Abstract
Endocytosis is a process by which extracellular material such as macromolecules can be incorporated into cells via a membrane-trafficking system. Although universal among eukaryotes, endocytosis has not been identified in Bacteria or Archaea. However, intracellular membranes are known to compartmentalize cells of bacteria in the phylum Planctomycetes, suggesting the potential for endocytosis and membrane trafficking in members of this phylum. Here we show that cells of the planctomycete Gemmata obscuriglobus have the ability to uptake proteins present in the external milieu in an energy-dependent process analogous to eukaryotic endocytosis, and that internalized proteins are associated with vesicle membranes. Occurrence of such ability in a bacterium is consistent with autogenous evolution of endocytosis and the endomembrane system in an ancestral noneukaryote cell.
Keywords: eukaryotes, evolution, endocytosis, bacteria, planctomycetes
A major unsolved problem in biology is how the many unique characteristics of the eukaryote cell evolved, including endomembranes and their dynamic features, such as endocytosis (1). Endocytosis is known as a eukaryote-specific process by which cells internalize molecules from the plasma membrane and recycle them back to the surface or sort them to lysosomes for degradation (2). There is evidence that endocytosis must have been present in cells as far back as the last eukaryotic common ancestor (LECA) (3, 4), but it has never been reported to occur in members of the domains Bacteria and Archaea. Recent hypotheses have placed the origin of endocytosis-like mechanisms as a primary step toward evolution of compartmentalization (3, 4). Surprisingly, subcellular compartmentalization is not a unique feature of the eukaryote cell: it has been discovered that a restricted group of bacteria belonging to the phyla Planctomycetes and Verrucomicrobia possess such compartmentalization (5–8). The planctomycetes are classified in Bacteria on phylogenetic and genomic grounds (9–11) and form a divergent group, reported to branch deeply within the Bacteria when slowly evolving alignment positions are used for generating 16S rRNA-based phylogenetic trees, although not necessarily when other methods are used. Phylogenetic and other data indicate that the planctomycetes are related most closely to bacterial phyla Verrucomicrobia and Chlamydiae (12, 13, 14). However, planctomycetes possess unusual properties such as budding reproduction (15–17), sterol biosynthesis (18), permanently condensed nucleoids different from uncondensed “corraline” nucleoids of other bacteria but similar to those of chlamydial elementary bodies (19), absence of cell wall peptidoglycan (20–22), and most strikingly, formation of intracellular membrane-bounded compartments (7). Some planctomycetes such as Gemmata obscuriglobus are compartmentalized into a nucleoid containing DNA, a ribosome-containing cytoplasm (riboplasm), and a ribosome-free cytoplasm (paryphoplasm) (5). A single intracytoplasmic membrane (ICM) separates the riboplasm from the paryphoplasm, with the paryphoplasm defined as a compartment between the cytoplasmic membrane and ICM (5). Planctomycetes are also exceptional among Bacteria because they carry genes homologous to those coding for membrane coat (MC) proteins central to eukaryotic endocytosis (23). Taken together, the unusual characteristics of planctomycetes suggest that these bacteria might possess a simple form of endocytosis.
Results and Discussion
G. obscuriglobus Cells Are Able to Internalize GFP via an Energy-Dependent Process.
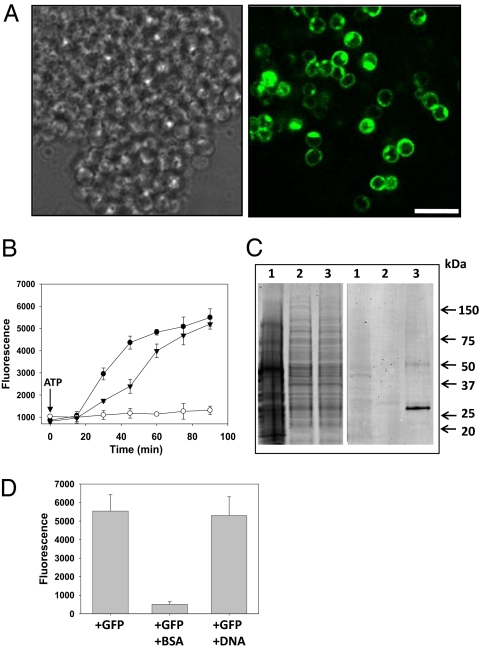
To investigate the possibility of an endocytosis-like mechanism in the planctomycete bacterium G. obscuriglobus, we incubated cells with GFP and examined them via confocal laser scanning microscopy (CLSM). Remarkably, the protein was detected inside the cells (Fig. 1A), usually within 5 min. Location of GFP inside the cells rather than on the cell surface was indicated by the optical sectioning inherent with CLSM. The uptake of GFP seemed to be energy-dependent, because if incubated at 0 °C or in the presence of sodium azide, an inhibitor of respiration and oxidative phosphorylation, cells did not take up GFP (Fig. 1B and Fig. S1A). Temperature sensitivity of GFP uptake was also demonstrated by its inhibition at 37 °C (Fig. S1A), a temperature above the maximum growth temperature of G. obscuriglobus (24). ATP suppressed the inhibitory effect of sodium azide (Fig. 1B and Fig. S1B), confirming that GFP uptake is an energy-dependent process. The uptake of full-length GFP by cells of G. obscuriglobus was confirmed by Western blot analysis (Fig. 1C). As expected, nonplanctomycete bacterium Escherichia coli, acting as a negative control, did not take up GFP (Fig. 1C). Such an uptake process is saturable, as expected for a receptor-mediated process, the concentration of proteins at which the uptake process reaches saturation corresponding to ≈10 μg/mL (Fig. S2). This finding demonstrates the internalization of proteins in a bacterium. Internalization of intact external macromolecules, and more specifically proteins, has not been recorded in members of domains Bacteria or Archaea. Some bacteria, such as Lactococcus lactis, can use proteins only by degrading them at the cell surface using proteases and incorporating the resulting peptides (25). A 35-aa oligopeptide is so far the longest shown to bind to a bacterial receptor (26).
Fig. 1.
G. obscuriglobus cells uptake proteins but not DNA in an energy-dependent process. (A) Bright field microscopy and CLSM images of G. obscuriglobus cells incubated with GFP (10 μg/mL) for 60 min. Left and Right microscopy images correspond to bright field and GFP fluorescence, respectively. Because of optical sectioning effects, only some of the cells in the clump positive for GFP signal can be visualized. (Scale bar, 4 μm.) (B) Time-course experiment with GFP alone (filled circles), with GFP and 1 mM sodium azide (open circles), or with GFP and both 1 mM sodium azide and 5 mM ATP (filled triangles). After washing of G. obscuriglobus cells, GFP fluorescence was quantitatively analyzed by microplate reader assay. Error bars represent the SD from three experimental replicates. (C) Western blot analysis showing uptake of full-length GFP (≈27 kDa) by G. obscuriglobus. Cells were incubated with GFP for 1 h. After washing, cell lysates were electrophoretically separated on an SDS gel (Left), then blotted and probed with an anti-GFP antibody (Right). Lane 1, E. coli DH5α incubated with GFP; lane 2, G. obscuriglobus incubated without GFP; lane 3, G. obscuriglobus incubated with GFP. (D) Competition uptake experiment with GFP, together with either BSA or DNA. G. obscuriglobus cells were incubated for 1 h with 10 μg/mL of GFP alone, with GFP and BSA (100 μg/mL), or with GFP and plasmid DNA (100 μg/mL). Cells were then washed and analyzed for fluorescence emission by microplate reader assay. Error bars represent the SD from three experimental replicates.
We next tested whether the mechanism of uptake is receptor mediated. Coincubation of GFP with unequal ratios of Cy3-labeled Ig resulted in dominant internalization of the protein in excess, whereas equal internalization was detected when a 1:1 ratio was used (Fig. S3). This implied that different proteins can be internalized by G. obscuriglobus and that they are competing for the same surface receptor. Such assumption was further supported by the fact that an excess of BSA, ovalbumin, or GST greatly diminished uptake of GFP (Fig. 1D and Fig. S4). Additionally, plasmid DNA did not affect GFP uptake (Fig. 1D). A Cy3-labeled 25-mer oligonucleotide was not internalized by G. obscuriglobus (Fig. S5), demonstrating that in contrast to proteins, DNA is not internalized by this bacterium. Considered together, these results are consistent with a receptor-mediated protein uptake mechanism in G. obscuriglobus.
Proteins Internalized by G. obscuriglobus Are Compartmentalized and Degraded in the Paryphoplasm Cell Region.
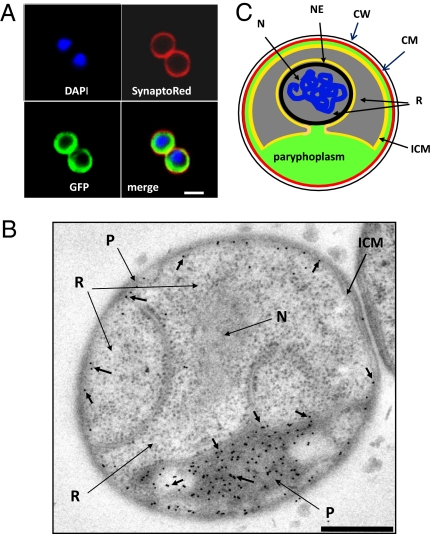
To gain more information about the mechanism of uptake, we investigated the subcellular localization of different proteins after their internalization. Coincubation of Cy5-labeled fluorescent streptavidin or Cy3-labeled Ig with GFP resulted in colocalization of the two distinct proteins to the same compartment (Fig. S6). The proteins were found to be restricted to an outer region of the cell cytoplasm, within the external cytoplasmic membrane indicated by SynaptoRed stain, and distinct from the DAPI-stained region containing DNA (Fig. 2A). Transmission electron microscopy (TEM) examination of thin sections of G. obscuriglobus cells preincubated with GFP defined this region as the paryphoplasm (Fig. 2B and schematic diagram Fig. 2C). This suggests a functional role for paryphoplasm as a protein-segregating compartment where exogenous proteins can be accumulated. We next tested whether the internalized proteins are proteolysed in this compartment. Cells were incubated with DQ Green BSA (DQBSA), fluorescence of which appears only upon proteolysis, and with IgG-Cy3, the latter added shortly before examination and serving as a marker for the paryphoplasm. The fluorescent DQBSA proteolysis product colocalized with IgG-Cy3 in the paryphoplasm (Fig. S7A), illustrating the role of this compartment in degradation of internalized proteins. The confirmation that internalized proteins are degraded was obtained by first incubating G. obscuriglobus with a mouse IgG and then monitoring its fate—proteolysis of the Ig was detected by Western blot analysis (Fig. S7B). GFP was not suitable for this assay because it is highly resistant to proteolysis (27).
Fig. 2.
Internalized proteins in G. obscuriglobus cells are localized to paryphoplasm. (A) G. obscuriglobus cells were incubated with GFP and then stained with DAPI and SynaptoRed. A GFP-containing region is seen in the cytoplasm bounded by the cytoplasmic membrane as defined by the SynaptoRed staining and is separated from the nuclear body (DAPI staining). (B) TEM image of a section of high-pressure frozen cryosubstituted cells of G. obscuriglobus, immunogold-labeled to detect GFP via anti-GFP antibody and secondary antibody conjugated with 10 nm colloidal gold. Gold particles (short arrows) labeling internalized GFP are only associated with paryphoplasm (P). Gold particles were excluded from both the double membrane-bounded nucleoid and the riboplasm. The riboplasm (R), fibrillar nucleoid (N), and intracytoplasmic membrane (ICM) are indicated. (Scale bar, 500 nm.) (C) Diagram representing the functional compartmentalization of G. obscuriglobus. N, nucleoid; NE, nuclear envelope; ICM, intracytoplasmic membrane; R, riboplasm; CM, cytoplasmic membrane; CW, cell wall.
Internalized Proteins Are Associated with Vesicles.
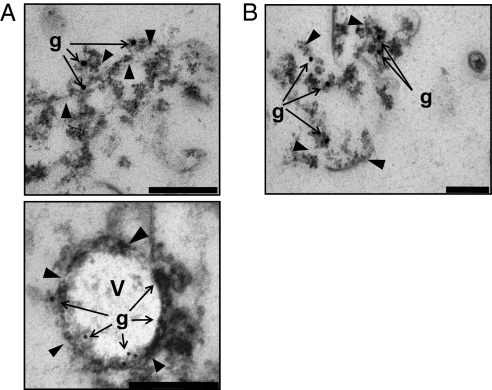
We next used immunogold labeling of high-pressure frozen cells to investigate whether the internalized GFP was associated with any specific intracellular structures. Gold particles indicating presence of GFP in the paryphoplasm were often seen lining vesicle-like entities ca. 50–200 nm wide bounded by a single bilayer membrane (Fig. 3A, Insets 2–4). In some cases, gold particles were seen lining an infolding of the cytoplasmic membrane (Fig. 3B), perhaps representing an initial stage in the process of GFP internalization. An invagination of cytoplasmic membrane was also seen during uptake of HRP-conjugated antibody, the reaction product of which was surrounded by an infolding membrane showing continuity with the cytoplasmic membrane (Fig. 3C). These results suggest formation of vesicle structures in the cells allowing extracellular molecules to be internalized. Remarkably, proteins possessing structural features of eukaryotic MC proteins have been recently described in planctomycetes (23). MC proteins are related to the clathrin and coat protein families, all members of which are associated with vesicle formation or membrane curvature, and some of which (e.g., clathrin) are necessary for receptor-mediated endocytosis. The similarity in secondary and tertiary structure between gp4978, an MC-like protein of G. obscuriglobus, and clathrin heavy chain of yeast is illustrated in Fig. S8. An antibody had been previously raised against G. obscuriglobus gp4978 (23). In our study this anti-gp4978 antibody reacted with vesicle-like structures within the paryphoplasm (Fig. 3D). In some cases, gp4978-reactive material was associated with cytoplasmic membranes apparently in the first stages of membrane invagination (Fig. 3D, Inset 3).
Fig. 3.
TEM of high-pressure frozen cryosubstituted G. obscuriglobus cells showing association of GFP and MC protein with membranes and vesicles. (A) Cells were immunogold-labeled to detect GFP, which is seen only in the paryphoplasm. A1 and the enlargements of boxed areas 2–4 show association of several gold particles with the same membrane. Arrowheads, membranes; V, vesicles. (Scale bars, 1 μm in A1, 50 nm in A2–A4.) (B) A section showing infolding of cytoplasmic membrane (arrowheads) associated with GFP (gold particles). (Scale bar, 50 nm.) (C) Cells were incubated with HRP-conjugated rabbit anti-mouse antibody. Peroxidase activity was detected via cytochemistry using diaminobenzidine substrate, yielding an electron-dense product. An example is shown of a region where the cytoplasmic membrane (CM) seems to have invaginated (arrowheads) together with HRP-conjugated antibody, as indicated by the dense reaction product. (Scale bar, 50 nm.) (D) G. obscuriglobus cells preincubated with GFP were immunogold-labeled to detect MC protein. D1 and enlargements of boxed areas 2 and 3 show association of several gold particles with membrane vesicle. Arrowheads, membranes; V, vesicles. (Scale bars, 500 nm in D1, 200 nm in D2, 50 nm in D3.)
To confirm that the internalized proteins are associated with membranes and vesicles, G. obscuriglobus cells were lysed by sonication and subjected to subcellular fractionation. After centrifugation at 100,000 × g, GFP was detected in the insoluble pellet consisting of membranes, cell walls, and other insoluble cell components, but it was excluded from the supernatant (Fig. S9A, step 1). The GFP-positive fraction was then separated via a series of sucrose steps and linear density gradient ultracentrifugations (Fig. S9). The maximum GFP signal was found to be associated with one fraction of ca. 60% sucrose density (Fig. S9A, step 3). TEM examination of this fraction demonstrated that it consisted of broken membranes and small membrane-bounded vesicles. Immunogold labeling showed that some membranes and vesicles in this preparation were positive for GFP (Fig. 4A) and MC-like protein (Fig. 4B). The association of GFP and MC-like protein with fractionated membranes from lysed cells is consistent with the invariable association of GFP with membranes in the paryphoplasm of sectioned G. obscuriglobus cells. It is unlikely that properly folded GFP could pass through a membrane pore because the size limit for molecules that can enter via bacterial membrane pores is much smaller [e.g., 600 Da for E. coli (28)]. It is also unlikely that GFP could pass through a membrane pore as an unfolded polypeptide because (i) this protein forms a highly stable structure of 30–40 Å diameter that is resistant to unfolding owing to a high folding/unfolding energy barrier (27, 29), and (ii) cytoplasmic components for protein secretion and/or associated ribosomal machinery are not present in the external milieu. However, to exclude the possibility of uptake of unfolded GFP through a membrane pore, we incubated G. obscuriglobus with a cross-linked protein (mouse IgG) that is expected to retain much of its tertiary structure during the uptake. This experiment demonstrated that the cross-linked protein was taken up by G. obscuriglobus and that it colocalized with MC-like protein in the same step gradient membrane fraction (Fig. S10). TEM of negatively stained fractions demonstrated their purity (Fig. S11).
Fig. 4.
Colocalization of GFP and MC proteins to the same density gradient membrane fractions of lysed G. obscurigobus cells. These cells had been incubated with GFP. (A and B) Immunogold labeling of the material obtained from fraction 9 of the 40–70% sucrose density gradient centrifugation (Fig. S9) showing cofractionation of GFP and MC protein and association of both proteins with membranes. (A) Fraction 9 was immunolabeled with anti-GFP antibody and secondary anti-mouse-10 nm gold antibody to visualize GFP. Gold particles (g) indicate that GFP is associated with broken membranes (Upper) and the vesicle membrane (Lower) rather than the vesicle interior. Arrowheads, membranes; V, vesicle. (Scale bar, 200 nm.) (B) Fraction 9 was immunolabeled with anti-gp4978 antibody and protein A-15 nm gold to visualize MC-like gp4978 protein. Arrowheads (membranes) and gold particles (g) indicate that MC protein is associated with broken membranes. (Scale bar, 200 nm.)
In summary, the observed results are compatible with an endocytosis-like mechanism but not with a channel-mediated mechanism in which unfolding of proteins is needed for import. An endocytosis-like mechanism is also consistent with our immunogold labeling results showing both GFP protein and MC protein to be associated with the vesicle membrane in initial infolding and later vesicle stages (Fig. 3 B and D).
Implications for Evolution.
Receptor-mediated endocytosis is universal among eukaryotes: it has been established in animal cells (30, 31), plant cells (32), fungi (33), and protozoa (34). Our discovery of a similar mechanism in a bacterium is of major significance to understanding evolution of cell complexity. There are two major types of hypotheses concerning the origin of eukaryote cell organization and its endomembranes. One postulates an ancient endosymbiosis involving archaea and bacteria, another proposes an autogenous origin involving internal membrane invagination and digestion of engulfed material (1, 35, 36). Our finding of an endocytosis-like process in a bacterium provides support for the latter possibility.
On the basis of our findings, an evolutionary scenario of autogenous endomembrane system development can be considered in which endocytosis may have first evolved before eukaryote separation as a distinct lineage. That is consistent with phylogenetic studies indicating that many components needed for endocytosis must have evolved before the LECA (3). Development of endocytosis could have been followed by or be simultaneous with development of an endomembrane sorting system correlated with formation of the nucleus. If the evolution of endocytosis predated the formation of the three Domains, retention of such a system might have occurred only in Domain Eucarya members and some isolated phyla in Bacteria. In this view, planctomycete endocytosis may represent a related process conserved in both eukaryotes and some bacteria during evolution from a common ancestor.
Endocytosis as found in planctomycetes may also be an example of a parallel evolutionary development of an analog of the eukaryote process. Evolution of a eukaryote-like cell plan may have occurred more than once from this point of view. The event which gave rise to the compartmentalized planctomycete cell plan enabling endocytosis-like abilities could be unrelated to eukaryote evolution in a direct phylogenetic sense. Planctomycete compartmentalization and endocytosis-like processes are nevertheless significant for models of eukaryote origins, because their occurrence makes it unnecessary to invoke endosymbiosis between an archaeon and a bacterium to explain eukaryogenesis.
The possibility of horizontal gene transfer (HGT) events as an explanation for endocytosis in a bacterium is not excluded and would be somewhat consistent with evidence that eukaryote genes of significance to cell biology (e.g., actin and profilin) have apparently been transferred to bacteria over evolutionary time (37). However, given the high complexity of the endocytotic system, it seems unlikely that HGT from eukaryotes is responsible for this phenomenon in G. obscuriglobus; the possibility of recent HGT is unlikely on the basis of codon usage and GC content (23). The remoteness of the detected homology of Gemmata MC-like proteins and eukaryote MC proteins including clathrins, distant enough to be revealed only by secondary and tertiary structure, is also consistent with the improbability of HGT as an explanation. It is also significant for consideration of potential origins of an endomembrane system that G. obscuriglobus possesses an analog of the eukaryote nucleus, because the nuclear envelope is formed from endoplasmic reticulum in metazoans and most likely in other eukaryotes.
In view of our findings with planctomycetes, an endomembrane system and compartmentalized cell organization in an ancestral protoeukaryote could have developed without the need for contributions from cells of other Domains of life. Our results imply that fusion between Archaea and Bacteria is not necessary to account for complex cell structure; internal membrane systems could have developed within a simple cell and without involvement of fusion of unrelated cells. G. obscuriglobus may be viewed as a snapshot of a possible stage in the autogenous origin of eukaryotes. This bacterium may exhibit an ancestral stage in the evolution of endocytosis and represent a unique in vivo model for a major stage in the evolution of the eukaryotic cell. The nature and extent of the endocytotic machinery in this organism should now be investigated at both structural and functional levels.
Materials and Methods
Material.
GFP was expressed from proviral vectors in tobacco leaves (38) and purified by using anion exchange chromatography. Sheep anti-mouse IgG-HRP was from Amersham Biosciences, Cy3-labeled Ig was from Sigma-Aldrich (catalog no. C2306), and Cy5-labeled streptavidin was from GE Healthcare (catalog no. PA 92005). Cy3-DNA (Sigma-Proligo) was a 25-mer oligonucleotide (5′-ATGGTGAGCAAGGGCGAGGAGCTGT-3′) labeled with Cy3 at the 5′ end and hybridized with its unlabeled antisense oligonucleotide (5′-ACAGCTCCTCGCCCTTGCTCACCAT-3′). Cy3 was from Molecular Probes. Mouse monoclonal anti-GFP antibody (Roche, catalog no. 11814460001) was used for dot-blots, and mouse monoclonal anti-GFP antibody (Clontech, catalog no. 632381) was used for immunogold labeling. Antibody to gp4978 was an affinity-purified (acid-elution) rabbit polyclonal antibody raised against recombinantly expressed and purified gp4978 protein of an ORF identified as a eukaryotic MC coatomer protein (National Center for Biotechnology Information reference sequence: ZP_02732338.1) (23).
Bacteria and Culture Conditions.
The planctomycete G. obscuriglobus was grown on plates containing M1 agar medium (39) and incubated aerobically at 28 °C for 4–7 d. M1 agar medium plates were prepared as follows: after pouring and allowing agar to set, plates were dried for 1 h with lids open in a biohazard cabinet. After inoculation by streaking with a sterile plastic loop charged with G. obscuriglobus grown on the same M1 agar medium, plates were sealed with parafilm before aerobic incubation. These standardized methods for medium preparation, inoculation, and incubation were necessary for optimal demonstration of protein uptake. E. coli strain DH5α was cultured on plates containing LB medium incubated at 37 °C for 16 h; plates were prepared in the same way as for M1 agar plates.
Uptake Experiments.
Protein uptake by G. obscuriglobus cells is dependent on pH, with optimum uptake occurring at pH 7.5. No protein uptake was observable at pH 5.8 or pH 8.8; therefore, for uptake experiments we used 10 mM Tris, pH 7.5, referred to as “incubation buffer” (IB). For all uptake experiments, cells were picked up directly from M1 agar plates using a 10-μL sterile plastic loop and resuspended in 50 μL of IB. In uptake experiments with GFP, Cy3-labeled Ig, Cy5-labeled streptavidin, and HRP-conjugated antibody, the final concentration of the proteins was 10 μg/mL, except if specified otherwise. After addition of proteins, cells were incubated at 28 °C for 60 min, except where specified otherwise. Cy3-DNA and Cy3 were added to the cells at a final concentration of 2 μM.
CLSM, TEM, and Image Analysis.
After incubation with fluorescent proteins, 5 μL of cell suspension was placed on a coverslip and the coverslip inverted onto a glass slide to make a wet mount for CLSM. A Zeiss LSM501 Meta (Carl Zeiss) confocal laser scanning microscope was used with 10× dry, and 20× water-immersion objectives or with a 100× oil-immersion objective. Fluorescent molecules were visualized with an argon laser and detected with band-path filter, with settings according to the manufacturer's recommendations. All high-pressure frozen and cryosubstituted sections were viewed using a JEOL 1010 transmission electron microscope operated at 80 kV. Images were captured using a SoftImaging Megaview III digital camera. The resulting files were annotated and resolution adjusted for final image production using Photoshop CS.
Time-Course Experiments.
All experiments were carried out in triple replicates using the same batch of G. obscuriglobus cells. Cells were resuspended in 2.5 mL of IB and aliquoted in three Eppendorf tubes, each containing 800 μL of cell suspension. Sodium azide (1 mM) was added 15 min before addition of GFP. ATP (5 mM) was added simultaneously with GFP. The samples were continuously mixed on a vertical rotating wheel. Aliquots of 100 μL from each tube were sampled at the indicated times and immediately cooled on ice. Upon completion of the experiment, all samples were centrifuged for 2 min at 5 °C at maximum speed (20,000 × g). After careful removal of the supernatant, cells were resuspended in 200 μL of ice-cold IB and centrifuged again. The supernatant was removed, and cells were resuspended in 100 μL of ice-cold IB. The cells were transferred into a black 96-well plate (Greiner Cellstar), and GFP fluorescence was measured using a fluorescence plate reader, POLARStar OPTIMA (Imgen Technologies) (excitation filter set to A-405, emission filter set to 520).
Competition Experiments.
The experiments were carried out in three replicates using the same batch of G. obscuriglobus cells. Cells grown on plates were resuspended in 450 μL of IB and aliquoted in four Eppendorf tubes, each containing 100 μL of cell suspension. One of the aliquots was not incubated with GFP and served to monitor the fluorescence background. BSA (100 μg/mL) or plasmid pET-15b DNA (100 μg/mL) were added simultaneously with GFP (10 μg/mL) to cell suspensions, and samples were continuously mixed on a vertical rotating wheel. After 90 min of incubation, the samples were immediately cooled on ice. The samples were then treated in the same way as for the time-course experiment. The reported fluorescence data were obtained by subtracting background values corresponding to autofluorescence of cells.
Detection of Proteins by Dot and Western Blots.
Total extracts of G. obscuriglobus or E. coli were resolved by SDS/PAGE (4–20% gradient gel) or dot-blotted and characterized by Western blot analysis using Alexa Fluor 680 goat anti-mouse antibody (Molecular Probes) as secondary antibody. Detection was done by using an Odyssey infrared imaging system (Li-COR).
High-Pressure Freezing and Cryosubstitution.
Bacteria cultures were high-pressure frozen with liquid nitrogen using a Leica EMPACT 2 high-pressure freezer. The frozen samples were kept and stored in a 2-mL tube containing liquid nitrogen before cryosubstitution was carried out. The frozen samples were transferred to a microfuge tube containing 0.2% uranyl acetate and 5% H2O in acetone and cryosubstituted in a Leica AFS at −85 °C for 48 h, warmed up to −50 °C at 3 °C/h, and washed 2 × 20 min in acetone at −50 °C. Cryosubstituted samples were embedded in Lowicryl HM20 resin by infiltration with 50% Lowicryl HM20 for 2 h, 75% for 2 h, and 3 × 100% for 12 h. Finally the samples in Lowicryl HM20 were polymerized under UV for 48 h at −50 °C and 48 h at 20 °C. The sample-containing Lowicryl blocks were ultrathin-sectioned using a Leica Ultracut Ultramicrotome UC61. The cut sections were placed onto formvar-carbon-coated copper grids. Sections were stained with 5% uranyl acetate in ethanol for 20 s and then with 1% lead citrate for 20 s. After immunolabeling, sections were incubated with 1% glutaraldehyde for 2 min before staining.
Immunolabeling.
Ultrathin sections of high-pressure frozen and cryosubstituted G. obscuriglobus and E. coli cells on formvar-carbon-coated copper grids were floated onto drops of Block solution containing 0.2% (wt/vol) fish skin gelatin, 0.2% (wt/vol) BSA, 200 mM glycine, and 1× PBS on a sheet of Parafilm, and treated for 1 min at 150 W in a Biowave microwave oven. For detection of GFP, the grids were then transferred onto 8 μL of primary antibody, mouse monoclonal anti-GFP antibody (Clontech, catalog no. 632381) diluted 1:25 in blocking buffer, and treated in the microwave at 150 W, for 2 min with microwave on, 2 min off, and 2 min on. The grids were then washed on drops of Block solution three times and treated in the microwave at 150 W each time for 1 min before being placed on 8 μL of goat anti-mouse IgG Fc (γ)-specific antibody conjugated with 10 nm gold (British Biocell International, catalog no. EM GAM10) diluted 1:50 in Block solution and treated in the microwave at 150 W, for 2 min with microwave on, 2 min off, and 2 min on. Then grids were washed three times in 1× PBS, each time being treated for 1 min each in the microwave at 150 W, and four times in water for 1 min each in the microwave at 150 W. The grids were examined via transmission electron microscope either not stained (for quantitation of gold particles) or stained with uranyl acetate and lead citrate after treatment with 1% glutaraldehyde. A negative control was performed, with no antibody of any type used in place of the primary antibody. Two replicates per species were performed for the immunogold labeling experiment. For detection of Gemmata MC-like protein, we used anti-gp4978 as primary antibody and protein A conjugated to 10-nm or 15-nm gold, for detection of the primary antibody. High-pressure frozen G. obscuriglobus cells preincubated with GFP were used for statistical analysis of GFP and MC-like protein distribution in different cellular compartments (Fig. S12).
HRP Cytochemistry.
Cells of G. obscuriglobus were grown, collected, and incubated with HRP-conjugated rabbit anti-mouse antibody as described for GFP uptake experiments. After 1 h of incubation, cells were washed three times with IB by repeated centrifugations in a microfuge and resuspensions of the pellet, and the final pellet was resuspended in IB containing 1.8% H2O2 and 0.1 mg/mL of DAB. After 5 min of incubation, cells were washed three times with IB, and the final pellet was high-pressure frozen for electron microscopy and processed as described above; resulting sections were not stained before examination. A negative control was used, whereby cells were incubated in IB only in place of HRP-conjugated rabbit anti-mouse antibody, with all other steps identical to above.
Supplementary Material
Acknowledgments
We thank Michael P. Rout at The Rockefeller University (RU) (New York) for donation of gp4978 antibody, arranging use of bioimaging at RU during a visit of J.A.F. to his laboratory, and valuable discussions and hospitality; Alison North of RU and members of the Rout laboratory, especially Ben Timney and Jaclyn Tetenbaum-Novatt; Rob Sullivan (Queensland Brain Institute, St. Lucia, Australia) for providing fluorophore-labeled proteins; Peter O'Donoghue and Vitalia Sagulenko for reading the manuscript and providing comments; Icon Genetics (Princeton, NJ) for providing viral vectors allowing expression of GFP in tobacco, from which the GFP used in this study was derived; and the School of Biological Sciences at The University of Queensland for providing a fluorescent plate reader. J.A.F. was supported by Australian Research Council (ARC) Discovery Project Grant DP0881485 and E.S. by a fellowship funded by the same ARC Discovery Project grant. T.G.A.L. was supported by a fellowship funded by ARC funding to B.J.C. D.P.D. was supported by the European Molecular Biology Laboratory. J.D.F. was supported by National Institutes of Health Grant F32 GM082029.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 12739.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001085107/-/DCSupplemental.
References
- 1.de Duve C. The origin of eukaryotes: A reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 2.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 3.Dacks JB, Poon PP, Field MC. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci USA. 2008;105:588–593. doi: 10.1073/pnas.0707318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- 5.Lindsay MR, et al. Cell compartmentalisation in planctomycetes: Novel types of structural organisation for the bacterial cell. Arch Microbiol. 2001;175:413–429. doi: 10.1007/s002030100280. [DOI] [PubMed] [Google Scholar]
- 6.Fuerst JA, Webb RI. Membrane-bounded nucleoid in the eubacterium Gemmatata obscuriglobus. Proc Natl Acad Sci USA. 1991;88:8184–8188. doi: 10.1073/pnas.88.18.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst JA. Intracellular compartmentation in planctomycetes. Annu Rev Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 8.Lee KC, et al. Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 2009;9:5. doi: 10.1186/1471-2180-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glöckner FO, et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchsman CA, Rocap G. Whole-genome reciprocal BLAST analysis reveals that planctomycetes do not share an unusually large number of genes with Eukarya and Archaea. Appl Environ Microbiol. 2006;72:6841–6844. doi: 10.1128/AEM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brochier C, Philippe H. Phylogeny: A non-hyperthermophilic ancestor for bacteria. Nature. 2002;417:244. doi: 10.1038/417244a. [DOI] [PubMed] [Google Scholar]
- 13.Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Teeling H, Lombardot T, Bauer M, Ludwig W, Glöckner FO. Evaluation of the phylogenetic position of the planctomycete ‘Rhodopirellula baltica’ SH 1 by means of concatenated ribosomal protein sequences, DNA-directed RNA polymerase subunit sequences and whole genome trees. Int J Syst Evol Microbiol. 2004;54:791–801. doi: 10.1099/ijs.0.02913-0. [DOI] [PubMed] [Google Scholar]
- 15.Tekniepe BL, Schmidt JM, Starr MP. Life-cycle of a budding and appendaged bacterium belonging to morphotype IV of the Blastocaulis-Planctomyces group. Curr Microbiol. 1981;5:1–6. [Google Scholar]
- 16.Lee KC, Webb RI, Fuerst JA. The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol. 2009;10:4. doi: 10.1186/1471-2121-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuerst JA. The planctomycetes: Emerging models for microbial ecology, evolution and cell biology. Microbiology. 1995;141:1493–1506. doi: 10.1099/13500872-141-7-1493. [DOI] [PubMed] [Google Scholar]
- 18.Pearson A, Budin M, Brocks JJ. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA. 2003;100:15352–15357. doi: 10.1073/pnas.2536559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieber A, Leis A, Kushmaro A, Minsky A, Medalia O. Chromatin organization and radio resistance in the bacterium Gemmata obscuriglobus. J Bacteriol. 2009;191:1439–1445. doi: 10.1128/JB.01513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.König E, Schlesner H, Hirsch P. Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol. 1984;138:200–205. [Google Scholar]
- 21.Liesack W, König H, Schlesner H, Hirsch P. Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch Microbiol. 1986;145:361–366. [Google Scholar]
- 22.Stackebrandt E, Wehmeyer U, Liesack W. 16S ribosomal RNA- and cell wall analysis of Gemmata obscuriglobus, a new member of the Order Planctomycetales. FEMS Microbiol Lett. 1986;37:289–292. [Google Scholar]
- 23.Santarella-Mellwig R, et al. The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 2010;8:e1000281. doi: 10.1371/journal.pbio.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzmann PD, Skerman VB. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie van Leeuwenhoek. 1984;50:261–268. doi: 10.1007/BF02342136. [DOI] [PubMed] [Google Scholar]
- 25.Doeven MK, Kok J, Poolman B. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Mol Microbiol. 2005;57:640–649. doi: 10.1111/j.1365-2958.2005.04698.x. [DOI] [PubMed] [Google Scholar]
- 26.Detmers FJ, et al. Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc Natl Acad Sci USA. 2000;97:12487–12492. doi: 10.1073/pnas.220308797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, Moss LG, Phillips GNJ., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 28.Jap BK, Walian PJ. Structure and functional mechanism of porins. Physiol Rev. 1996;76:1073–1088. doi: 10.1152/physrev.1996.76.4.1073. [DOI] [PubMed] [Google Scholar]
- 29.Huang JR, Craggs TD, Christodoulou J, Jackson SE. Stable intermediate states and high energy barriers in the unfolding of GFP. J Mol Biol. 2007;370:356–371. doi: 10.1016/j.jmb.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 30.Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- 31.Roth MG. Clathrin-mediated endocytosis before fluorescent proteins. Nat Rev Mol Cell Biol. 2006;7:63–68. doi: 10.1038/nrm1783. [DOI] [PubMed] [Google Scholar]
- 32.Etxeberria E, Baroja-Fernandez E, Muñoz FJ, Pozueta-Romero J. Sucrose-inducible endocytosis as a mechanism for nutrient uptake in heterotrophic plant cells. Plant Cell Physiol. 2005;46:474–481. doi: 10.1093/pcp/pci044. [DOI] [PubMed] [Google Scholar]
- 33.Mulholland J, Konopka J, Singer-Kruger B, Zerial M, Botstein D. Visualization of receptor-mediated endocytosis in yeast. Mol Biol Cell. 1999;10:799–817. doi: 10.1091/mbc.10.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuhaus EM, Almers W, Soldati T. Morphology and dynamics of the endocytic pathway in Dictyostelium discoideum. Mol Biol Cell. 2002;13:1390–1407. doi: 10.1091/mbc.01-08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavalier-tsmith T. The origin of nuclei and of eukaryotic cells. Nature. 1975;256:463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- 36.Cavalier-Smith T. Predation and eukaryote cell origins: A coevolutionary perspective. Int J Biochem Cell Biol. 2009;41:307–322. doi: 10.1016/j.biocel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Guljamow A, et al. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr Biol. 2007;17:R757–R759. doi: 10.1016/j.cub.2007.06.063. [DOI] [PubMed] [Google Scholar]
- 38.Marillonnet S, et al. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlesner H. The development of media suitable for microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst Appl Microbiol. 2004;17:135–145. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.