Abstract
The role of geographic isolation in marine microbial speciation is hotly debated because of the high dispersal potential and large population sizes of planktonic microorganisms and the apparent lack of strong dispersal barriers in the open sea. Here, we show that gene flow between distant populations of the globally distributed, bloom-forming diatom species Pseudo-nitzschia pungens (clade I) is limited and follows a strong isolation by distance pattern. Furthermore, phylogenetic analysis implies that under appropriate geographic and environmental circumstances, like the pronounced climatic changes in the Pleistocene, population structuring may lead to speciation and hence may play an important role in diversification of marine planktonic microorganisms. A better understanding of the factors that control population structuring is thus essential to reveal the role of allopatric speciation in marine microorganisms.
Keywords: allopatric speciation, dispersal, marine cosmopolitan planktonic microorganisms, population structure, microsatellites
Allopatric speciation models are challenging for marine planktonic microorganisms because the huge population sizes and high passive dispersal potential of these organisms, as well as the lack of apparent dispersal barriers in the ocean, may allow extensive gene flow at large geographic scales (1–3). Therefore, marine environments are believed to sustain only a limited number of cosmopolitan planktonic microbial species that lack spatial population genetic structuring. However, accumulating molecular data provide evidence for extensive genetic diversity and the prevalence of cryptic species in many alleged cosmopolitan planktonic microorganisms (4–8). The concept of planktonic superspecies has been introduced to describe these constrained morphological monophyletic entities, which comprise multiple closely related species that may be geographically restricted or adapted to different ecological niches (9). The apparent paradox of high cryptic microbial species diversity in the absence of opportunities for allopatric speciation, in combination with fossil evidence for gradual patterns of evolution in sympatry (reviewed in ref. 10) has lead to the paradigm that sympatric processes and/or genomic changes are the prevalent mode of speciation in planktonic marine microbial organisms (11–14). On the other hand, based on the restricted geographic distributions of several cryptic species, it has been hypothesized that isolation by physical and/or ecological barriers is an important driver in allopatric processes, even in high dispersal marine microbes (15–18).
Allopatric speciation results from geographic partitioning of genetic diversity due to historical (e.g., dispersal limitation) and/or contemporary environmental processes (e.g., divergence resulting from local selection) (19). The relative importance of these factors in structuring microbial systems is, however, still poorly understood (3). To gain insight in the mechanisms of microbial speciation in the sea, it is essential to identify the factors that affect population structuring at the different spatiotemporal scales at which gene flow may operate. Population genetic surveys using high-resolution markers that allow differentiation among individuals, are greatly improving our understanding of the mechanisms that cause genetic diversity within and gene flow between populations (20). To date, population genetic studies of high-dispersal organisms in the marine environment have largely focused on pelagic or benthic animals with planktonic larval stages (21–24). Population genetic structuring of planktonic microorganisms has been much less explored due to difficulties with species delineation and lack of fine-scale genetic markers for these organisms (25–28).
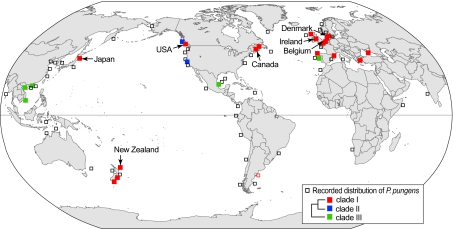
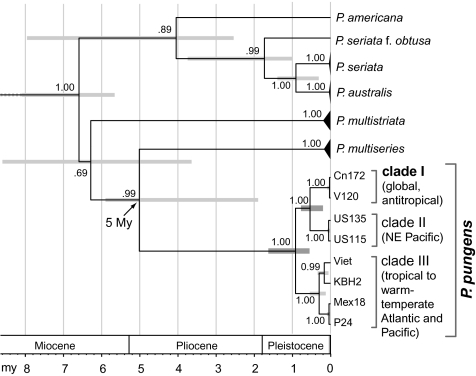
Diatoms form a major component of the plankton and despite their relatively young age (≤240 Ma), they are the most diverse group of marine phytoplankton (29, 30). We used the obligate sexual planktonic pennate diatom Pseudo-nitzschia pungens sensu lato to study gene flow on a global scale because (i) the species complex has been reported worldwide (31); (ii) it is known to form extensive blooms (32, 33) and thus large population sizes that could facilitate gene flow at macrogeographic scales; and (iii) its life history, taxonomy, and biogeography have been well studied (34, 35). Within P. pungens, both nuclear rDNA internal transcribed spacer (ITS) and plastid-encoded rbcL sequences have revealed the existence of three distinct clades (I–III), which are also distinguishable by subtle but clear differences in frustule ultrastructure. Although these clades are currently recognized as subspecific taxa (varieties), the observed genetic and morphological discontinuities suggest that these clades behave as independent evolutionary lineages (36), hence justifying their recognition as discrete species-lineages within a planktonic superspecies P. pungens (35, 37). To avoid confusion, we refer to these three taxa as clades I–III. The clades differ markedly in their geographical distribution (35) (Fig. 1). Clade I (P. pungens var. pungens) has a cosmopolitan distribution in temperate waters of the Atlantic and Pacific Oceans. Clade II (P. pungens var. cingulata) has only been found in the northeastern Pacific, where it cooccurs with clade I. Clade III (P. pungens var. aveirensis) occurs in the tropical to warm-temperate waters of the Atlantic and Pacific Oceans. Although clades I and II are genetically well-differentiated, occasional hybrids between both clades have been recorded in the northeastern Pacific (38–40), suggesting incomplete reproductive isolation. Within clade I, all individuals have identical ITS and rbcL sequences and isolates from geographically widely separated populations reproduce sexually under laboratory conditions, suggesting the potential for gene flow between distant, or even transoceanic, populations (35, 39). Therefore, clade I is well-suited to explore patterns of population genetic structuring and to evaluate gene flow over a range of geographical scales. For example, recent microsatellite analyses showed that at regional scale, North Sea populations constitute a highly diverse genetic continuum with extensive gene flow (41, 42).
Fig. 1.
Geographic distribution of P. pungens. Morphology-based records are indicated by open squares and verified isolates based on DNA sequencing are shown as filled squares. Sampling locations for the microsatellite study of the P. pungens clade I are indicated by arrows. Redrawn and updated from Trainer et al. (32).
This study assesses global patterns of gene flow in P. pungens clade I by means of six highly polymorphic nuclear microsatellite loci. We analyzed isolates from different localities in the Atlantic and Pacific Ocean to test the assumption that clade I represents a single globally panmictic population (35). Using a calibrated phylogenetic analysis we estimated the maximal divergence times between different populations of this clade, providing a historical framework to interpret patterns of gene flow and population differentiation.
Results and Discussion
Our analysis refutes the hypothesis that P. pungens clade I constitutes a single globally panmictic population. Instead, it shows significant geographical genetic structuring.
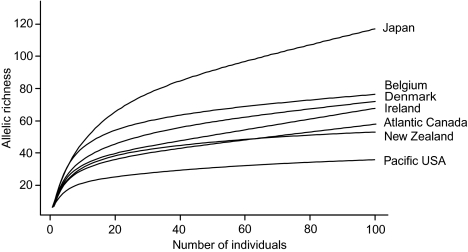
Allelic diversity was significantly higher on a global scale than on a regional scale in the North Sea (41, 42). Globally, we recorded 118 alleles in 242 isolates (19.7 alleles per locus on average) (Tables S1 and S2), whereas in the North Sea a maximum of 77 alleles were found in almost twice as many isolates (12.5 alleles per locus on average) (41, 42), indicating that the higher global allelic richness is not a matter of sample size but due to additional alleles outside the North Sea. Highest and lowest allelic richness and heterozygosity (He 0.83 vs. 0.53) were found on opposite sides of the northern Pacific, in Japan and the Pacific United States respectively (Fig. 2). Other populations showed intermediate He values that are comparable with those of clade I in the North Sea [0.69 and 0.73 (41, 42)] and of the marine planktonic diatom Ditylum brightwellii in the northwestern Pacific (0.71) (43).
Fig. 2.
Comparison of allelic richness between P. pungens clade I predefined populations from different areas. Allelic richness was inferred from multilocus genotypes (six microsatellite loci) and extrapolated beyond the sample size using ARES.
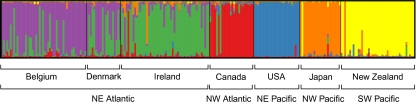
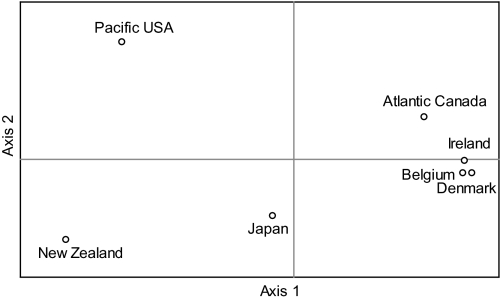
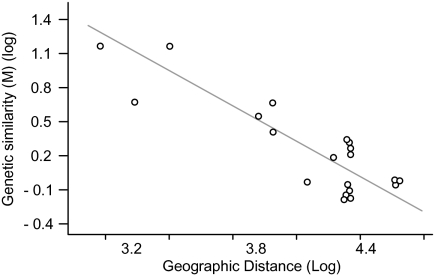
There was a large overlap in allele size classes for the six loci, although allele frequency distributions differed among predefined populations (Fig. S1). Allelic and genotypic differentiation tests (for each locus separately and over all loci) gave the same results (Table 1). Pairwise FST values between populations were relatively high (ranging from 0.06 to 0.76) and all, except Belgium vs. Denmark, were significant (Table 1). In agreement with the results based on the predefined populations, the STRUCTURE analyses revealed six genetic clusters (Fig. 3, Fig. S2, and SI Text), indicating that genetic structuring was strong enough to assign isolates to different groups. Whereas northeastern Atlantic isolates showed a more admixed background, northwestern Atlantic and Pacific isolates clustered mainly according to their geographic origin, suggesting that migration rates and/or gene flow between the sampling locations is insufficient to make the sampling locations act as a single panmictic population (44). This is in line with the PCA and isolation by distance (IBD) analyses (Figs. 4 and 5). The PCA of pairwise FST values separated Pacific and Atlantic populations along the first axis, and indicated that populations from the Atlantic are less differentiated than those from the Pacific (Fig. 4). There was a strong IBD signal (Fig. 5) (Z = 16.8975, r = −0.8707, one-sided P = 0.0044), although even at the largest geographic scale, the STRUCTURE analysis detected some admixing (Fig. 3). This suggests that long-distance dispersal may occur, but not frequently enough to counteract population differentiation. Whether this long-distance dispersal is natural and may involve introductions of new genotypes through low-abundance populations [eukaryotic “rare biosphere” (45)] or rather reflect anthropogenic introductions (e.g., via ballast water or translocation of aquaculture stocks), cannot be deduced from our data. Historical distribution records, data on divergence times between the populations and testing the observed geographic patterns with oceanic current models (46, 47) may resolve this question. Whereas the age of population differentiation in clade I is uncertain, it must be more recent than the split between clades I and II, which has occurred in the Middle to Lower Pleistocene (200–800 kya) (Fig. 6).
Table 1.
Estimates of standardized multilocus Fst values among pairs of predefined populations (above diagonal); the loci that were significantly different among populations in the single locus tests (allelic and genotypic tests) are given below the diagonal
| Belgium | Denmark | Ireland | Atlantic Canada | Pacific United States | Japan | New Zealand | |
| Belgium | 0.06 | 0.18 | 0.31 | 0.74 | 0.59 | 0.73 | |
| Denmark | 6 | 0.06 | 0.18 | 0.73 | 0.49 | 0.74 | |
| Ireland | 1, 2, 3, 4, 6 | 1, 4 | 0.23 | 0.70 | 0.47 | 0.76 | |
| Canada | 1, 2, 3, 4, 6 | 1, 2, 3, 4, 6 | 1, 2, 3, 4, 6 | 0.57 | 0.50 | 0.74 | |
| USA | all 6 | all 6 | all 6 | all 6 | 0.66 | 0.65 | |
| Japan | all 6 | all 6 | all 6 | all 6 | all 6 | 0.57 | |
| New Zealand | all 6 | all 6 | all 6 | all 6 | all 6 | all 6 |
Significant values (after sequential Bonferroni correction) are shown in bold.
Fig. 3.
Structure plot for K = 6. Each individual is depicted by a vertical line that is partitioned into K colored sections, with the length of each section proportional to the estimated membership coefficient (qind) of the isolate to each cluster.
Fig. 4.
Principal component analysis of pairwise FST values among the seven predefined populations of P. pungens clade I. The first and second principal components account for 59.47% and 24.58% of the total variation, respectively.
Fig. 5.
Isolation by distance. Scatter plot of genetic similarity versus geographic distances (in km) among seven P. pungens clade I populations, showing significant correlation between geographic and genetic distance. Mantel test for matrix correlation between log(genetic similarity (M)) and log(geographic distance): Z = 16.8975, r = −0.8707, one-sided P = 0.0044. Regression analyses: r2 = 0.758, 95% CI: 0.450–0.889.
Fig. 6.
Divergence time estimates (Mya) among Pseudo-nitzschia species, based on a Bayesian relaxed molecular clock applied to a concatenated alignment of rbcL, LSU rDNA and ITS sequences, and calibrated based on Sorhannus et al. (48). Uncertainty of divergence times are indicated by gray bars on internal nodes, corresponding to the 95% highest posterior density of node ages. The chronogram shows that within P. pungens the clade III diverged first (around 0.6 and 1.6 Mya), followed by the divergence of clades I and II, between 200 and 800 kya.
In contrast to the panmictic clade I population in the North Sea (41, 42), our results suggest that at larger geographic scales gene flow is more restricted. The limited population mixing at global scale may be due to pre- or postcolonization barriers (49). Precolonization barriers are a result of restricted dispersal abilities and generally scale with distance. In contrast, postcolonization barriers act in a distance-independent manner and reduce population admixing if new colonizers fail to become established because of different local environmental factors and selection pressures. Our PCA and IBD analyses indicate that there are limits to the dispersal ability of clade I so that geographic genetic differentiation is prevalent in this taxon. Hence, our data strongly suggest that isolation by large distances allows for genetic divergence, even in high-dispersal organisms like planktonic diatoms. This is in line with other marine organisms, where similar patterns of significant IBD were observed from regional to global scales (23, 50–52).
Apart from distance, various other isolating mechanisms, including present-day ecological and physical barriers, may be at work in the supposedly homogeneous marine environment. Clade I is hitherto only known from temperate coastal regions. Its apparently antitropical distribution suggests that geographic variation in seawater temperature may be an ecological barrier for transequatorial gene flow, as has been shown in some cool-water fishes and foraminifers (16, 53–55). This hypothesis remains to be tested, particularly in the light of the possibility that seasonal upwelling of cold waters along the western margins of the American and African continents may act as a corridor for transequatorial dispersal, as has been suggested for anti-tropical and bipolar planktonic foraminifers (54, 56). Overall, transoceanic dispersal of clade I seems less plausible than previously thought because coastal and open sea environments have very different physical and biological characteristics (53, 57). In addition, several key features of the life history of Pseudo-nitzschia appear to preclude easy dispersal over the open ocean. Firstly, it does not produce cysts needed to survive inhospitable open ocean conditions. Secondly, its obligate sexual life cycle is controlled by specific environmental conditions during seasonal blooms that are promoted by high levels of inorganic nutrients introduced by coastal upwelling. Importantly, in the onset of reproduction, mating cells attach to dense colonies of surf-zone diatoms (40). Together, these findings imply that P. pungens requires specific coastal conditions to complete its life cycle, which probably constrain transoceanic dispersal. Similar conservatism in reproductive habit has been invoked to explain restricted dispersal in planktonic foraminifers (55).
Flow patterns of tidal or oceanic currents may also contribute to the genetic isolation among global populations by hindering dispersal (22, 23, 28, 58, 59). For example, there were no significant FST values between populations from the North Sea and a connected Danish inland fjord, yet populations from the Irish Sea and the adjacent North Sea showed significant differentiation. The Irish Sea and the North Sea are separated by sea fronts that limit exchange between both areas (60). At a macrogeographic scale, ocean circulation and continental land masses may act as effective barriers to gene flow among oceanic basins and among oceanic circulation centers (61). Such barriers may explain the reduced gene flow of clade I between Atlantic and Pacific populations, and between the northern and southwestern Pacific Ocean. Similarly, oceanic gyres and continental land masses are barriers to gene flow between two globally distributed oceanic copepod sister species (62), even if the impact of specific barriers on population genetic structuring varied between the two species. In the same sense, ecological and geographic barriers to gene flow are species-specific in marine foraminifers (17), reflecting the unique evolutionary histories and ecological requirements of even closely related species (17, 62, 63).
Given enough time and appropriate geographic and environmental circumstances, the population structure in clade I may provoke allopatric speciation. This is illustrated by a phylogenetic analysis, which shows that the split of P. pungens into three distinct clades probably occurred during the Pleistocene (Fig. 6), a period characterized by repeated glacial cycles. To date we can only speculate on how glacial cycles have promoted divergence within this superspecies, but a number of possible scenarios can be forwarded. During periods of glacially lowered sea level when coastal seas drained or large lagoons became landlocked (61), populations of P. pungens may have experienced prolonged geographical isolation. In such environments, populations are likely to experience demographic declines that will amplify the effects of genetic drift, which in concert or interaction with local adaptation to specific environmental conditions may lead to speciation (19, 64). In addition, creation and colonization of empty niches associated with Pleistocene cycles of sea level rise-and-fall (e.g., as a consequence of local extinctions) likely accelerated speciation rates. Extended periods of isolation or colonization events possibly led to the differentiation into the warm-water clade III and the cool-water clades I and II. Increased cooling and upwelling along the equatorial divergences in the Pacific and the Atlantic possibly allowed the latter to disperse from one hemisphere to the other, resulting in an antitropical distribution, similar to evolutionary scenarios proposed for bipolar mollusks and foraminifers (54–56, 65). The diversification and geographic distribution of clades I and II, and their regional cooccurrence in the northeastern Pacific can be explained by two scenarios: (i) the two clades evolved sympatrically along the northeastern Pacific coast, or (ii) a secondary contact arose after para- or allopatric differentiation of the two clades, either along the same coast or in distant geographic regions. Both scenarios are difficult to separate (66), but a number of clues favor the second hypothesis in which clade II originated separately in the northeastern Pacific and came later in secondary contact with clade I, producing limited genetic admixture. First, individuals from both clades occur in the same samples (38), indicating limited ecological differentiation. This argues against sympatric speciation through ecological mechanisms (19). Second, reproduction in both clades is induced by similar environmental cues and hybridization is common (38–40). This suggests incomplete reproductive isolation, which is indicative for allopatric divergence where reproductive isolation in the absence of gene flow is expected to arise more gradually than in sympatry (67). Third, assuming that centers of origin show a higher genetic diversity than newly invaded areas (68), the highest microsatellite allelic richness and He in Japan suggests that clade I has spread from the northwestern Pacific to other regions. A similar phylogeographic pattern, in which planktonic microorganisms have spread from a NW Pacific center has been suggested for the dinoflagellate Alexandrium catenella (26). The lowest allelic richness and He in the northeastern Pacific may then be due to founder effects caused by recent range expansions. This hypothesis is supported by the much higher allelic richness and He of clade II in the northeastern Pacific (comparable with clade I in the northwestern Pacific) than clade I (38) (Fig. S3). The biogeographical asymmetry between clades I and II (Fig. 1) may result from a wider ecological tolerance of clade I, but may also be related to specific historical or environmental conditions in the northeastern Pacific (69), as endemism there is not unique to clade II but is also observed in other planktonic groups (e.g., 14, 55). Finally, it should be noted that the possibility of abrupt genomic changes (e.g., genomic duplications) as a mechanism for sympatric genetic diversification (14) remains to be tested for P. pungens.
In conclusion, our results show that within the cosmopolitan marine planktonic diatom P. pungens clade I, significant population differentiation exists at macrogeographic scales. Our data indicate that dispersal limitation by geographic distance may be an important factor in genetic differentiation, even in high dispersal marine microorganisms. The globally distributed clade I therefore behaves as an incipient planktonic superspecies (9) in which allopatric populations may eventually diverge into cryptic species. Our results oppose earlier assumptions that uninterrupted gene flow and range expansions of marine planktonic microbes produce global, homogeneous populations and limit opportunities for geographic speciation (1, 3, 13) and thus challenge the current paradigm that sympatric speciation is the prevalent mode of speciation in marine planktonic microorganisms (9, 14). Instead, we suggest that persistent restricted gene flow between geographically separated populations can, given the right geographic and environmental conditions, lead to population differentiation and high diversity in planktonic organisms, despite the dampening effect of large population sizes.
Materials and Methods
Isolation of P. pungens and Molecular Analysis.
Sampling was performed in seven areas in the Atlantic and Pacific Ocean (Fig. 1 and Table S1). P. pungens cells were isolated and monoclonal cultures established as described in Casteleyn et al. (35, 42). Identification of isolates (clades I−III) was based on nrDNA ITS sequencing as described in Casteleyn et al. (35). For clade I isolates, six nuclear microsatellite loci (PP1, PP2, PP3, PP4, PP5, and PP6) were amplified with primers developed by Evans and Hayes (70) (SI Text). Two hundred and forty-two isolates were successfully genotyped, except for a single Canadian isolate that had a missing genotype at locus PP4.
Microsatellite Data Analysis.
Initial data analysis included identification of matching multilocus genotypes (MLGs) and verification of possible scoring errors due to stuttering or large allele dropout (SI Text). For measurement of genetic diversity the seven sampling areas were arbitrarily regarded as populations. Probability of identity (P(ID) = 3.75 × 10−9) and the probability of identity for siblings (P(ID)sib = 1.95 × 10−3) calculated with GIMLET v1.3.3 (71) were within the threshold values suggested by Waits et al. (72), indicating that our six microsatellite markers had enough resolving power to distinguish between individuals. Genetic diversity was measured by numbers of alleles, allele frequencies, numbers of genotypes, observed heterozygosity (Ho), and expected heterozygosity (He) per population and per locus (SI Text). Extrapolation of allele numbers beyond the sample size was done with ARES v1.2.2 (73). Departures from Hardy-Weinberg Equilibrium (HWE) at each locus and across loci in every population, and linkage disequilibrium (LD) between all pairs of loci per population were tested using exact tests (SI Text). Multiple test problems were dealt with by calculating corrected P values using the sequential Bonferroni technique.
Population Structure.
Population differentiation was evaluated with allelic and genotypic exact tests in GENEPOP v4.0 (74) using default settings. Tests were run for each locus between all pairs of populations and combinations of P values across loci were obtained according to Fisher's method. The degree of genetic differentiation between pairs of populations was quantified using Weir and Cockerham’s (75) estimate of FST in FSTAT v2.9.3 (76). Permutation tests were used to determine whether FST values were significantly different from zero. FST values depend on allelic diversities, therefore standardized FST values were calculated using RECODEDATA v0.1 (77). In all cases, P values were corrected using a sequential Bonferroni technique. A principal component analysis (PCA) was performed to visualize differentiation among populations (FST) using GENALEX v6 (78). To infer population structure without a predefined population subdivision, the Bayesian clustering program STRUCTURE v2.2 (44) was used (SI Text). The most likely number of populations was estimated to be K = 6 following the recommendation of Pritchard et al. (79) (Fig. S2). For the selected K value, we evaluated the individual membership coefficient (qind) to the inferred clusters. CLUMPP v1.1.1 (80) was used to line up the cluster labels across runs and to estimate the degree of congruence between independent runs. Visualization of the results from the Bayesian analyses was done with DISTRUCT v1.1 (81).
Isolation by Distance.
Correlations between genetic and geographic distances among populations (IBD) were evaluated with the program IBDWS v3.16 (82) using nonparametric Mantel tests. Geographic distance between sampling areas was measured as the shortest continuous water surface distance using Google Earth, even if these sea connections do not necessarily reflect the actual dispersal distance in complex oceanic circulations. Genetic similarities among population pairs were computed using Slatkin’s (83) similarity measure, M = [(1/FST) − 1]/4. The significance of the major axis regression was assessed by 10,000 permutations of the data.
Phylogenetic Analysis.
A dated phylogeny of Pseudo-nitzschia clade I [as presented in Lundholm et al. (6)] was based on Bayesian inference of rbcL, large subunit rDNA and rDNA ITS sequence data (Table S3). Divergence times were estimated under a relaxed molecular clock using an uncorrelated lognormal model in BEAST v1.4.6 (84). The Markov chain Monte Carlo analysis was run for 10 million generations, of which the last 5 million were used for generating summary statistics and trees. The single calibration point was the estimated split of P. pungens with its closest relative P. multiseries at 5 Mya (48). Of course, the chronogram has to be interpreted with care, as different molecular clock studies have shown variation in divergence times between major diatom lineages (48).
Supplementary Material
Acknowledgments
We thank Victor Chepurnov, (Ghent University, Ghent, Belgium) Veronique Creach, (Centre for Environment, Fisheries & Aquaculture Science, Lowestoft, UK) Stephen Bates, (Fisheries & Oceans Canada, Moncton, Canada) Claude Léger, (Fisheries & Oceans Canada) Nick Adams, (NOAA-Fisheries Service, Seattle) and the Flemish Marine Institute (VLIZ), for providing isolates. We thank Andy Vierstraete (Ghent University) for allele fragment electrophoresis. We thank two anonymous reviewers and the editor for insightful suggestions that greatly improved the manuscript. Funding was provided by the Flanders agency for Innovation by Science and Technology (IWT) (PhD fellowship to G.C.), Research Foundation (FWO) Flanders (Grants G.0197.05, G.0208.08, G.0292.00, G.0404.07, and G.0208.08, and a postdoctoral fellowship to F.L.) and Special Research Fund (BOF) of Ghent University (GOA 12050398). Northeastern Pacific samples were collected as part of the Ecology and Oceanography of Harmful Algal Blooms in the Pacific Northwest (ECOHAB-PNW) project funded by National Science Foundation ECOHAB Project OCE-0234587 and National Oceanic and Atmospheric Administration ECOHAB Grant NA16OP1450.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001380107/-/DCSupplemental.
References
- 1.Norris RD. Pelagic species diversity, biogeography, and evolution. Paleobiology. 2000;26:236–258. [Google Scholar]
- 2.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 3.Cermeño P, Falkowski PG. Controls on diatom biogeography in the ocean. Science. 2009;325:1539–1541. doi: 10.1126/science.1174159. [DOI] [PubMed] [Google Scholar]
- 4.de Vargas C, Norris R, Zaninetti L, Gibb SW, Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc Natl Acad Sci USA. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez AG, et al. Pseudo-cryptic speciation in coccolithophores. Proc Natl Acad Sci USA. 2003;100:7163–7168. doi: 10.1073/pnas.1132069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundholm N, et al. Inter- and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses. J Phycol. 2006;42:464–481. [Google Scholar]
- 7.Šlapeta J, López-García P, Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol Biol Evol. 2006;23:23–29. doi: 10.1093/molbev/msj001. [DOI] [PubMed] [Google Scholar]
- 8.Kooistra WH, et al. Global diversity and biogeography of Skeletonema species (bacillariophyta) Protist. 2008;159:177–193. doi: 10.1016/j.protis.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 9.de Vargas C, Sáez AG, Medlin LK, Thierstein HR. Super-Species in the calcareous plankton. In: Thierstein HR, Young JR, editors. Coccolithophores: From Molecular Processes to Global Impact. Berlin: Springer-Verlag; 2004. pp. 271–298. [Google Scholar]
- 10.Benton MJ, Pearson PN. Speciation in the fossil record. Trends Ecol Evol. 2001;16:405–411. doi: 10.1016/s0169-5347(01)02149-8. [DOI] [PubMed] [Google Scholar]
- 11.Bierne N, Bonhomme F, David P. Habitat preference and the marine-speciation paradox. Proc R Soc B Biol Sci. 2003;270:1399–1406. doi: 10.1098/rspb.2003.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sexton PF, Norris RD. Dispersal and biogeography of marine plankton: Long-distance dispersal of the foraminifer Truncorotalia truncatulinoides. Geology. 2008;36:899–902. [Google Scholar]
- 14.Koester JA, Swalwell JE, von Dassow P, Armbrust EV. Genome size differentiates co-occurring populations of the planktonic diatom Ditylum brightwellii (Bacillariophyta) BMC Evol Biol. 2010;10:1. doi: 10.1186/1471-2148-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vargas C, et al. A molecular approach to biodiversity and biogeography in the planktonic foraminifer Globigerinella siphonifera (d'Orbigny) Mar Micropaleontol. 2002;45:101–116. [Google Scholar]
- 16.Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darling KF, Wade CA. The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes. Mar Micropaleontol. 2008;67:216–238. [Google Scholar]
- 18.Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kucera M. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Mol Ecol. 2009;18:1692–1706. doi: 10.1111/j.1365-294X.2009.04136.x. [DOI] [PubMed] [Google Scholar]
- 19.Sobel JM, Chen GF, Watt LR, Schemske DW. The biology of speciation. Evolution. 2010;64:295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 20.Balloux F, Lugon-Moulin N. The estimation of population differentiation with microsatellite markers. Mol Ecol. 2002;11:155–165. doi: 10.1046/j.0962-1083.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- 21.Knutsen H, et al. Bathymetric barriers promoting genetic structure in the deepwater demersal fish tusk (Brosme brosme) Mol Ecol. 2009;18:3151–3162. doi: 10.1111/j.1365-294X.2009.04253.x. [DOI] [PubMed] [Google Scholar]
- 22.White TA, Stefanni S, Stamford J, Hoelzel AR. Unexpected panmixia in a long-lived, deep-sea fish with well-defined spawning habitat and relatively low fecundity. Mol Ecol. 2009;18:2563–2573. doi: 10.1111/j.1365-294X.2009.04218.x. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda N, et al. Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Mol Ecol. 2009;18:1574–1590. doi: 10.1111/j.1365-294X.2009.04133.x. [DOI] [PubMed] [Google Scholar]
- 24.Galarza JA, et al. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA. 2009;106:1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iglesias-Rodriguez MD, et al. Intraspecific genetic diversity in the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae): The use of microsatellite analysis in marine phytoplankton population studies. J Phycol. 2006;42:526–536. [Google Scholar]
- 26.Masseret E, et al. Unexpected genetic diversity among and within populations of the toxic dinoflagellate Alexandrium catenella as revealed by nuclear microsatellite markers. Appl Environ Microbiol. 2009;75:2037–2045. doi: 10.1128/AEM.01686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCauley LAR, et al. Biogeographic analysis of the globally distributed harmful algal bloom species Alexandrium minutum (Dinophyceae) based on rRNA gene sequences and microsatellite markers. J Phycol. 2009;45:454–463. doi: 10.1111/j.1529-8817.2009.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai S, et al. Genetic structuring and transfer of marine dinoflagellate Cochlodinium polykrikoides in Japanese and Korean coastal waters revealed by microsatellites. Mol Ecol. 2009;18:2337–2352. doi: 10.1111/j.1365-294X.2009.04193.x. [DOI] [PubMed] [Google Scholar]
- 29.Kooistra WHCF, Gersonden R, Medlin LK, Mann DG. The origin and evolution of the diatoms: Their adaptation to a planktonic existence. In: Falkowski PG, Knoll AH, editors. Evolution of Planktonic Photoautotrophs. Burlington: Academic Press; 2007. pp. 207–249. [Google Scholar]
- 30.Armbrust EV. The life of diatoms in the world's oceans. Nature. 2009;459:185–192. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- 31.Hasle GR. Are most of the domoic acid-producing species of the diatom genus Pseudo-nitzschia cosmopolites? Harmful Algae. 2002;1:137–146. [Google Scholar]
- 32.Trainer VL, et al. Domoic acid production by Pseudo-nitzschia pungens. In: Reguera B, Blanco J, Fernández ML, Wyatt T, editors. Harmful Algae. Paris: Xunta de Galicia and the IOC of UNESCO; 1998. pp. 337–340. [Google Scholar]
- 33.Marchetti A, et al. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature. 2009;457:467–470. doi: 10.1038/nature07539. [DOI] [PubMed] [Google Scholar]
- 34.Chepurnov VA, et al. Sexual reproduction, mating system, chloroplast dynamics and abrupt cell size reduction in Pseudo-nitzschia pungens from the North Sea (Bacillariophyta) Eur J Phycol. 2005;40:379–395. [Google Scholar]
- 35.Casteleyn G, et al. Pseudo-nitzschia pungens (Bacillariophyceae): A cosmopolitan diatom species? Harmful Algae. 2008;7:241–257. [Google Scholar]
- 36.De Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 37.Churro CI, et al. Diversity and abundance of potentially toxic Pseudo-nitzschia Peragallo in Aveiro coastal lagoon, Portugal and description of a new variety, P. pungens var. aveirensis var. nov. Diatom Res. 2009;24:35–62. [Google Scholar]
- 38.Adams NG, et al. Genetic population structure of Pseudo-nitzschia pungens (Bacillariophyceae) from the Pacific Northwest and the North Sea. J Phycol. 2009;45:1037–1045. doi: 10.1111/j.1529-8817.2009.00746.x. [DOI] [PubMed] [Google Scholar]
- 39.Casteleyn G, et al. Natural hybrids in the marine diatom Pseudo-nitzschia pungens (Bacillariophyceae): Genetic and morphological evidence. Protist. 2009;160:343–354. doi: 10.1016/j.protis.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Holtermann KE, et al. Mass sexual reproduction in the toxigenic diatoms Pseudo-nitzschia australis and P. pungens (Bacillariophyceae) on the Washington coast, USA. J Phycol. 2010;46:41–52. [Google Scholar]
- 41.Evans KM, Kuhn SF, Hayes PK. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzschia pungens (Bacillariophyceae) populations. J Phycol. 2005;41:506–514. [Google Scholar]
- 42.Casteleyn G, et al. Lack of population genetic structuring in the marine planktonic diatom Pseudo-nitzschia pungens (Bacillariophyceae) in a heterogeneous area in the Southern Bight of the North Sea. Mar Biol. 2009;156:1149–1158. [Google Scholar]
- 43.Rynearson TA, Armbrust EV. Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol Ecol. 2005;14:1631–1640. doi: 10.1111/j.1365-294X.2005.02526.x. [DOI] [PubMed] [Google Scholar]
- 44.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoeck T, et al. Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 2009;7:72. doi: 10.1186/1741-7007-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dawson MN, Sen Gupta A, England MH. Coupled biophysical global ocean model and molecular genetic analyses identify multiple introductions of cryptogenic species. Proc Natl Acad Sci USA. 2005;102:11968–11973. doi: 10.1073/pnas.0503811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolch CJS, de Salas MF. A review of the molecular evidence for ballast water introduction of the toxic dinoflagellates Gymnodinium catenatum and the Alexandrium “tamarensis complex” to Australasia. Harmful Algae. 2007;6:465–485. [Google Scholar]
- 48.Sorhannus U. A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar Micropaleontol. 2007;65:1–12. [Google Scholar]
- 49.Marshall DJ, Monro K, Bode M, Keough MJ, Swearer S. Phenotype-environment mismatches reduce connectivity in the sea. Ecol Lett. 2010;13:128–140. doi: 10.1111/j.1461-0248.2009.01408.x. [DOI] [PubMed] [Google Scholar]
- 50.Nishida M, Lucas JS. Genetic differences between geographic populations of the Crown-of-thorns starfish throughout the Pacific region. Mar Biol. 1988;98:359–368. [Google Scholar]
- 51.Palumbi SR, Metz EC. Strong reproductive isolation between closely related tropical sea urchins (genus Echinometra) Mol Biol Evol. 1991;8:227–239. doi: 10.1093/oxfordjournals.molbev.a040642. [DOI] [PubMed] [Google Scholar]
- 52.Pogson GH, Taggart CT, Mesa KA, Boutilier RG. Isolation by distance in the Atlantic cod, Gadus morhua, at large and small geographic scales. Evolution. 2001;55:131–146. doi: 10.1111/j.0014-3820.2001.tb01279.x. [DOI] [PubMed] [Google Scholar]
- 53.Bowen BW, Grant WS. Phylogeography of the sardines (Sardinops spp): Assessing biogeographic models and population histories in temperate upwelling zones. Evolution. 1997;51:1601–1610. doi: 10.1111/j.1558-5646.1997.tb01483.x. [DOI] [PubMed] [Google Scholar]
- 54.Darling KF, et al. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405:43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- 55.Darling KF, Kucera M, Wade CM. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc Natl Acad Sci USA. 2007;104:5002–5007. doi: 10.1073/pnas.0700520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Norris RD, de Vargas C. Evolution all at sea. Nature. 2000;405:23–24. doi: 10.1038/35011162. [DOI] [PubMed] [Google Scholar]
- 57.Nuwer M, Frost B, Armbrust EV. Population structure of the planktonic copepod Calanus pacificus in the North Pacific Ocean. Mar Biol. 2008;156:107–115. [Google Scholar]
- 58.Nagai S, et al. Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. J Phycol. 2007;43:43–54. [Google Scholar]
- 59.Peijnenburg KT, Fauvelot C, Breeuwer JA, Menken SBJ. Spatial and temporal genetic structure of the planktonic Sagitta setosa (Chaetognatha) in European seas as revealed by mitochondrial and nuclear DNA markers. Mol Ecol. 2006;15:3319–3338. doi: 10.1111/j.1365-294X.2006.03002.x. [DOI] [PubMed] [Google Scholar]
- 60.Pingree RD, Griffiths DK. Tidal fronts on shelf seas around British Isles. J Geophys Res. 1978;83:4615–4622. [Google Scholar]
- 61.Palumbi SR. Genetic divergence, reproductive isolation and marine speciation. Annu Rev Ecol Syst. 1994;25:547–572. [Google Scholar]
- 62.Goetze E. Global population genetic structure and biogeography of the oceanic copepods Eucalanus hyalinus and E. spinifer. Evolution. 2005;59:2378–2398. [PubMed] [Google Scholar]
- 63.Planes S, Fauvelot C. Isolation by distance and vicariance drive genetic structure of a coral reef fish in the Pacific Ocean. Evolution. 2002;56:378–399. doi: 10.1111/j.0014-3820.2002.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 64.Dawson MN, Hamner WM. Rapid evolutionary radiation of marine zooplankton in peripheral environments. Proc Natl Acad Sci USA. 2005;102:9235–9240. doi: 10.1073/pnas.0503635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crame JA. Bipolar mollusks and their evolutionary implications. J Biogeogr. 1993;20:145–161. [Google Scholar]
- 66.Savolainen V, et al. Sympatric speciation in palms on an oceanic island. Nature. 2006;441:210–213. doi: 10.1038/nature04566. [DOI] [PubMed] [Google Scholar]
- 67.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 68.Dlugosch KM, Parker IM. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- 69.Briggs JC. Marine centres of origin as evolutionary engines. J Biogeogr. 2003;30:1–18. [Google Scholar]
- 70.Evans KM, Hayes PK. Microsatellite markers for the cosmopolitan marine diatom Pseudo-nitzschia pungens. Mol Ecol Notes. 2004;4:125–126. [Google Scholar]
- 71.Valiere N. GIMLET: A computer program for analysing genetic individual identification data. Mol Ecol Notes. 2002;2:377–379. [Google Scholar]
- 72.Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol Ecol. 2001;10:249–256. doi: 10.1046/j.1365-294x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- 73.Van Loon EE, Cleary DFR, Fauvelot C. ARES: Software to compare allelic richness between uneven samples. Mol Ecol Notes. 2007;7:579–582. [Google Scholar]
- 74.Rousset F. GENEPOP'007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 75.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 76.Goudet J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices 2.9.3. [Google Scholar]
- 77.Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- 78.Peakall R, Smouse PE. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pritchard JK, Wen X, Falush D. 2009. STRUCTURE 2.2. [Google Scholar]
- 80.Jakobsson M, Rosenberg NA. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 81.Rosenberg NA. DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- 82.Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genet. 2005;6:13. doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 84.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.