in four lines of BAC Tg mice and the four derived lines of ΔLTR isogenic Tg mice were analyzed by Southern blots following pulsed-field gel electrophoresis (Fig. 1B and Fig. S2). In all four lines, the sizes of the Southern bands generated by the BAC and the corresponding ΔLTR lines were identical (Fig. 1B and Fig. S2B), indicating that the human globin gene loci in each set of BAC and ΔLTR lines were integrated into the same host sites. Note that the 10-kb band generated by the LTR probe was present in the BAC lines but not in the ΔLTR lines, confirming deletion of the ERV-9 LTR in the ΔLTR lines (Fig. 1B, LTR blot). In Tg lines carrying two copies of the human globin gene locus, identical numbers and sizes of Southern bands were generated by the BAC and the ΔLTR mice (line 16, Fig. 1; line 6, Fig. S2). This result indicated that the two copies of the human globin gene locus in the BAC lines were integrated not in tandem and did not undergo cre-mediated recombination between them, which otherwise would have produced a single Southern band of a different size in the ΔLTR lines.
Long-Range Function of the ERV-9 LTR in Regulating Globin Gene Switching.
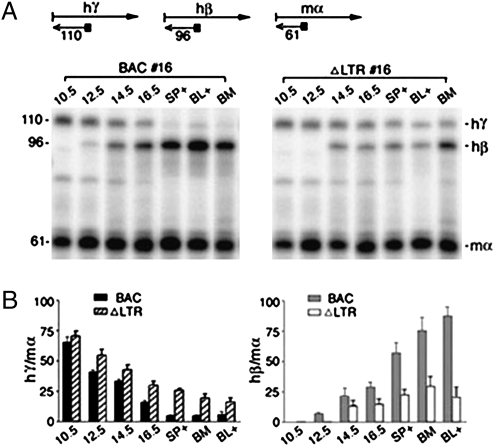
To determine the effect of the ERV-9 LTR on transcription of the globin genes, we analyzed the globin mRNA levels in erythroid cells of BAC and ΔLTR Tg mice at different developmental stages: yolk sac and fetal livers of 10.5- to16.5-d postcoitum embryos and adult spleen, bone marrow, and blood (Fig. 2A). Quantification of globin mRNA bands showed that LTR deletion drastically reduced transcription of the β-globin gene by 50–80% in adult erythroid cells of ΔLTR Tg mice (Fig. 2B). Interestingly, the fetal γ-globin gene, normally silenced in adult erythroid cells, was reactivated by 2- to 5-fold (compare γ-globin mRNA levels in adult spleen, bone marrow, and blood of BAC and ΔLTR Tg mice, Fig. 2; see Table S1 for statistical analysis of data). Analysis of globin mRNAs by RNase protection assay (line 16) and primer extension of two additional lines, Tg 6 and 10 (Figs. S3 and S4 and Table S1), showed the same long-range effects of LTR deletion in reprogramming transcription of the globin genes. Note that in all three lines of ΔLTR Tg mice, reactivation of the γ-gene was clearly observed only in later stages of erythroid development when the transcriptionally active β-globin gene was drastically suppressed. Moreover, in the fourth line, Tg 7, LTR deletion suppressed the β-globin gene by ∼50% as the other lines, without, however, the accompanying reactivation of γ-globin gene (Fig. S4B and Table S1). Thus, β-globin gene appeared to be the primary target of transcriptional regulation by the LTR.
Fig. 2.
ERV-9 LTR deletion suppressed transcription of the β-globin gene and reactivated the γ-globin gene in adult erythroid cells. (A) (Upper) Scheme of globin mRNA analysis by primer extension. Right arrows, human γ- and β- and mouse α-globin mRNAs; left arrows, cDNAs synthesized from the mRNAs with gene-specific primers (solid squares); numbers, sizes in bases of the cDNAs. (Lower) Polyacrylamide gel electrophoresis of globin cDNAs synthesized from globin mRNAs in 10.5 d postcoitum (dpc) embryonic yolk sac; 12.5, 14.5, and 16.5 dpc fetal livers; adult spleen; and blood erythroid cells of phenylhydrazine (PHZ)-injected adult Tg mice (SP+ and BL+) and bone marrow (BM) of non-PHZ-injected adult Tg mice. (B) Quantification of primer extension bands by PhosphorImager. hγ/mα and hβ/mα show intensities of the hγ and hβ bands normalized with respect to those of the mα bands set at 100. Values are mean ± SEM of three determinations from two RNA preparations.
ERV-9 LTR Assembles an LTR/NF-Y/GATA-2 Complex and Selectively Transfers the Associated Proteins to the β-Globin Gene 70 kb Away.
To investigate the molecular mechanism of long-range ERV-9 LTR function in mediating globin gene switching, we carried out locus-wide chromatin immunoprecipitation (ChIP) to examine the effect of the LTR on occupancies of transcription factors at sites throughout the human globin gene locus in adult erythroid cells of BAC and ΔLTR Tg mice. We focused on three key transcription factors: the ubiquitous NF-Y and the erythroid GATA-1 and -2. NF-Y binds to the recurrent CCAAT motifs in the ERV-9 LTR and serves as a scaffold to assemble the LTR complex; GATA-2 is recruited by NF-Y to the neighboring GATA motifs; and GATA-1, highly expressed in adult erythroid cells, can be recruited to the LTR through protein–protein interactions with GATA-2 (19, 20). We also investigated CBP, a coactivator, which interacts with NF-Y and the GATA factors (21, 22) and may mediate communication between the LTR complex and the basal transcription factors bound at the globin promoters (22, 23), and pol II, which has been reported to be recruited by the LCR and transferred by an undefined, long-range mechanism to the distant globin promoter (24).
NF-Y is a trimeric complex composed of NF-YA, -YB, and -YC subunits; YB and YC first associate through their histone fold domains to form a dimer, which then recruits YA; the trimeric NF-Y through a DNA-binding domain in YA binds to the consensus sequence (A/G)CCAATC(A/G)G(C/A)(G/A) with specificity and affinity among the highest for DNA-binding transcription factors (19, 25, 26). In the human globin gene locus, the ERV-9 LTR contains a uniquely high density of NF-Y binding sites, 10 sites in 0.7 kb of the LTR enhancer and promoter region, whereas the LCR contains no NF-Y binding sites in the core HS sites and only 4 NF-Y sites distributed across 15 kb of LCR DNA; the rest of the locus contains an average of 1 CCAAT motif per 0.5 kb DNA, many of which are nonconsensus NF-Y sites except for the γ-globin promoter, which contains 2 tandem NF-Y binding CCAAT motifs within 40 bases of DNA (Fig. 3A and Table S2).
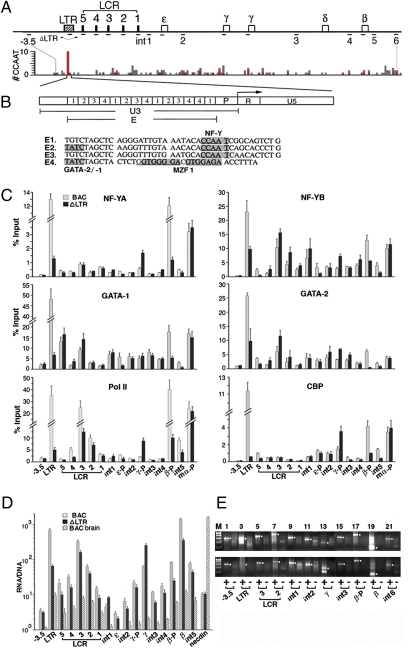
Fig. 3.
Occupancies of transcription factors in the human globin gene locus. (A) (Upper) Locations of DNA regions, marked by short horizontal lines, analyzed in ChIP assays; −3.5, DNA region at −3.5 kb 5′ of the ERV-9 LTR; int 1–5, intergenic regions 1–5 (see SI Materials and Methods for locations of PCR primer pairs). (Lower) Locus-wide distribution of CCAAT motifs. y axis, the number of CCAAT motifs in unit DNA lengths of 500 bases; gray bars, total number of CCAAT motifs/500 bp; red bars, NF-Y binding CCAAT motifs/500 bp (Table S2). (B) (Upper) Enlarged map of the ERV-9 LTR: U3 region spanning LTR enhancer (E) and promoter (P) and R and U5 regions. Angled arrow, direction of retroviral transcription initiated from the LTR promoter (15); boxes labeled 1, 2, 3, and 4, enhancer subunits E1–4. (Lower) DNA sequences of E1–4. Shaded bases show binding sites for NF-Y, GATA-2/-1, and MZF-1, respectively (19). (C) Locus-wide ChIP assays of adult spleen erythroid cells from PHZ-injected line 16 BAC and ΔLTR Tg mice. γ-P, β-P, and mα-P are human γ- and β-globin promoters and mouse α-globin promoter, respectively; for designations of other DNA regions see A. y axis shows relative levels of respective transcription factors and pol II associated with the globin locus; ChIP values were determined by real-time PCR and expressed as percentages of the input chromatin (SI Materials and Methods). ChIP values of preimmune IgG for separate DNA regions were 0.1–0.3% of Input (Fig. S5). ChIP values were averages of two independent antibody pull-down assays. Similar ChIP results were obtained with line 6 BAC and ΔLTR Tg mice (Fig. S5B). (D) Transcriptome analysis of the human globin gene locus in adult spleen erythroid cells of PHZ-injected line 16 BAC and ΔLTR Tg mice and brain cells of line 16 BAC Tg mice determined by real-time RT-PCR. Abundance of the RNAs was calculated from the PCR cycle numbers at the crossing-over points (in the range of 20–35 PCR cycles) and normalized with respect to the abundance of β-actin (29). RNA/DNA: Abundance of the RT-PCR products was further normalized with respect to abundance of the DNA products amplified by the same primer pair to correct for differences in amplification efficiencies of different primer pairs; primer sequences were selected to amplify RNAs transcribed from the human but not the mouse globin gene locus. Necdin: a murine neuronal gene. (E) (Upper and Lower) Directional RT-PCR (29) of RNAs isolated from the adult erythroid cells of line 16 BAC and ΔLTR Tg mice, respectively. + and − lanes show RT-PCR products of sense and antisense RNAs, which were reverse transcribed into cDNAs, respectively, by the reverse and forward primer of each primer pair. The cDNAs were then amplified with each primer pair by 44 PCR cycles. White dots in the margins show anticipated sizes of the RT-PCR bands. M: 100- to 500-bp DNA size markers.
Consistent with the distribution of NF-Y binding sites, the ERV-9 LTR bound NF-YA at a high level, whereas the LCR and the intergenic regions bound NF-YA at very low levels in adult erythroid cells of BAC Tg mice (Fig. 3C Top Left, NF-YA, BAC lanes). Surprisingly, the β-globin promoter bound much more NF-YA than the γ-globin promoter (Fig. 3C Top Left, NF-YA, β-P and γ-P lanes), even though the β-globin promoter contains a very weak NF-Y site and the γ-globin promoter contains two strong NF-Y sites, as shown by in vitro gel shift assays (27). Thus, the in vivo context of the locus enabled the weak NF-Y site in the β-globin promoter to bind NF-YA at a high level comparable to that of the LTR.
The occupancies of NF-YB and -YC, GATA-1 and -2, and CBP were similar to those of NF-YA (Fig. 3C and Fig. S5): high at the LTR and the β-globin promoter but low at the LCR, γ-globin promoter, and intergenic regions, except for HS3, which bound relatively high levels of some of these factors (Fig. 3C, BAC HS3 lanes). Although the LTR contains recurrent MZF1 binding sites (Fig. 3B), it did not bind a significant level of MZF1 (Fig. S5B), perhaps because the myeloid MZF1 was expressed at an extremely low level in adult erythroid cells (Fig. S6). However, GATA-2 was expressed also at a very low level in adult erythroid cells (Fig. S6). Yet, the LTR bound GATA-2 at a level comparable to that of the abundant GATA-1 (Fig. 3C Middle, GATA-1 and -2, LTR lanes). This was probably due to the LTR-bound NF-Y, which was able to recruit GATA-2 from a dilute pool and concentrate it at the LTR. Thus, the LTR recruited high concentrations of NF-YA and GATA-2 present in low abundance in adult erythroid cells (Fig. S6) to assemble an LTR/NF-Y/GATA-2 and -1 complex.
In the absence of the LTR in ΔLTR Tg mice, the β-globin promoter bound drastically lower levels of all six proteins (Fig. 3C, compare BAC and ΔLTR lanes of β-P), indicating that the LTR transferred the associated proteins to the β-globin promoter 70 kb away to enable it to assemble a functional promoter complex that could efficiently activate transcription of the β-globin gene in adult erythroid cells. In contrast, in the absence of the LTR, the LCR and the γ-globin promoter bound some or all of the six proteins at higher levels (Fig. 3C, compare BAC and ΔLTR lanes of the LCR and γ-P), indicating that the ERV-9 LTR did not transfer the associated proteins to the LCR and γ-promoter but competed with them for binding these proteins. Hence, in the absence of competition from the LTR, the γ-globin promoter with two tandem NF-Y sites could bind higher levels of NF-Y, and also GATA-2 and CBP, to reactivate transcription of the γ-globin gene in the adult erythroid cells of ΔLTR Tg mice. The switch in levels of occupancies of NF-Y, GATA-2, and CBP between the β- and the γ-globin promoter, due to LTR deletion, correlated with the switch in globin gene transcription: suppression of the β-globin gene and reactivation of the γ-globin gene. This correlation indicated that the γ- and β-globin promoters competed for binding transcription activators including NF-Y and GATA-2 present in limiting amounts and that the LTR was able to recruit and concentrate these limiting proteins and selectively transfer them to the β-globin promoter to activate transcription of the β-globin gene in adult erythroid cells.
RNA Polymerase II Recruited by the LTR Complex Synthesizes Sense, Intergenic RNAs Through the Globin Gene Locus.
Locus-wide pol II occupancies in Tg mice showed that LTR deletion significantly reduced pol II occupancy at both the LCR and the β-globin promoter (Fig. 3C Bottom Left, pol II), suggesting that the LTR-bound pol II transcribed through the LCR to potentially modulate transcription of the globin genes. To examine this possibility, we mapped the transcriptome of the globin gene locus in adult erythroid cells of both BAC and ΔLTR Tg mice. Quantitative RNA analysis by real-time RT-PCR showed that in BAC Tg mice the ERV-9 LTR, LCR, globin genes, and the intergenic regions were transcribed. However, the intergenic regions were transcribed at one to two orders of magnitude lower levels than the LTR, the LCR, and the γ- and β-globin genes (Fig. 3D). LTR deletion reduced transcription levels of the locus except for the region spanning the γ-globin gene (Fig. 3D), consistent with the reciprocal switch in γ- and β-globin gene transcription due to LTR deletion determined by primer extension (Fig. 2). The transcriptome analysis suggested that pol II recruited by the LTR transcribed through the LCR and multiple sites in intergenic DNA to ultimately reach and transcribe the β-globin gene. In support of this interpretation, directional RT-PCR showed that the locus in BAC Tg mice was transcribed in a sense direction colinear with the direction of transcription initiated from the LTR (Fig. 3E Upper). In the absence of the LTR, the locus was still transcribed in the sense direction (Fig. 3E Lower), indicating that pol II recruited by the LCR (Fig. 3C Bottom Left, pol II) also initiated sense transcription. However, sense transcription of the locus was generally lower in the absence of the LTR (Fig. 3E Upper and Lower, compare band intensities). Thus, pol II recruited by the LTR appeared to track and transcribe the LCR and intergenic DNA sites synthesizing sense, intergenic RNAs at very low levels to reach and activate transcription of the β-globin gene 70 kb away.
ERV-9 LTR Loops with Downstream, Intergenic DNA Sites and the Globin Genes.
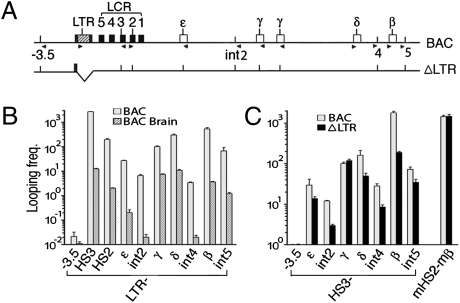
Assuming that the transcribing pol II enzyme recruited by ERV-9 LTR was associated with a stable LTR complex assembled by NF-Y tightly bound to the LTR DNA, we anticipated that the LTR DNA in the LTR/pol II complex should also physically track through and thus interact and loop with multiple sites in the LCR and intergenic DNA to ultimately reach and loop with the far downstream β-globin gene. To examine this possibility, we performed chromosome conformation capture (3C) assays (28, 29). In spleen erythroid cells of BAC Tg mice, the LTR DNA indeed looped with multiple sites in the locus, but at very different frequencies: It looped with the LCR HS3 site at 20 times higher frequency and with the β-globin gene at 3 times higher frequency than the looping frequency with the inactive γ-globin gene; in contrast, the LTR looped with the intergenic DNAs at one to two orders of magnitude lower frequencies and with the DNA upstream of it at nearly four orders of magnitude lower frequency than the looping frequency of the LTR with the γ-globin gene (Fig. 4 A and B). The very low looping frequencies of the LTR with the intergenic DNAs were, however, one to three orders of magnitude higher than those in the inactive globin gene locus in the brain cells of BAC Tg mice (Fig. 4B). In ΔLTR Tg mice, LTR deletion greatly reduced the looping frequencies of the LCR HS3 site with the β-globin gene and the intergenic DNAs, but did not significantly change the looping frequency of HS3 with the reactivated γ-globin gene (Fig. 4C). Thus, the LTR complex looped transiently with multiple, intergenic DNA sites to ultimately loop/interact with the β-globin gene to transfer the associated proteins. Moreover, presence of the LTR facilitated looping of the LCR with the β-globin gene.
Fig. 4.
The ERV-9 LTR interacted/looped with the globin genes and multiple sites in the intergenic DNA. (A) Locations of restriction enzyme sites and primers used in 3C assays. Vertical bars show BglII and BamHI sites used in 3C assays; arrowheads show locations and 5′ → 3′ direction of 3C primers (see SI Materials and Methods for primer sequences). (B) Looping frequencies of the ERV-9 LTR with the LCR, globin genes, and intergenic DNAs in adult spleen erythroid and brain cells of line 16 BAC Tg mice. y axis shows looping frequencies of the LTR with upstream and downstream DNA sites, shown on the x axis. Equivalent amounts of spleen erythroid and brain DNAs were used for 3C, as determined by PCR amplification of the human and mouse β-globin genes (see Fig. S7 for other 3C controls and SI Materials and Methods and ref. 29 for calculating looping frequencies). Looping frequency of the LTR with the γ-gene in erythroid cells was set at 100; values were averages of two independent 3C experiments. (C) Looping frequencies of the LCR HS3 site with the globin gene locus in spleen eythroid cells of line 16 BAC and ΔLTR Tg mice. Looping frequency of HS3-γ in BAC Tg mice was set at 100 and that of mouse HS2 with mouse β-globin gene served as a positive control.
Expression of ERV-9 LTR in Erythroid Progenitor Cells and Germinal Cells.
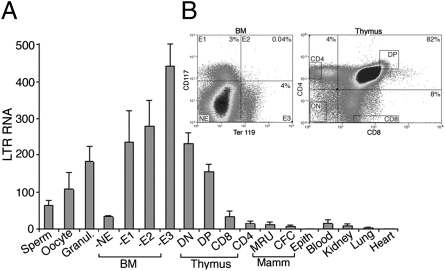
ERV-9 LTR retrotransposons are found in primates but not in rodents. Yet ERV-9 LTR function was tested in Tg mice. To determine if ERV-9 LTR was similarly expressed in Tg mice as in humans, we analyzed the transcription levels of ERV-9 LTR in various Tg mouse tissues. In humans, ERV-9 LTR is transcribed in the progenitor cells of erythroid and a number of other tissues and in oocytes and follicular granulosa cells (17). Similarly in Tg mice, ERV-9 LTR was transcribed at high levels in female germinal cells and erythroid and thymus progenitor cells (Fig. 5, BM-E1-3 and thymus DN and DP lanes) but at very low levels in mammary progenitor cells and cells of differentiated tissues including blood, kidney, lung, and heart (Fig. 5). An exception was mouse sperms, which expressed ERV-9 LTR at a high level (Fig. 5), in contrast to human sperms (17). The generally similar expression patterns of the ERV-9 LTR in Tg mice and humans indicated that mouse transgenics can serve as an appropriate system to test the function of primate ERV-9 LTRs.
Fig. 5.
Expression of the ERV-9 LTR in germinal cells and hematopoietic progenitor cells. (A) Transcription levels of the ERV-9 LTR (R-U5 region) in various tissues of line 16 Tg mice (2–4 wk old) determined by real-time RT-PCR. NE, E1, E2, and E3: nonerythroid cells and early-, mid-, and late-stage erythroid progenitor cells, respectively, from bone marrow (BM) sorted by FACS with antibodies to stem cell factor (CD117) and glycophorin A-associated protein (Ter 119) (38). Thymus DN, DP, CD8, and CD4 cells: double negative most immature progenitor cells, double positive more mature progenitor cells, CD8high, CD4low and CD4high, and CD8low mature thymocytes, respectively, sorted by FACS with CD4 and CD8 antibodies. MRU, CFC, and Epith: FACS-sorted, mammary stem, progenitor, and epithelial cells of female Tg mice. LTR RNA levels were normalized with respect to the level of β-actin RNA set at 100. Data were averages of two independent determinations. (B) FACS dot plots of sorted BM and thymus cells. Percentages are percentages of sorted cells in each quadrant.
Discussion
This study demonstrated that the ERV-9 LTR, spanning a uniquely high density of CCAAT motifs in the globin gene locus, competitively bound NF-Y and GATA-2 present in low abundance in adult erythroid cells and selectively transferred the associated proteins to the β-globin promoter to activate transcription of the β-globin gene 70 kb away (Fig. 6). Activation of the β-globin gene is normally accompanied by repression of the γ-globin gene and vice versa. This reciprocal globin gene switching suggests that γ- and β-globin genes compete for binding certain limiting factors in erythroid cells (30). Indeed, many factors have been reported to reactivate the γ-globin gene and thus regulate globin gene switching (30–32). However, NF-Y and GATA-2 are prime examples of transcription factors, which directly bind to DNA motifs in the γ- and β-globin promoters, and due to their low abundance in adult erythroid cells, showed reciprocal occupancies at the γ- and β-globin promoters in correlation with reciprocal switching of γ- and β-globin gene transcription. In the monkey genome, before retrotransposition of the ERV-9 LTR into the β-globin gene locus in the higher primates (15), globin genes undergo switching in the absence of the LTR, although NF-Y bound at the CCAAT motif in the γ-globin promoter is critical to transcriptional activation and developmental switching of the γ-globin gene in the lower primates (33, 34). Thus, during primate evolution different mechanisms may have evolved to modulate NF-Y recruitment and globin gene switching.
Fig. 6.
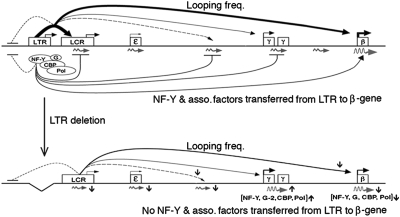
Long-range function of the ERV-9 LTR. Horizontal lines, the human globin gene locus; ovals, proteins associated with the LTR; G, GATA-2 and -1; angled arrows, direction of transcription initiated from the LTR, LCR, and globin genes. Curved arrows above the horizontal line show looping frequencies of the LTR with downstream sites; thickness of solid, dashed, and dotted lines shows high to very low levels of transcription and looping frequencies. Curved arrow below the horizontal line shows factor transfer from the LTR. Curved lines ending in horizontal bars show no interaction/looping with the LTR or no transfer of factors from the LTR. Squiggly arrows, intergenic and globin RNAs transcribed in the sense direction by the ERV-9 LTR complex; brackets, proteins whose levels changed reciprocally at the γ- and β-globin promoters due to LTR deletion; G-2, GATA-2. Vertically up and down arrows, up- or down-regulation in looping frequencies, transcription factor occupancies, and transcription due to LTR deletion.
A tracking and transcription mechanism initiated by the LTR/pol II complex appeared to mediate the long-range transfer of proteins from the ERV-9 LTR to the β-globin gene. Our 3C results indicated that the LTR complex did not randomly sample and loop with DNA sites in the surrounding nucleoplasm to finally loop with the β-globin gene by chance encounter, because the LTR DNA looped not with upstream DNA but only with downstream DNA. As the globin locus was transcribed in a direction from the LTR to the β-globin gene, the LTR complex was likely guided by a unidirectional, tracking and transcription mechanism (29): It transcribed and looped transiently with multiple, downstream intergenic sites, synthesizing sense intergenic RNAs at low levels, to ultimately loop with and transfer the associated proteins to the β-globin gene. Although the LTR complex also looped with the γ-globin gene and at a very high frequency with the LCR HS3 site, it did not transfer the associated proteins to these sites, probably because the HS3 site and the γ-globin promoter contain distinct combinations of DNA sequence motifs and assembled spacio-specific transcription complexes that were unable to interact productively with the LTR complex to allow transfer of LTR-associated proteins.
The ERV-9 LTR was transcribed at high levels in erythroid and thymus progenitor cells, indicating that the ERV-9 LTR enhancer and promoter initiated RNA synthesis preferentially in hematopoietic progenitor cells. This result is consistent with recent genome-wide transcriptome analysis showing that retrotransposons in the human and mouse genomes initiate synthesis of noncoding, intergenic RNAs in a tissue-specific manner (35). As 60% of the annotated human promoters contain CCAAT motifs that may bind NF-Y (36), the ERV-9 LTRs in the human genome may competitively bind NF-Y present in limiting amounts and selectively transfer NF-Y to target promoters to modulate transcription of distant, cis-linked genes, such as the globin, MHC, axin, and selectin genes. The ERV-9 LTR through its high-affinity binding of NF-Y may coordinate the transcriptional networks of these cis-linked genes during hematopoiesis. Thus, at least some of the 4,000 copies of the ERV-9 LTR retrotransposons distributed across the human chromosomes may serve a beneficial host function and may not be junk DNAs.
Materials and Methods
Construction of a BAC Clone Spanning the Human β-Globin Gene Locus with Floxed ERV-9 LTR.
The ERV-9 LTR flanked by loxP sites was introduced into a BAC clone spanning the 100-kb human β-globin gene locus (37) by Chi-RecBCD-mediated homologous recombination in Escherichia coli as described (18).
Generation of BAC and ΔLTR Tg Mice.
Purified, floxed BAC DNA was injected into the mouse zygotes by the Medical College of Georgia Transgenic Core. ΔLTR Tg lines were generated by crossing the parental BAC Tg mice with isogenic Cre Tg mice.
RNA analyses, ChIP, and 3C were carried out as described (29). For protocol details and other methods, see SI Text.
Supplementary Material
Acknowledgments
We thank Dr. T. Ley (Washington University) for the BAC clone of the human globin gene locus, Drs. J. Jenssen and H. Shizuya for advice on BAC cloning, Dr. P. Koni (Medical College of Georgia, Augusta, GA) for cre Tg mice, Dr. K. Terui for demonstrating dissection of mouse embryos, Dr. L. Ignatowicz for instruction on fractionation and biology of thymocytes, and Drs. W. Dynan and V. Ganapathy for critical reading of the manuscript. This work was supported by National Institutes of Health Grants HL 62308 and 73453.
Footnotes
This article is a PNAS Direct Submission.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004139107/-/DCSupplemental.
References
- 1.Henikoff S, et al. Gene families: The taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J, Stoye J. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. pp. 343–436. [PubMed] [Google Scholar]
- 4.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 5.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 8.Sela N, et al. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkison D, Mager D, Leong J. In: The Retroviridae. Levy J, editor. Vol. 3. New York: Plenum; 1994. pp. 465–535. [Google Scholar]
- 10.Henthorn PS, Mager DL, Huisman TH, Smithies O. A gene deletion ending within a complex array of repeated sequences 3′ to the human beta-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:5194–5198. doi: 10.1073/pnas.83.14.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löwer R, Löwer J, Kurth R. The viruses in all of us: Characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Temin HM. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981;27:1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- 13.La Mantia G, et al. Identification and characterization of novel human endogenous retroviral sequences prefentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 1991;19:1513–1520. doi: 10.1093/nar/19.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long Q, Bengra C, Li C, Kutlar F, Tuan D. A long terminal repeat of the human endogenous retrovirus ERV-9 is located in the 5′ boundary area of the human β-globin locus control region. Genomics. 1998;54:542–555. doi: 10.1006/geno.1998.5608. [DOI] [PubMed] [Google Scholar]
- 15.Ling J, et al. The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol. 2002;76:2410–2423. doi: 10.1128/jvi.76.5.2410-2423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svensson AC, Setterblad N, Sigurdardóttir S, Rask L, Andersson G. Primate DRB genes from the DR3 and DR8 haplotypes contain ERV9 LTR elements at identical positions. Immunogenetics. 1995;41:74–82. doi: 10.1007/BF00182316. [DOI] [PubMed] [Google Scholar]
- 17.Pi W, et al. The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc Natl Acad Sci USA. 2004;101:805–810. doi: 10.1073/pnas.0307698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jessen JR, et al. Modification of bacterial artificial chromosomes through chi-stimulated homologous recombination and its application in zebrafish transgenesis. Proc Natl Acad Sci USA. 1998;95:5121–5126. doi: 10.1073/pnas.95.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, et al. The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2. J Biol Chem. 2005;280:35184–35194. doi: 10.1074/jbc.M508138200. [DOI] [PubMed] [Google Scholar]
- 20.Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, et al. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blobel GA. CBP and p300: Versatile coregulators with important roles in hematopoietic gene expression. J Leukoc Biol. 2002;71:545–556. [PubMed] [Google Scholar]
- 23.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 24.Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 25.Bi W, Wu L, Coustry F, de Crombrugghe B, Maity SN. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- 26.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:16–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 27.Liberati C, Ronchi A, Lievens P, Ottolenghi S, Mantovani R. NF-Y organizes the gamma-globin CCAAT boxes region. J Biol Chem. 1998;273:16880–16889. doi: 10.1074/jbc.273.27.16880. [DOI] [PubMed] [Google Scholar]
- 28.Dekker J. The three ‘C’ s of chromosome conformation capture: Controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X, et al. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mabaera R, et al. A cell stress signaling model of fetal hemoglobin induction: What doesn't kill red blood cells may make them stronger. Exp Hematol. 2008;36:1057–1072. doi: 10.1016/j.exphem.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 33.Johnson RM, Gumucio D, Goodman M. Globin gene switching in primates. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:877–883. doi: 10.1016/s1095-6433(02)00205-2. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Fang X, Han H, Stamatoyannopoulos G. The minimal promoter plays a major role in silencing of the galago γ-globin gene in adult erythropoiesis. Proc Natl Acad Sci USA. 2004;101:8096–8101. doi: 10.1073/pnas.0402594101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki Y, et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman R, Pham C, Ley T. Transgenic analysis of a 100-kb human beta-globin cluster-containing DNA fragment propagated as a bacterial artificial chromosome. Blood. 1999;94:3178–3184. [PubMed] [Google Scholar]
- 38.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.