Abstract
A 64-year-old male patient was diagnosed with 3 consecutive non-small cell lung carcinomas (NSCLC). In the current study, we applied whole-genome gene expression analysis to control, primary and locally recurrent cancer, and supposed metastasis samples of a single patient. According to our knowledge, there are no published papers describing the gene expression profiles of a single patient's squamous cell lung cancers. As the histology and differentiation grade of the primary cancer and the supposed metastasis differed minimally, but local recurrence was poorly differentiated, molecular profiling of the samples was carried out in order to confirm or reject the hypothesis of second primary cancer. Principal component analysis of the gene expression data revealed distinction of the local recurrence. Gene ontology analysis showed no molecular characteristics of metastasis in the supposed metastasis. Gene expression analysis is valuable and can be supportive in decision-making of diagnostically complicated cancer cases.
Key Words: Local recurrence, Non-small cell lung cancer, NSCLC, Metastasis, Gene expression profile
Case Report
Clinical Data and Histology
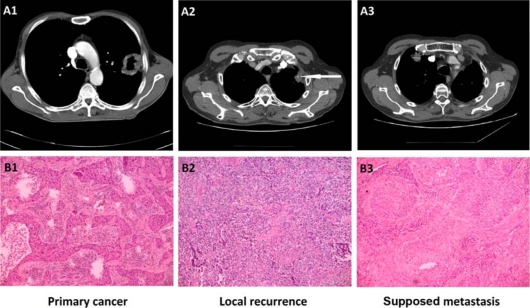
A 64-year-old male patient with a smoking history of 46 pack-years was diagnosed with peripheral tumour of the left upper lobe of the lung and underwent typical left upper lobectomy. As the upper lobe had near the primary tumour slight adhesions to the sixth segment, the upper lobe was resected anatomically and from the lower lobe atypically as one specimen. An Ethicon linear cutter was used for resection. Resection lines were macroscopically and microscopically cancer free and squamocellular cancer G2, in some areas G3 (fig. 1) was diagnosed histologically. TNM (tumour, node, metastasis) diagnosis after the specimen and resection line inspection was pT2N0M0R0 G2 stage IB.
Fig. 1.
Preoperative CT scans and histological staining of the resected tumours (HE, ×100).
34 months later, on the sixth postoperative visit, a recurrent tumour of the left lung was diagnosed radiologically (CT scan). After removal of the cancer, the previous resection line in the middle of the tumour (fig. 1A2 arrow) could be seen, which clearly indicates local recurrence. Pleural invasion was present in the apical part of the removed parietal pleura. The recurrence had anaplastic cancer features with a very low differentiation grade (G4; fig. 1B2) and differed from the primary tumour mainly in its lack of tissue pattern and in immunohistochemical features (CK5/6 negative). Radiation therapy with 50 Gy was administered to the pleural contact site. Polychemotherapy with cisplatin, etoposide and Vepesid was applied after the second surgical treatment.
15 months after the second operation, a metastasis-like tumour of the right upper lobe was diagnosed radiologically (fig. 1A3). Squamocellular cancer G2 with keratinization in some areas was histologically confirmed (fig. 1B3).
Tumour histology and stage were estimated according to the WHO/IASLC Histological Classification of Lung and Pleural Tumours [1] and TNM staging according to the UICC (International Union Against Cancer) classifications, respectively [2]. Control samples for the gene expression analysis were obtained from the same cancer patient at a site distant from the tumour and were approved as control samples by the pathologist. Histological evaluation was carried out on formalin-fixed, paraffin-embedded tumour specimens.
Analysis of Gene Expression and Ontology
Postsurgical tissue specimens for gene expression analysis were immediately cut to an appropriate size and submerged in RNAlaterh (Ambion, Catalog No. AM7021) to inhibit RNA degradation. The samples were stored at −80°C until further processing. Total cellular RNA from tissue specimens of 50 mg was extracted and purified using Ribopure Kit (Ambion, Catalog No. AM1924) according to the manufacturer's instructions. For tissue disruption, the IKA Ultra-Turrax T8 homogenisator was used. RNA quantity and quality were assessed using the NanoDrop-1000 spectrophotometer and the Agilent Bioanalyzer lab-on-a-chip technology (Agilent RNA 6000 Nano Kit, catalog No. 5067-1511) respectively. The Illuminah TotalPrep RNA Amplification Kit (Ambion, Catalog No. AMIL1971) was used for RNA amplification and labelling. Amplifications were carried out according to the manufacturer's instructions using 300 ng of total RNA as a template. The Illuminah BeadChip platform and the corresponding whole-genome HumanHT-12 v3 Expression BeadChip were used for the gene expression analysis. Experiments were carried out according to the manufacturer's instructions. Illumina internal controls and BeadStudio software were used for data consistency and quality control of the hybridization raw data. Further data analysis was performed with R-software and Bioconductor package. Gene ontology (GO) enrichments were calculated using the g:Profiler web toolkit [3]. The GO analysis was performed with the gene lists representing at least 2-fold change of expression between the samples.
As the gene expression difference between the primary control (obtained during the primary resection) and the recurrent cancer control (obtained during the supposed metastasis resection) samples was minimal, the further gene analyses were based on the comparison of samples of interest with the primary tumour control gene expression values. At least 2-fold change of expression between the samples was applied. The highest number of up- and downregulated genes (2,577 genes altogether) was identified between the locally recurrent cancer and the control sample (table 1). Surprisingly, the number of deregulated genes in the supposed metastasis sample (1,710) was substantially smaller than in primary cancer (2,095).
Table 1.
The up- and downregulated genes between different samples analysed
| Primary cancer | Recurrent cancer | Supposed metastasis | Primary control | |
|---|---|---|---|---|
| Primary cancer | ↑ 1,079 | ↑ 676 | ↑ 951 | |
| Recurrent cancer | ↓ 1,026 | ↑ 1,109 | ↑ 1,274 | |
| Supposed metastasis | ↓ 769 | ↓ 951 | ↑ 822 | |
| Primary control | ↓ 1,144 | ↓ 1,303 | ↓ 888 |
The GO analysis of 676 up- and 769 downregulated genes (at least 2-fold difference in expression was applied) between the supposed metastasis and primary cancer samples were performed in order to uncover the biological processes hindered within.
As a result of the analysis, the biological processes associated with system and organ development, adhesion, oxidative stress, homeostasis as well as ossification came forth in the supposed metastasis sample. The known biological hallmarks of metastasis-associated processes like dedifferentiation, extensive metabolism, DNA synthesis and inflammation were not noted. GO analysis of genes downregulated in supposed metastasis revealed deactivation of processes like cellular localization, cytoskeleton and organelle organization, glucose catabolism and locomotion indicative of the more active nature of primary cancer compared with metastasis.
Altogether these gene expression-based GO analyses did not support the hypothesis of metastasis. The molecular phenotype of the supposed metastasis was only minimally different from the primary cancer sample and the GO analysis revealed no activation of processes like matrix remodelling, metastasis, dedifferentiation, mitosis, etc., which are characteristic of metastatic cancers. Therefore, the evolvement of new primary cancer instead of metastasis is more probable.
Correlations and Principal Component Analysis
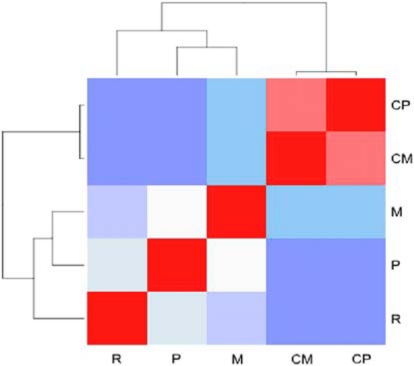
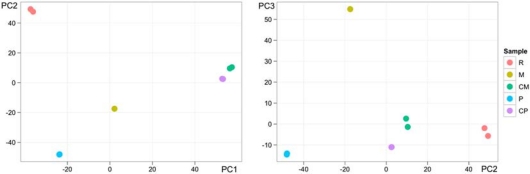
To visualize the gene expression data, a correlation heatmap (fig. 2) and principal component analysis (PCA) were performed (fig. 3). The correlation analysis as well as PCA revealed similar behaviour of the genes of the primary tumour control and the supposed metastasis control samples although there were 2 chemotherapy treatments between the sample collections. This indicates relatively minor changes in the gene expression profiles of histologically normal lung tissues collected prior to and after chemotherapy treatment. In the PC1/PC2 analysis, the recurrent cancer samples were most distant from controls and the supposed metastasis sample located in the middle section. Interestingly, the difference that can be measured as distance in the PCA between the control and the supposed metastasis samples is significantly smaller than distances between the control and the primary cancer samples as well as between the control samples and the recurrent cancer sample. Thus, according to the molecular profiles, one could suggest to reevaluate the status of the metastasis to a second primary cancer that should also be less aggressive than a primary and locally revived one.
Fig. 2.
Correlation heatmap representing the behaviour of gene expression changes between the samples. P = Primary cancer; R = recurrent cancer; M = supposed metastasis; CP = control sample obtained during the primary resection; CM = control sample obtained during the metastasis resection. The intensity of the red colour represents the positive gene expression correlation between the samples and the intensity of blue represents the discorrelation of the gene expression.
Fig. 3.
PCA of primary and recurrent cancer, supposed metastasis, and control samples of primary cancer and of supposed metastasis gene expression data. There were replicate array data available for all except the supposed metastasis sample. R = Recurrent cancer; M = supposed metastasis; CM = control sample obtained during the supposed metastasis resection; P = primary cancer; CP = control sample obtained during the primary resection.
The results of PC2/PC3 analysis show that the supposed metastasis sample has the most distinct pattern.
Discussion
Approximately 7% of all cancer patients experience a new primary cancer later in life [4]. In the current case we have analysed and compared the histology and whole-genome mRNA expression levels within primary, recurrent and supposed metastatic lung cancer specimens. PCA and GO analysis were applied to reveal the molecular variance and activities of the tissues.
As the histological patterns, clinical manifestation and performance of the patient were unexpected, the strong clinical suggestion of second primary disease instead of metastasis emerged. Supposed metastases and primary tumour had relatively similar histological differentiation. Clinically, the supposed metastasis had no common histological signs of previous chemotherapy like vacuolization, increased amount of apoptotic cells or inflammatory infiltration but had clear upregulation of genes as a response to a chemical stimulus which can be explained with previous chemotherapy (see online suppl. table 1, www.karger.com/doi/10.1159/000318010). The fact that supposed metastasis followed by chemotherapy was more similar to the primary tumour makes second primary cancer more likely.
As a result of PCA of gene expression data, a clear distinction of recurrent cancer was seen, whereas similarities between supposed metastases and the primary tumour were noted. The recurrence sample was histologically classified as undifferentiated and the clear distinction was also seen in PCA. In comparison of primary tumour and local recurrence samples, upregulation of processes like neural development, axonogenesis, neural degeneration processes and different developmental processes was seen in the recurrence sample (online suppl. table 1). The difference between the primary tumour and local recurrence could be explained by micrometastasis and the cancer stem cell phenomenon [5].
In the current case, we have shown the correlation of gene expression-based analysis with the histology of different control and cancer samples of one patient. The gene expression profiles supported the hypothesis of second primary squamous cell cancer of the lung. For 1 year and 10 months, since the third operation, the patient has had a good performance and recurrence-free life as controlled by CT scan, which is uncommon in everyday practice. Survival data support the second primary cancer hypothesis. The exclusion of the metastasis possibility is crucial in choosing the optimal treatment strategy and also results in a better prognosis of the patient.
Supplementary Material
The up- and downregulated genes between different samples analysed
Acknowledgements
We would like to acknowledge Viljo Soo for technical assistance.
This study was supported by Targeted Financing from the Estonian Ministry of Education and Research (SF0180142s08), an Estonian Science Foundation grant ETF6465, an EU FP7 grant ECOGENE (#205419, EBC), and by the EU via the European Regional Development Fund grant to the Centre of Excellence in Genomics, Estonian Biocentre and University of Tartu.
Footnotes
Tõnu Vooder and Kristjan Välk contributed equally to this work and share the first authorship.
References
- 1.Brambilla E, et al. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059–1068. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- 2.Mountain CF. The international system for staging lung cancer. Semin Surg Oncol. 2000;18:106–115. doi: 10.1002/(sici)1098-2388(200003)18:2<106::aid-ssu4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Reimand J, et al. g:Profiler – a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engeland A, et al. Use of multiple primary cancers to indicate associations between smoking and cancer incidence: an analysis of 500,000 cancer cases diagnosed in Norway during 1953-93. Int J Cancer. 1997;70:401–407. doi: 10.1002/(sici)1097-0215(19970207)70:4<401::aid-ijc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene. 2001 doi: 10.1038/onc.2010.207. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The up- and downregulated genes between different samples analysed