Abstract
Many Gram-negative bacteria secrete specific proteins via the type II secretion systems (T2SS). These complex machineries share with the related archaeal flagella and type IV pilus (T4P) biogenesis systems the ability to assemble thin, flexible filaments composed of small, initially inner membrane-localized proteins called “pilins.” In the T2SS from Klebsiella oxytoca, periplasmic pseudopili that are essential for pullulanase (PulA) secretion extend beyond the bacterial surface and form pili when the major pilin PulG is overproduced. Here, we describe the detailed, experimentally validated structure of the PulG pilus generated from crystallographic and electron microscopy data by a molecular modeling approach. Two intermolecular salt bridges crucial for function were demonstrated using single and complementary charge inversions. Double-cysteine substitutions in the transmembrane segment of PulG led to position-specific cross-linking of protomers in assembled pili. These biochemical data provided information on residue distances in the filament that were used to derive a refined model of the T2SS pilus at pseudoatomic resolution. PulG is organized as a right-handed helix of subunits, consistent with protomer organization in gonococcal T4P. The conserved character of residues involved in key hydrophobic and electrostatic interactions within the major pseudopilin family supports the general relevance of this model for T2SS pseudopilus structure.
Keywords: molecular modeling, pilus assembly, protein secretion, pseudopilus, cysteine cross-linking
Gram-negative bacteria use several mechanisms to secrete toxins and hydrolases with important roles in pathogenesis and metabolism. The type II protein secretion systems (T2SS) are membrane protein complexes that transport folded proteins from the periplasm to the extracellular medium through the outer membrane channel formed by the protein secretin (reviewed in ref. 1). Between 12 and 15 proteins comprising the T2SS are essential for function, and many are highly similar to components of type IV piliation (T4P) systems (2). Beyond their similarities to T4P in composition and sequence, T2SS were found to assemble pili on the bacterial surface upon overproduction of the major pseudopilin (3), suggesting a common basic mechanism (2, 4, 5). The surface-exposed T2SS pili are regarded as artificially extended periplasmic filaments called “pseudopili” that play a crucial but poorly understood role in protein secretion. The current models propose that the inner membrane (IM) assembly platform (6) transduces the energy generated by cytoplasmic ATPase PulE to facilitate pilin extraction from the IM into the growing periplasmic filament (7). According to the piston hypothesis, pseudopilus polymerization promotes secretion by direct interaction with the substrate to push it through the secretin channel (4, 8).
The surface pili of the pullulanase T2SS from Klebsiella oxytoca consist essentially of the major pseudopilin subunit PulG (3). The dimensions, flexibility, and bundling propensity of extended pseudopili are indistinguishable from those of T4P (4, 9). To understand the pilus assembly, it is essential to describe the interprotomer contacts at the atomic level and determine its high-resolution structure. In a previous study, Köhler et al. (9) analyzed the nonbundling PulG6xHis pili by scanning transmission electron microscopy (STEM) and metal shadowing. The measured helical parameters and the PulG crystal structure with a polylalanine helix replacing the missing transmembrane segment (TMS) were used to build a left-handed pseudopilus model (9). However, the highly conserved TMS is essential for several pilus biogenesis steps, from pilin membrane insertion via the signal recognition particle and Sec pathways (10), its maturation by the prepilin peptidase, and incorporation into pili through the IM assembly complex (11), to pilus stabilization. Here, the STEM data (9) and the 1.6-Å resolution PulG structure completed by modeling were used in a molecular dynamics approach to build a pseudoatomic model of the T2SS pilus. A detailed structure of the gonococcal T4P derived from a cryo-EM electron density map and the pilin crystal structure has allowed Craig et al. (7) to propose a model for T4P biogenesis. The T2SS pilus model generated here has the advantage of providing testable predictions that allowed us both to validate the structure and to define interactions key for pilus assembly and stability. It provides a framework for further dissecting pilus polymerization, dynamics, and function in protein secretion and for designing antibacterial agents targeting the T2SS. This approach is generally applicable in determining the structure of other protein polymers.
Results
PulG Structure and the Pilus Model Building.
The basis of the revised PulG homopolymer model was the structure of the PulG protomer, derived from the x-ray crystal structure of the PulG periplasmic domain (9). The 20 C-terminal residues of PulG were modeled by exploiting the close homology to GspG (70% identity in this segment) from enterohemorrhagic Escherichia coli (pdb 3G20), to include the calcium-binding site in the β2–β3 loop (12). The TMS, which is highly conserved and partially interchangeable between T4P and T2SS major pilins (9), was modeled by homology to the full-length group a T4 pilin (T4aP), PilA, of Pseudomonas aeruginosa (13) (pdb 1OQW), with a kink at Pro22 (48% identity, 84% similarity in the TMS; Fig. S1). The N-terminal methyl group was included in the model of the pilin (14).
The full-length PulG model then was used to generate molecular models of the pilus by a multistage minimization and molecular dynamics procedure (details are given in Materials and Methods, SI Materials and Methods, and Movie S1). Only the biophysical properties measured by STEM were used as initial parameters (i.e., repeat unit of 17.5 nm, helix turn of 4.38 nm) (9). Only one protomer was explicitly modeled, and 15 neighbors on each side were added during the minimization and molecular dynamics “on the fly” by using the appropriate symmetry operators. The monomer was placed close to the pilus axis, with the α-helix approximately parallel to the pilus axis and the globular domain pointing away from it. Initially, only a soft, “constant force” restraint was used to attract the protomer to the pilus axis, with a salt-bridge restraint to favor interactions between oppositely charged residues. The backbone of the initial structure was maintained by adaptive and soft distance restraints. One thousand pilus structures were calculated and clustered with respect to structural differences, position, and orientation. The main clusters were analyzed with respect to structural quality, energetic contributions of residues to pilus stability using the molecular mechanics/generalized Born surface area (MMGBSA) method, and residue–residue interactions (details are given in SI Materials and Methods). Consistently, the main cluster (around 2.5 Å rmsd in atomic positions, including protomer structure, orientation, and position) was far bigger than the other clusters and was considered to represent the correct model. In this modeling procedure, in addition to the overall flexibility of the backbone, the side chains and the two N-terminal residues were completely flexible, allowing the protomers to explore conformations that would be missed in a “rigid” procedure.
The type II secretion (T2S) pseudopilus structure determined in this study closely resembles that of the gonococcal T4aP (7). Despite differences in the structure of the pilins and in the measured helicity (37.8 Å helical pitch and 3.6 subunits per turn in T4P vs. 43.8 Å and 4.25, respectively, in PulG pili), their axial rise is almost identical (10.5 Å vs. 10.4 Å). The outer diameter, 65 Å, is similar to that of T4aP (∼60 Å). Both types of pili have a rough surface with profound grooves separating the helix strands and a very narrow hydrophobic central cavity (Fig. S2C).
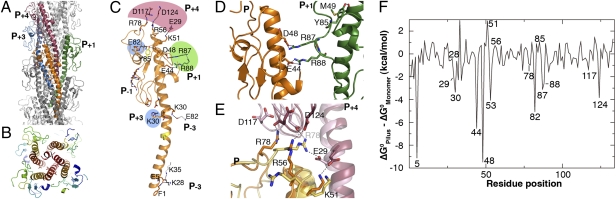
The tight packing within the pilus produces numerous specific interactions between neighboring protomers such that each protomer (P) interacts directly with three upper (P+1, P+3, P+4) and three lower (P−1, P−3, P−4) protomers in the filament, through hydrophobic (P−1 and P+1) and electrostatic (all six) interactions (Fig. 1). Overall, the N-terminal α-helix (Nα-helix) has seven negatively charged and six positively charged residues, including the N-terminal Phe1, considerably more than in the T4P. However, intra- or intersubunit salt bridges can neutralize each charge.
Fig. 1.
The PulG pilus structure. (A) Ribbon view of the pilus model with the central protomer (P) in orange and the upper partner protomers in green (P+1), blue (P+3), and red (P+4); (B) Top view of a 30-Å slice of a pilus model in ribbon view, rainbow-colored, from red (N terminus) to blue (C terminus). Residues Ile10 and Leu16 are shown as sticks. (C) Residues involved in key intermolecular interactions are shown as sticks, and their positions are indicated by the color code used in A. (D) Asp48–Arg87, and Glu44–Arg88 salt bridges between P and P+1. (E) P–P+4 interaction interface shown in ribbon view. Yellow and pink are alternative conformations of orange and purple protomers P and P+4, respectively. (F) Contribution of PulG residues to filament stability, calculated using the MMGBSA method as the difference between the Gibbs free energy changes in the filament and in the monomer (details are given in SI Materials and Methods).
Model Validation: Electrostatic Interactions.
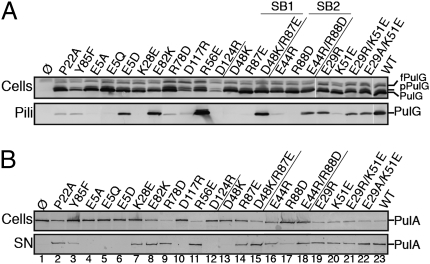
According to MMGBSA calculations, few dominant residues are predicted to add substantially to the stability of the pilus (Fig. 1F). Among them, two pairs, Asp48–Arg87 and Asp44–Arg88, were consistently predicted to form intermolecular salt bridges (Fig. 1D). To test the model predictions, site-directed mutagenesis was used to replace PulG residues 44, 48, 87, and 88 individually by residues of the opposite charge. We tested the ability of the resulting PulG variants to assemble pili in the presence of the complete set of pul genes encoding T2SS on a compatible plasmid (Fig. 2A). Although they were produced at normal levels, the single-residue substitution variants PulGD48K, PulGR87E, PulGE44R, and PulGR88D were strongly or completely defective in pilus assembly. Strikingly, combining two charge inversions in variants PulGD48K/R87E and PulGE44R/R88D restored normal piliation levels, indicating salt-bridge restoration (Fig. 2A, lanes 13–18). We also tested the effects of the substitutions on the secretion of a nonacylated variant of pullulanase (PulA) (15) at low pul gene expression levels (4). Although PulA secretion was mildly impaired with variants PulGE44R and PulGR87E, these substitutions rescued the secretion-negative phenotypes of variants PulGR88D and PulGD48K, respectively (Fig. 2B, lanes 13–18). These results demonstrate the predictive power of the models and the essential role of the two salt bridges in PulG assembly and in PulA secretion.
Fig. 2.
Assembly of PulG variants into pili (A) and their function in PulA secretion (B). Cell and pili or supernatant (SN) fractions from pulG mutants were analyzed by SDS/PAGE and immunoblot using anti-PulG (A) or anti-PulA (B) antibodies. Equivalent amounts of cells (Upper) and supernatant fractions (Lower) were analyzed. fPulG, FLAG-prePulG; pPulG, prePulG; SB, salt bridge.
Arg87 and Arg88 involved in the above salt bridges are located in a loop that is connected to the top of the long PulG Nα-helix via a hydrogen bond between the Tyr85 and Met49 (Fig. 1D). Variant PulGY85F, in which this bond is absent, was almost as abundant as native PulG but was assembly deficient. This substitution strongly reduced PulA secretion, consistent with the structural role of Tyr85 in the correct positioning of Arg87 and Arg88 for salt-bridge formation (Fig. 2, lane 3).
Despite the predicted contribution of Glu82 to the pilus stability (Fig. 1 C and F), its charge could be inverted without destabilizing the filament (Fig. 2A, lane 8). Because Glu82 is exposed in a flexible loop on the pilus surface (Fig. S2E), it is possible that substitution Glu82Lys may not induce a clash required to disrupt the intricate PulG complex.
Residues Lys51, Arg56, and Arg78 form a positive patch on top of protomer P that faces a negative patch on protomer P+4 including Glu29 and the two Ca2+ binding-site residues, Asp117 and Asp124 (Fig. 1 C and E). Among the 200 main cluster models, examples of electrostatic interaction heterogeneity were observed at this interface. Such alternative interactions would increase both the stability and the flexibility of the pilus by reducing the entropic loss of side chains involved in salt bridges and offering them alternate positions. Arg78 may interact with Asp124 (in 44% of the models of this cluster), with Asp117 (28%), and/or with Glu29 (37%). Charge inversions Asp124Arg and Asp117Arg affected PulG monomer stability (Fig. 2A, lanes 10, 12) and led to strong piliation and secretion defects in these mutants, consistent with the role of Ca2+ in stabilizing the β2–β3 loop of PulG (12). Nevertheless, Arg78 charge inversion abolished piliation, supporting an electrostatic interaction (Fig. 2A, lane 9). Glu29 on subunit P+4 was predicted to interact with P-subunit residues Lys51 (41%), Arg78 (37%), and/or Arg56 (8%) (Fig. 1E). The Arg56Glu substitution had no effect on piliation or secretion (Fig. 2, lane 11), probably because of the low frequency of interaction with Arg56. Neither of the charge inversions nor an alanine substitution at position 29 had an effect on pilus assembly (Fig. 2A, lanes 19–22). Lys51Glu substitution abolished piliation, but pilus assembly was rescued by a removal of negative charge at position 29 (variant PulGE29A/K51E), suggesting a clash between glutamate residues at both positions. This result implies the proximity of Lys51 and Glu29, in agreement with the models.
Double-Cysteine Substitutions in PulG Result in Covalently Cross-Linked Pili.
Current T4P structural models propose the helical organization of pilins stabilized mainly by the hydrophobic contacts between the TMSs tightly packed in the filament core (7, 16). Double-cysteine scanning was used to probe and validate contacts between TMSs in the assembled pili. Such cysteine residues should be protected from oxidation in the membrane-embedded, unassembled state. Upon PulG assembly, however, they could form disulfide bonds depending on their relative orientation and distance (Fig. 3E), providing the means to probe interprotomer interactions.
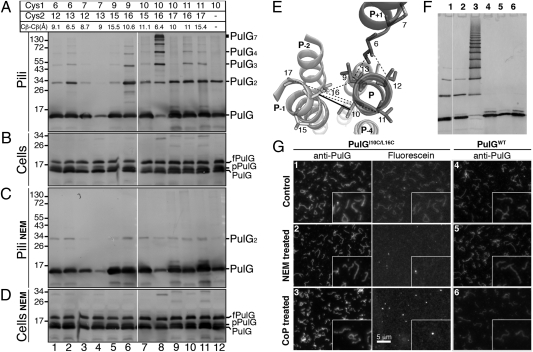
Fig. 3.
Model validation by double-cysteine scanning. Position of pairs of cysteine substitutions in mature PulG and their average Cβ–Cβ distances (calculated from 200 models) are indicated at the top of A. Sheared pili (A and C) and cell (B and D) fractions were incubated for 60 min at 30 °C (A and B) or were treated with NEM (C and D). fPulG, FLAG-prePulG; pPulG, prePulG; PulGn, PulG n-mer. (E) A top-down ribbon view of pilus segments Ile6–Ala17. Positions allowing cysteine cross-linking are linked by solid lines for residues 10 and 16 and by dashed lines for all other pairs. (F) Purified PulGI10C/L16C or PulGWT pili (lanes 1 and 4, respectively), after NEM (lanes 2 and 5), or after CuoP (lanes 3 and 6) treatment analyzed by SDS/PAGE and immunodetection with anti-PulG antibodies. (G) Fluorescence microscopy of the pili samples in F revealed by anti-PulG antibodies followed by Alexa 555-coupled IgG (Anti-PulG) or labeled with fluorescein-maleimide (Fluorescein). Insets show a part of each field enlarged 2-fold. Lane numbers in F correspond to image numbers in G.
Residues Ile10 (P) and Leu16 (P−1) are particularly close in the proposed models. A series of PulG variants with cysteine substitutions at positions indicated in Fig. 3A was tested for the ability to form pili. PulG protein levels were comparable for all variants, indicating that the cysteine substitutions did not affect the protein production or stability (Fig. 3 B and D). The only variant in which piliation was affected was PulGM7C/L13C (Fig. 3 A and C, lane 4), although neither single-cysteine substitution had this effect when combined with cysteine substitutions at other positions (Fig. 3A, lanes 2 and 3). Variants with cysteines at positions 6–13, 9–16, 10–16, 11–16, and 11–17 produced detergent-resistant multimers of PulG (lanes 2, 6, 8, 10, and 11). In contrast, variants PulGV9C/V15C, PulGI10C/V15C, and PulGI10C/A17C and the single-cysteine variant PulGI10C gave rise only to monomers and dimers (lanes 5, 7, 9, and 12, respectively), indicating position-specific multimerization.
Cross-linking essentially occurred in pilus fractions (Fig. 3 A and B). Treatment with N-ethylmaleimide (NEM) inhibited cross-linking, confirming cysteine specificity (Fig. 3 A and C). The cross-linking was reversible by treatment with DTT (Fig. S3). Strikingly, pili assembled by variant PulGI10C/L16C were by far the most efficiently cross-linked (Fig. 3A, lane 8). The average distance between γ-carbons of residues 10 and 16 in assembled pili therefore is close to the distance required to form a disulfide bridge (3–4.5 Å) (17). The cross-linking efficiency dropped drastically when the cysteine substitutions were shifted to the neighboring positions, suggesting an increased distance between the thiol groups in these variants (e.g., 10–17 or 10–15). However, PulGV11C/A17C with more distant β-carbons formed cross-links almost as efficiently as PulGV9C/L16C. Along with the lack of disulfide bond formation between positions 9 and 15, this result favors the model of directional iris-like sliding of the TMS in the filament core during pilus assembly. This movement is consistent with the more central position of protomer P TMS with respect to protomer P+1 (Fig. 1B and Fig. S2F). Cysteines at positions 6 and 7 appeared to be less reactive, because variants carrying these substitutions formed fewer dimers than the single-cysteine variant PulGI10C (Fig. 3A, lanes 1–4). Nevertheless, the multimerization level of PulGI6C/L13C was comparable with that of PulGV9C/L16C and was efficient, considering the lack of reactivity in the lower TMS. These results validate filament models with distances between TMS residues 10 and 16 in protomers P and P−1, respectively, in the range of 6.4 (±1.4) Å.
Cysteine Accessibility and Oxidation in the Assembled Pili.
Equal amounts of PulGWT and PulGI10C/L16C pili were purified at 4 °C and examined by SDS/PAGE (Fig. 3F) and fluorescence microscopy (Fig. 3G). Similar to PulGWT, cold-purified PulGI10C/L16C pili were mostly monomeric (Fig. 3F, lanes 1 and 4). Labeling by cysteine-specific fluorescein maleimide probe yielded a filament pattern in the green fluorescence range superimposable on that revealed by anti-PulG antibodies (compare anti-PulG and fluorescein in Fig. 3G, panel 1). Blocking of sulfhydryl groups by NEM prevented the fluorescein labeling (Fig. 3G, panel 2, and Fig. 3F, lane 2), as did the pilus oxidation by Cu-orthophenanthroline (CuoP) (Fig. 3G, panel 3), which promoted efficient PulGI10C/L16C cross-linking through the formation of disulfide bonds, indicated by the formation of detergent-resistant PulG multimers (Fig. 3F, lane 3). The electrophoretic pattern analysis indicated that the cross-linked products were integral homomultimers of the PulG protomer (Fig. S4). These treatments did not affect filament appearance as visualized by immunofluorescence (Fig. 3G) and had no effect on PulGWT pili (Fig. 3F, lanes 4–6, and G, panels 4–6). In conclusion, cysteine residues introduced in the TMS of PulGI10C/L16C can be maintained in a reduced state, alkylated, or oxidized to form interprotomer disulfide bonds, without a major effect on the filament aspect. Thus, cysteine-mediated PulG cross-linking occurs upon protomer assembly into pili (Fig. S3), and cysteine residues placed in the TMS are partially accessible to solvent in the pilus core (Fig. S2D).
Taken together, these results biochemically validate the pilus structural models, the large majority of which predict the demonstrated salt bridges. The results also suggest that, as predicted by most of the models, in the native pilus the nearest neighbors, Ile10 and Leu16 β-carbons, are separated by more than 5 Å, and that the closer contact necessary to form a disulfide bridge is of transient nature.
Model Refinement and Functional Implications.
With no constraints other than the symmetry parameters (9) and the attraction toward the pilus axis, more than 20% of the modeling runs converged to the described cluster from random orientation around the long axis of the protomer. This cluster represented more than 50% of the models when additional distance restraints based on biochemical data were imposed in a conservative manner (salt bridge 48–87, distance restraints between the Cβ atoms of residues 10–16, 9–16, and 6–13). Different combinations of these restraints did not alter the protomer arrangement in the filament in the first cluster. The rmsd of the original cluster and between the constrained and unconstrained PulG protomers were comparable.
Because previous work suggested a left-handed pilus organization (9), we tested the possibility of modeling such helices independently. Such models were not compatible with the above biochemical data and consistently showed a higher interaction energy (i.e., were less stable). Furthermore, a wide panel of solutions was explored during the modeling process, before converging toward a right-handed helix (Movie S1).
Although the Nα-helix in the starting model shows only one kink near Pro22, the “sigmoid” shape as in the T4 pilins appeared in many of the structures during the modeling. The kink induced by Pro22 is an important structural element that seems to facilitate the Nα-helix bundle packing. This observation is consistent with the phenotypes of pulGP22A mutant, which was significantly less piliated than the wild type and showed reduced PulA secretion (Fig. 2, lane 2). This result argues against the second largest cluster of models (2%) containing straight Nα-helices.
The totally conserved Glu5 side chain and Phe1 N-terminal ammonium both point toward the filament center and can form an intersubunit salt bridge, as proposed in T4P models based on the pilus axial rise (7). However, in analyzing intermolecular distances between β-carbons from 200 independent models, we found this salt bridge in only 18% of the models. Other salt bridges form more frequently: Intersubunit interaction of Glu5 with the side chains of residues Lys35 of P−3 (73%) or with both Lys35 and Lys28 of P−3 (55%) leaves the N-terminal charge free to interact with Glu5 (intramolecularly) or with Asp53 (P−4). The Lys28Glu substitution abolished piliation and reduced PulA secretion, favoring a possible interaction with Glu5. However, this interaction could not be confirmed by double-charge inversion, because Glu5 is crucial for function (14). Although conserving the negative charge at this position allowed partial pili assembly, PulGE5D did not complement secretion (Fig. 2, lanes 4–7).
Interestingly, the two alternative predicted salt bridges involving Glu5 are correlated with two extreme TMS configurations found in the modeling runs. The Glu5(P)–Phe1(P+1) distance was negatively correlated with the tested Val11–Ala17(P−1) distance (correlation coefficient ρ = −0.34; n = 200) and positively with the Val9–Leu16 (P−1) proximity (ρ = 0.44). Inversely, the Glu5–Lys35(P−3) distance is negatively correlated with the short Val9–Leu16(P−1) distance (ρ = −0.35). This result suggests that the TMS mobility reflected by low-level cysteine cross-linking of PulGV9C/L16C, PulGV11C/L16C, and PulGV11C/A17C is associated with formation of alternative electrostatic interactions. It would be of interest to determine whether they form sequentially or stochastically.
Discussion
We report a structural model of the T2SS pseudopilus at pseudoatomic resolution, based on the high-resolution structure of the PulG monomer completed by modeling and the STEM analysis of single PulG filaments (9). The initial models were generated using only the STEM data and served to predict intermolecular contacts, which then were validated experimentally. The contacts were used as restraints in a conservative manner for subsequent modeling. Construction of models with pseudoatomic resolution from EM data and high-resolution structures for the building blocks is limited to relatively high-resolution EM data (in general better than 10 Å) (18). We show that one can compensate for the lack of resolution in the EM data by adding a molecular mechanics energy function and a few key distance restraints derived experimentally to distinguish between models or to refine models. In this study, the restraints were obtained from cysteine cross-linking and charge inversion experiments, but other sources are possible (e.g., FRET, NMR). The models converged, despite the ambiguity in the restraints caused by the symmetry of the system, because of the use of an adapted modeling strategy.
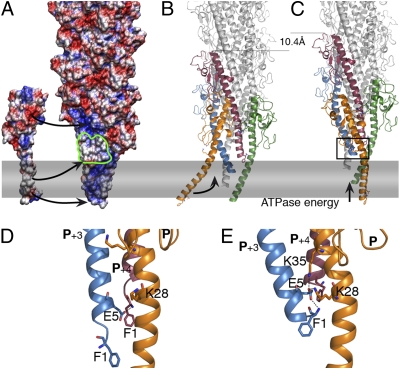
The models support the overall right-handed organization of T2SS pilus, consistent with the gonococcal T4aP structure, and confirm the fundamental similarities between these two systems. Interestingly, as shown in the representation of the electrostatic surface of the pilus (Fig. 4A), they reproduce the surface features with the left-handed groove observed previously by EM (9). The piston hypothesis implies that pseudopilus assembly drives protein secretion. In the assembly process, Glu5 plays a crucial role that is not reduced to its charge. This residue was shown to be essential for pilin methylation (14) which may be required at an earlier step, possibly involving pilin interactions with the assembly factors, as suggested previously (19).
Fig. 4.
The pseudopilus assembly model. (A) Electrostatic envelope of the pilus and PulG monomer, as calculated by APBS (31), inserted in the membrane (gray rectangle). Arrows indicate the PulG tethering via Asp48–Arg87 and Glu44–Arg88 salt bridges, the hydrophobic patch (outlined in green), and Glu5. (B) Ribbon view of the pilus and the incoming protomer, color-coded as in Fig.1. (C) The incoming protomer (P) incorporation is associated with the membrane extraction of the P+1, driven by the PulE ATPase, adding 10.4 Å to the pilus. (D and E) Zoom views of the rectangle in C showing two alternative interactions of Glu5 of P+3 with Phe1 of P+4 (D) or Lys35 and Lys28 of P, stabilizing the pilus in its optimal state (E).
Based on the functional analysis of pulG mutants and the network of alternative interactions, the models presented here suggest a mechanism for T2S pilus assembly. Crucial interactions involve adjacent protomers, giving a biological meaning to the one-start helix model. In the first step the protomer P is tethered to the nascent filament via electrostatic interactions with protomer P+1 (Asp48–Arg87, Glu44–Arg88, and Glu5–Phe1). The hydrophobic patch on protomers P+3 and P+4 exposed to aqueous environment (Fig. 4 A, B, and D) could contribute to these contacts or be masked by an assembly factor (PulF/L/M). Upon insertion, Lys35 and Lys28 of protomer P could interact with Glu5 of P+3, stabilizing the P+3 in its final state (Fig. 4 B, C, and E) and promoting the most frequently modeled TMS arrangement.
Interestingly, charge inversions at the P–P+4 interface that blocked piliation (Arg78Asp and Lys51Glu) had no effect on PulA secretion, suggesting a specific role for this interface in pilus stabilization and possibly in controlling pseudopilus length. This flexible interaction surface also is predicted to participate in heterologous contacts with minor pseudopilins, as suggested by the recent structural and biochemical data (20, 21), which place the homologs of PulK, -I, -J, and -H at the tip of the periplasmic pseudopilus. As already noted (22), the minor pseudopilins in the GspJ-I-K tip also are arranged in a right-handed helix.
The charge complementarity of residues forming the demonstrated salt bridges is highly conserved (87% for 48–87) in the major pseudopilin family, with charges naturally inverted relative to PulG in 6% of the 281 pseudopilins aligned (Table S1). The highly conserved TMS involved in hydrophobic and van der Waals contacts and the overall structural conservation of the major T2SS pseudopilins (9, 12, 23) argue that our model should apply to all other T2S pseudopilus core structures.
The high cross-linking efficiency of the variant PulGI10C/L16C reflects the favorable relative positioning of the two residues but also their accessibility to water or oxidative agents, which is predicted by the pilus model (Fig. S2D). This accessibility to water is in agreement with the solvent accessibility of the TcpA subunit Nα-helix in the assembled Vibrio cholerae toxin coregulated pilus, a member of the group b T4P (16). Pseudopilus cross-linking into detergent-resistant multimers should provide a unique tool for studying the periplasmic pseudopilus structure and dynamics under physiological expression conditions.
The modeling approach developed in this study, combined with extensive biochemical and functional analyses, provides powerful tools for studying the structure of protein polymers. The information gained from this work will be exploited to build a model of the complete pseudopilus as a basis for further structure function analysis of this essential dynamic component of T2SS.
Materials and Methods
Modeling of the Protomer and Pilus.
To obtain a complete model of PulG, we best-fitted the 25 N-terminal residues of PilA from P. aeruginosa PAK PilA (13) to that of the PulG pilin. Side chains were replaced with those of PulG with SCWRL software (24). The 20 C-terminal residues were modeled by exploiting the close homology to GspG (pdb 3G20) in a similar way. The resulting structure was minimized with CNS software (25), modified for the use of symmetrically related ambiguous restraints (26) and electrostatic calculations with the generalized Born model (27, 28).
We used several different search and minimization strategies to identify likely orientations of the protomer in the pilus. The protomer was placed such that the N-terminal α-helix (residues 5–25) was aligned along the pilus axis. Starting orientations for the minimization were generated by randomly varying the rotations around the pilus axis (by 180°), the two axes perpendicular to it (by 10°), and the distance from the pilus axis (between 8 and 10 Å away from the pilus axis). All modeling was done with CNS software, with the CHARMM PARAM19 force field (29). The symmetry of the pilus was enforced throughout with noncrystallographic symmetry operators: Only one protomer is modeled explicitly, and 15 neighbors were generated in each direction “on the fly” as exact copies of the primary protomer. The minimization proceeded in a three-stage protocol. Standard molecular docking programs could not be used because they usually do not take the helical symmetry into account, do not include flexibility in a satisfactory way, and/or cannot use ambiguities in the restraints. The known helical parameters (10.4 Å, 84.5°) (9) were used and both right- and left-handed helices were tested, initially without any restraints on the modeling other than a packing term (a constant force pulling the helix toward the pilus axis). An ambiguous salt-bridge restraint (an attractive interaction between a charged residue and all residues of opposite charge on any protomer in the pilus) was used in all calculations. Structure pictures were created using MacPymol (http://www.pymol.org) and VMD software (30). Electrostatics calculations were performed with CNS software (for the energetic contributions per residue) and with the Adaptive Poisson-Boltzmann Solver (APBS) (31). Models and modeling protocols are available on request.
Bacterial Strains, Plasmids, and Molecular Biology Techniques.
E. coli K12 strains and plasmids used in this study are listed in Tables S2 and S3. Mutations in pulG were made using QuickChange (Stratagene) and were confirmed by sequencing (at GATC). Oligonucleotides (Sigma) are listed in Table S4.
Shearing Assay for Pilus Assembly.
Piliation was assayed in strain PAP9304 (Table S2). The pulG alleles carried on plasmid pCHAP7303 encode PulG containing a FLAG epitope fused to the N terminus of the PulG presequence. Shearing assay was performed as described (9). When appropriate, pilus fractions were mixed with 50 mM NEM (Fluka) or 100 μM CuoP (Sigma) and incubated at 30 °C with constant agitation for 30 min or 10 min, respectively. The equivalent of 0.05 A600 of each fraction was analyzed by SDS/PAGE on 12% Tris-tricine or 4–20% Tris-glycine gradient gels (BioRad). Proteins were transferred to nitrocellulose membranes, which were probed with anti-PulG antibodies (1:2,000) followed by HRP-coupled IgG (1:40,000; Amersham). Immunoblots were developed using ECL+ and Storm (GE Healthcare).
Secretion Assay.
PulA secretion was assayed in strain PAP5300 (Table S2). Expression of pulG alleles on pCHAP7303 and derivatives was induced by 1 mM isopropyl-β-D-thiogalactopyranoside. An equivalent of 0.005 A600 of bacterial and supernatant fractions was analyzed by 10% Tris-glycine SDS/PAGE and immunodetection using anti PulA antiserum (1:2,000).
Microscopy.
Immunofluorescence labeling of pili was performed as described (4), with anti-PulG antibodies (1:2,000) and secondary Alexa Fluor 455-coupled IgG (Molecular Probes). Samples were examined with an Axiovert 200 microscope (Zeiss). Images were taken with AxioVision (Zeiss) and processed simultaneously with Photoshop CS2. Pili from sheared fractions were ultracentrifuged for 45 min at 135,000 × g and labeled with 0.1 mM fluorescein 5′-maleimide (Pierce) in sodium phosphate buffer, pH 7.0, overnight at 4 °C. Pili were washed by ultracentrifugation for 45 min at 135,000 × g in the presence of 10 mM L-cysteine and were processed for immunofluorescence.
Supplementary Material
Acknowledgments
We thank Tony Pugsley, who initiated this project, for his generous support and input. We thank Sandy Niellèz for making the plasmid pCHAP7303 during her stay in the Unité de Génétique Moléculaire, Institut Pasteur; Nathalie Nadeau for excellent technical assistance; Tony Pugsley, Rémi Fronzes, and Evelyne Richet for comments on the manuscript; Hervé Waxin for the use of the microscope; and Ansgar Philippsen and all members of the Pugsley laboratory for helpful discussions and friendly support. This work was supported by the Institut Pasteur Projet Transversal de Recherche 339. M.C. was supported by a fellowship from the French Ministry of Research and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001703107/-/DCSupplemental.
References
- 1.Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: From structure to function. FEMS Microbiol Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 2.Hazes B, Frost L. Towards a systems biology approach to study type II/IV secretion systems. Biochim Biophys Acta. 2008;1778:1839–1850. doi: 10.1016/j.bbamem.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Sauvonnet N, Vignon G, Pugsley AP, Gounon P. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 2000;19:2221–2228. doi: 10.1093/emboj/19.10.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vignon G, et al. Type IV-like pili formed by the type II secreton: Specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol. 2003;185:3416–3428. doi: 10.1128/JB.185.11.3416-3428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durand E, et al. Type II protein secretion in Pseudomonas aeruginosa: The pseudopilus is a multifibrillar and adhesive structure. J Bacteriol. 2003;185:2749–2758. doi: 10.1128/JB.185.9.2749-2758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Py B, Loiseau F, Barras F. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2001;2:244–248. doi: 10.1093/embo-reports/kve042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig L, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Shevchik VE, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohler R, et al. Structure and assembly of the pseudopilin PulG. Mol Microbiol. 2004;54:647–664. doi: 10.1111/j.1365-2958.2004.04307.x. [DOI] [PubMed] [Google Scholar]
- 10.Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J Bacteriol. 2007;189:1783–1793. doi: 10.1128/JB.01230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Possot OM, Vignon G, Bomchil N, Ebel F, Pugsley AP. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J Bacteriol. 2000;182:2142–2152. doi: 10.1128/jb.182.8.2142-2152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korotkov KV, et al. Calcium is essential for the major pseudopilin in the type 2 secretion system. J Biol Chem. 2009;284:25466–25470. doi: 10.1074/jbc.C109.037655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig L, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 14.Strom MS, Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem. 1991;266:1656–1664. [PubMed] [Google Scholar]
- 15.Francetic O, Pugsley AP. Towards the identification of type II secretion signals in a nonacylated variant of pullulanase from Klebsiella oxytoca. J Bacteriol. 2005;187:7045–7055. doi: 10.1128/JB.187.20.7045-7055.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, et al. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. 2008;16:137–148. doi: 10.1016/j.str.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinchio M, Czaplewski C, Liwo A, Oldzej S, Scheraga H. Dynamic formation and breaking of disulfide bonds in molecular dynamics simulations with the UNRES force field. J Chem Theory Comput. 2007;3:1236–1248. doi: 10.1021/ct7000842. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Walz T. The advent of near atomic resolution in single particle electron microscopy. Annu Rev Biochem. 2009;78:723–742. doi: 10.1146/annurev.biochem.78.070507.140543. [DOI] [PubMed] [Google Scholar]
- 19.Aas FE, et al. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect type IV pilus assembly, dynamics and associated functions. Mol Microbiol. 2007;63:69–85. doi: 10.1111/j.1365-2958.2006.05482.x. [DOI] [PubMed] [Google Scholar]
- 20.Korotkov KV, Hol WG. Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol. 2008;15:462–468. doi: 10.1038/nsmb.1426. [DOI] [PubMed] [Google Scholar]
- 21.Douzi B, et al. The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J Biol Chem. 2009;284:34580–34589. doi: 10.1074/jbc.M109.042366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forest KT. The type II secretion arrowhead: The structure of GspI-GspJ-GspK. Nat Struct Mol Biol. 2008;15:428–430. doi: 10.1038/nsmb0508-428. [DOI] [PubMed] [Google Scholar]
- 23.Alphonse S, et al. Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J Struct Biol. 2010;169:75–80. doi: 10.1016/j.jsb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Canutescu AA, Dunbrack RL. SCWRL and MolIDE: Computer programs for side-chain conformation prediction and homology modeling. Nat Protoc. 2008;3:1832–1847. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Nilges M, Bardiaux B, Malliavin T. Encyclopedia of Magnetic Resonance. New York: John Wiley and Sons; 2010. Protein structure calculation using ambiguous restraints. 10.1002/9780470034590.emrstm1156. [Google Scholar]
- 27.Calimet N, Schaefer M, Simonson T. Protein molecular dynamics with the generalized Born/ACE solvent model. Proteins. 2001;45:144–158. doi: 10.1002/prot.1134. [DOI] [PubMed] [Google Scholar]
- 28.Moulinier L, Case DA, Simonson T. Reintroducing electrostatics into protein X-ray structure refinement: Bulk solvent treated as a dielectric continuum. Acta Crystallogr D Biol Crystallogr. 2003;59:2094–2103. doi: 10.1107/s090744490301833x. [DOI] [PubMed] [Google Scholar]
- 29.Brooks B, et al. CHARMM: A program for macromolecular energy and minimization and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 30.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.