Abstract
Microarray-based prediction of clinical endpoints may be performed using either a one-color approach reflecting mRNA abundance in absolute intensity values or a two-color approach yielding ratios of fluorescent intensities. In this study, as part of the MAQC-II project, we systematically compared the classification performance resulting from one- and two-color gene-expression profiles of 478 neuroblastoma samples. In total, 196 classification models were applied to these measurements to predict four clinical endpoints, and classification performances were compared in terms of accuracy, area under the curve, Matthews correlation coefficient and root mean-squared error. Whereas prediction performance varied with distinct clinical endpoints and classification models, equivalent performance metrics were observed for one- and two-color measurements in both internal and external validation. Furthermore, overlap of selected signature genes correlated inversely with endpoint prediction difficulty. In summary, our data strongly substantiate that the choice of platform is not a primary factor for successful gene expression based-prediction of clinical endpoints.
Keywords: MAQC-II, microarray, one-color, two-color, neuroblastoma, classification of clinical endpoints
Introduction
This study is part of the second phase of the microarray quality control (MAQC) project (MAQC-II).1 The main design of MAQC-II is laid out in Shi et al.1 (see also project summary in Supplemental Material and Supplemental Table S1). This study aimed at comparing the performance of classifiers for clinical endpoints that were trained on one-color or two-color microarray data.
Prediction of clinical endpoints from microarray-based gene-expression measurements can be performed using either of two experimental procedures: (1) a one-color approach, in which a single RNA sample is labeled with a fluorophore (such as phycoerythrin, cyanine-3 (Cy-3) or cyanine-5 (Cy-5)) and hybridized alone to a microarray, or (2) a two-color strategy, in which two samples (usually a sample and a reference) are labelled with different fluorophores (for example, Cy-3 and Cy-5) and are then hybridized together on a single microarray. The resulting data are fundamentally different: while two-color arrays yield ratios of fluorescence intensities (that is, sample fluorescence/reference fluorescence), one-color arrays result in absolute fluorescence intensities, which are assumed to be monotonically (if not linearly) related to the abundance of mRNA species complementary to the probes on the array.
There are pros and cons for both systems. While simplicity and flexibility of the experimental design might favor one-color analyses in a clinical setting, two-color measurements are often believed to be more robust due to the internal reference, which should cancel out biases related to single array measurements.2, 3, 4 However, two-color approaches may be affected by dye bias5 and, although dye influence can be corrected for by performing dye-flipped replicate hybridizations, this strategy substantially augments experimental costs. Therefore, most researchers refrain from following such an approach. Moreover, considerable logistic effort has to be undertaken for two-color analysis, since reference RNA (or cDNA) must be available in constant quality throughout the course of the measurements, even if the study may take years to complete. It is also not easy to choose an appropriate reference for each application, and there have been controversial discussions on the use of reference RNAs with divergent similarity to the sample RNAs of the study.6, 7 While some laboratories stick to mixtures of RNA prepared from various human tissues (far reference), it may be sometimes more appropriate to choose a pool of related cell lines or tissue samples as a source of reference RNA (near reference), which represent an average over all samples of the study.
Despite these essential differences, both one-color and two-color microarray analyses have been widely used with similarly convincing outcome. However, while consistency and correlation of primary measurements obtained with one- and two-color platforms in terms of identifying differentially expressed genes have been addressed previously,8, 9 to the best of our knowledge, a systematic comparison of the classification performance of these two different techniques has not been reported. Therefore, it is still unclear if either of these platforms is better suited for clinical applications that aim at predicting the endpoint of a disease. Consequently, preference of either experimental system is at present mostly motivated by criteria that are not primarily related to the experimental question, such as the local availability of a specific system, the financial effort of the intended survey, or presumed but unconfirmed belief of the superiority of either system.
In this study, we systematically compared for the first time the power of one-color and two-color measurements to predict clinical endpoints. To this end, we generated gene-expression profiles for 478 neuroblastoma tumor samples using both one-color and two-color microarrays of the same manufacturer (Agilent Technologies, Waldbronn, Germany). We have used neuroblastoma here as a model system for our primary question. While the two-channel data were already part of the main MAQC-II project, the one-channel data were generated exclusively for this study using RNA from the very same tumor samples. Subsequently, eight different classification algorithms were combined with other processing steps (for example, normalization, feature selection, etc.) and applied to predict four different clinical endpoints for these data sets, resulting in a total of 196 different models. Classification performance of these models was then compared between one-color and two-color data by area under the receiver operating-characteristics curve (AUC), Matthews correlation coefficient (MCC), accuracy and root mean squared error (RMSE) as defined by MAQC.1 Moreover, for models that were trained to choose only a small number of features for classification, the overlap of the resulting gene signatures was computed.
Materials and Methods
Sample and RNA preparation
This study comprised a total of 478 different neuroblastoma tumor samples for which both one-color and two-color gene-expression profiles were generated. Two-color profiles were the same as utilized for the MAQC-II (a total of 499 samples, subdivided in training set (n=246) and test set (n=253). For 478 of these samples, single-color profiles were generated according to a protocol of the same manufacturer (training set: n=244 (99.2% overlap) and test set: n=234 (92.9% overlap). Clinical co-variates of the patient cohort are given in Supplementary Tables S2 and S3.
Sample and RNA preparation was performed as described.10 In summary, tumor samples were checked by a pathologist before RNA isolation. Subsequently, samples with at least 60% tumor content were utilized and total RNA was isolated from ∼50 mg of snap-frozen neuroblastoma tissue obtained before chemotherapeutic treatment. After homogenization of tumor tissue by using the FastPrep FP120 cell disruptor (Qbiogene, Carlsbad, CA, USA) total RNA was isolated using the TRIzol reagent (Invitrogen, Karlsruhe, Germany). Integrity of the isolated RNA was assessed using the 2100 Bioanalyzer (Agilent Technologies) and only samples with an RNA integrity number of at least 7.5 were considered for further processing.
Neuroblastoma two-color gene-expression profiles
All two-color gene-expression of the MAQC-II training set were generated using a customized 2 × 11K neuroblastoma-related microarray.10 Furthermore, 20 patients of the MAQC-II two-color validation set were also profiled utilizing this microarray. Two-color profiles of the remaining patients of the MAQC-II validation set were performed using a slightly revised version of the 2 × 11K microarray. For this version V2.0 of the array, 100 oligonucleotide probes of the original design were removed due to consistent low expression values (near background) observed in the training set profiles, and 200 novel oligonucleotide probes were added. These minor modifications of the microarray design resulted in a total of 9986 probes present on both versions of the 2 × 11K microarray. The experimental protocol did not differ between both sets, and gene-expression profiles were performed as described.10 In summary, 1 μg of total RNA of each tumor sample was linearly amplified and labeled with Cy3 and Cy5, respectively, using Agilent's Low-RNA-Input LinearAmp Kit. Then, 500 ng of labeled cRNA was hybridized together with 500 ng of reverse color Cy-labeled cRNA of a total RNA pool of 100 neuroblastoma tumor samples on the 2 × 11K arrays using Agilent's in situ Hyb-Kit Plus following the manufacturer's protocol. Hybridization was performed for 17 h at 60 °C in a rotating hyb oven at a speed of 4 rounds per minute (r.p.m.). Washing was performed at room temperature following the manufacturer's protocol. After scanning, the resulting TIFF-images were processed using Agilent's Feature Extraction software (Versions 7.5–9.5.1).
Neuroblastoma single-color profiles
Single-color gene-expression profiles were generated for 478/499 neuroblastoma samples of the MAQC-II dual-color training and validation set (training set 244/246; validation set 234/253). For the remaining 21 samples no single-color data was available due to either shortage of tumor material of these patients (n=15), poor experimental quality of the generated single-color profiles (n=5) or correlation of one single-color profile to two different dual-color profiles for the one patient profiled with both versions of the 2 × 11K microarrays. Single-Color gene-expression profiles were generated using customized 4 × 44K oligonucleotide microarrays produced by Agilent Technologies (Palo Alto, CA, USA). These 4 × 44K microarrays included all probes represented by Agilent's Whole Human Genome Oligo Microarray and all probes of the version V2.0 of the 2 × 11K customized microarray that were not present in the former probe set. Labeling and hybridization was performed following the manufacturer's protocol. In brief, 1 μg total of tumor RNA was linearily amplified and labeled with Cy3 using Agilent's one-color Quick Amp Labeling Kit following the instructions of the protocol. Then, 1650 ng of Cy3-labeled cRNA was hybridized on the 4 × 44K arrays using Agilent's High-RPM Gene Expression Hyb Kit. Hybridization was performed for 17 h at 65 °C in a rotating hyb oven at 10 r.p.m. according to the company's recommendations. After washing and scanning, resulting TIFF-images were processed using Agilent's Feature Extraction software Version 9.5.1.
The expression profiling data are available within the in-house and MIAME compliant database iCHIP of the DKFZ (http://www.ichip.de). This comprises both the one-color as well as the two-color Agilent gene expression studies and includes raw as well as processed data. Comprehensive and actual patient information is associated with the related experiments, SOPs and protocols for treatment procedures are included according to the MIAME standard. In addition, data will be made available through Arrayexpress (Accession E-TABM-38, E-MTAB-161, E-MTAB-179), and through the MAQC web site (http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/).
Statistical analysis
Classifiers were trained as described.1 Classification was based on microarray data only, clinical data have not been taken into account. Briefly, data were normalized using the quantile algorithm from limma.11 Normalization by the vsn algorithm yielded similar results (data not shown). For normalization of the validation set, the training set was used as reference to ensure that data were on equal scales. Further details are given in Supplemental Methods.
As performance measures, accuracy (Acc), sensitivity (Sen), specificity (Spec), Matthews correlation coefficient (MCC), root mean squared error and AUC of a receiver-operating characteristics curve were used. For prediction, a classifier was trained on all available training data using variables that have been selected in 65% of all cross-validation runs;10 where no features were selected (endpoint N only), this threshold was lowered to 25%.
Sensitivity (SENS) and specificity (SPEC) are given by:
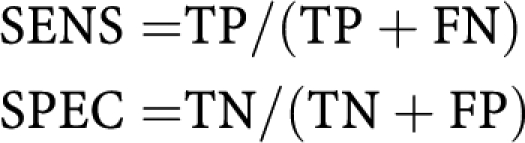 |
Where TP, TN, FP and FN are the number of true-positive, true-negative, false-positive and false-negative samples, respectively. Accuracy is calculated as
The Matthews Correlation Coefficient (MCC) is
Root mean squared error is defined as:
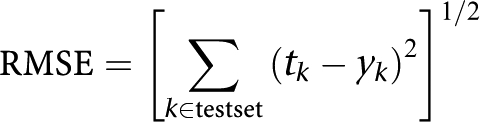 |
where tk are the prediction scores (binary or continuous), the yk are the true class of sample k and summation is over all samples k in the testset.
The area under an ROC curve12 is given as:13
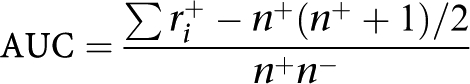 |
where ri+ is the rank of the ith positive sample, and n+ and n− are the number of samples in the positive and negative group, respectively.
Results
Data sets for comparative analysis
Two-color gene expression profiles for a total of 499 neuroblastoma (NB) tumor samples were provided to the MAQC-II consortium as a benchmark set for evaluation of classification workflows.1 These profiles were generated as dye-flipped replicates using a 2 × 11K oligonucleotide microarray manufactured by Agilent Technologies (Santa Clara, CA, USA).
To allow for an in-depth comparison of the classification performance of one-color vs two-color microarray measurements, we additionally performed one-color gene-expression profiling for 478/499 of the afore-mentioned neuroblastoma tumor samples. Following the basic design of the MAQC-II study, in which two-color profiles of neuroblastoma samples were divided into a training set (n=246) and an approximately equal-size-blinded validation set (n=253), one-color profiles of the same tumor samples were generated for 244/246 MAQC-II NB two-color training set samples (99% overlap) and 234/253 MAQC-II NB validation set samples (92.5% overlap). The reasons for lacking one-color profiles were: (i) shortage of RNA or tumor material (n=15), (ii) poor experimental quality for one-color profile (n=5) and (iii) correlation of one one-color profile to two different two-color profiles for one patient of the validation set who had been profiled with different versions of the 2 × 11K microarray (see Methods and Supplementary Tables S2 and S3). For the comparative analysis, only those 478 samples that had been profiled with both platforms were considered. It is interesting to note that Agilent Technologies no longer manufactured the 2 × 11K design used for generation of the two-color profiles. For this reason one-color profile had to be generated utilizing a 4 × 44K oligonucleotide microarray manufactured by the same company. This higher-density 4 × 44K microarray comprised all probes from the 2 × 11K oligonucleotide microarray. Thus, data from the resulting gene-expression profiles of both platforms was restricted to a common set of 9986 probes represented by both designs to allow for a reasonable comparison of classification performance of one- or two-color measurements. It is interesting to note that some parameters of the labelling and hybridization procedure had to be adjusted for the higher-density 4 × 44K format thus resulting in slight differences between the one-color and two-color experimental protocols (details are indicated in the Methods section and summarized in Supplementary Table S4).
Comparison of cross-validated classification performances (training set)
Following the basic approach of the MAQC-II study, classifiers from both one-color and two-color gene-expression measurements sets were generated to predict all four clinical endpoints that had been defined for the neuroblastoma data set (endpoints J, K, L and M, Table 1). To allow for a comprehensive comparison of the prediction performance of both experimental systems, eight different algorithms were applied to both data sets, including discriminant analysis, generalized linear models, logistic regression (LR), prediction analysis of microarrays (PAM14), partial least squares (PLS), partition tree (PT), radial basis machine (RBM) and support vector machines in combination with recursive feature elimination (SVM+RFE15). Combination of these algorithms with other processing steps (for example, normalization, feature selection, etc.) resulted in a total of 196 different classification models that were applied to both platforms and were compared with respect to classification performance (all results are summarized in Supplementary Table S5).
Table 1. Number of samples available for training and validation, and number of positive or negative cases for each clinical endpoint.
| Endpoint code | Endpoint description | Training set | Validation set | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of samples | Positives (P) | Negatives (N) | P/N Ratio | Number of samples | Positives (P) | Negatives (N) | P/N Ratio | ||
| J | OS_MO—overall survival milestone outcome (OS, 900-day cutoff) | 236 | 22 | 214 | 0.10 | 161 | 37 | 124 | 0.30 |
| K | EFS_MO—event-free survival milestone outcome (EFS, 900-day cutoff) | 237 | 59 | 178 | 0.33 | 174 | 78 | 96 | 0.80 |
| L | NEP_S—newly established parameter S. *The actual class label is the sex of the patient and unaware to data analysis teams. Used as a ‘positive' control endpoint. | 244 | 144 | 100 | 1.44 | 231 | 133 | 98 | 1.36 |
| M | NEP_R—newly established parameter R (NEP_R). **The actual class label is randomly assigned and unaware to the data analysis teams. Used as a ‘negative' control endpoint. | 244 | 143 | 101 | 1.41 | 231 | 132 | 99 | 1.33 |
Lower number of samples available for endpoints EFS_MO and OS_MO is because of missing values for these endpoints.
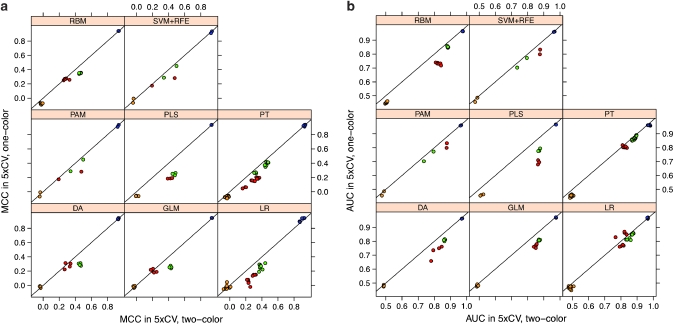
Cross-validated classification performance of both platforms was measured by MCC, a balanced measure composed of sensitivity and specificity,16 AUC, RMSE and accuracy. Then, values of these parameters for classifiers built from one-color data were plotted against values observed for classifiers built from two-color data (Figure 1, Supplementary Figure 1). As indicated in Figure 1, cross-validated predictive performance varied primarily between the different endpoints. This result was expected as two endpoints were introduced as positive (patients' sex, endpoint L) and negative (random selection, endpoint M) control endpoints, respectively. These endpoints were further characterized by a balanced group distribution of the samples (Table 1). For the positive endpoint L, nearly perfect classification performance was reached (MCC: 0.87–0.95, mean MCC: 0.93; AUC: 0.96–0.98, mean AUC: 0.97) while results for the negative endpoint M were close to random prediction (MCC: −0.09–0.11, mean MCC: −0.04; AUC: 0.44–0.57, mean AUC: 0.48). In contrast, endpoints J (overall survival) and K (event-free survival) were true clinical endpoints with medium classification difficulty.1 Results for endpoint J ranged from MCC values of −0.02 to 0.41 (mean 0.19) and from AUC values of 0.76–0.88 (mean 0.84), respectively. Results for endpoint K yielded MCC values ranging from 0.14 to 0.50 (mean 0.37) and AUC values of 0.70–0.90 (mean 0.85). As shown in Table 1, the latter two endpoints present highly unbalanced distribution among the samples.
Figure 1.
Comparison of results classification from one-color vs two-color training data using 10 iterations of 5 × cross validation. The data shown are the mean results from 10 independent runs. A set of 9986 common probes and 244 samples was used to train the classifiers. For each endpoint, eight different classification methods were applied, namely discriminant analysis, general linear model selection, logistic regression, partial least square (PLS), partition tree, radial basis machine, prediction analysis of microarrays (PAM) and support vector machines plus recursive feature elimination (SVM+RFE) were selected for prediction. Values for MCC (a) and AUC (b) of the prediction results using one-color and two-color data are plotted against each other. Endpoints are coded by color: OS_MO (endpoint J), red; EFS_MO (endpoint K), green; patient's sex (endpoint L), blue; random classes (endpoint M), orange.
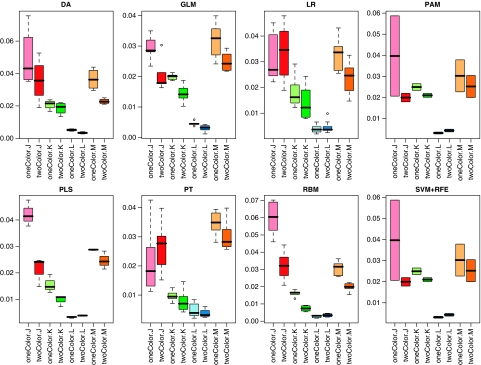
Second, as shown in Figure 1, minor differences in classification performance were observed between the different classification models applied to the data sets. However, as the most important finding with respect to the objective of our study, no substantial difference in classification performance was observed between the two different experimental platforms as indicated by a remarkably high correlation between one-color and two-color performance observed for all four classification criteria (Figure 1, Supplementary Figure 1). Furthermore, to assess the robustness of predictive models from one-color and two-color measurements, the variance of the performance was estimated by calculating the s.d. of the performance metrics MCC, AUC, RMSE and accuracy from repeated runs of the algorithms. As expected, s.d. depended on both the clinical endpoint and the classification algorithm used for prediction. However, apart from these effects, we also observed that the s.d. of performance metrics was consistently lower when classifiers were trained on two-color data. This observation is illustrated in Figure 2, in which s.d. of AUC are indicated for different endpoints for each of the classification algorithms. The only exception is classifiers for endpoint L using LR, PAM, PLS, RBM or SVM+RFE as classification algorithms (Figure 2). Thus, the variability of the prediction was in general lower when classifiers were trained from two-color data, whereas the average estimate of predictive performance was not different (Figure 1).
Figure 2.
Standard deviation of AUC for different endpoints for each of the classification algorithms as assessed from repeated runs of the cross-validation.
Comparison of classification performance on the validation set
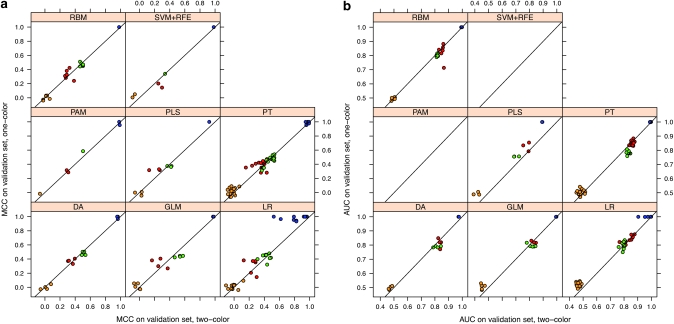
After internal validation, classifiers of each algorithm were trained on the entire training set using optimized parameters and—if applicable—a set of features frequently selected in cross-validation. Subsequently, the resulting final classifiers were applied to predict class labels for the validation set of samples for all endpoints. Again, classification performance was assessed by calculating MCC, AUC, RMSE and accuracy.
In line with the cross-validated results observed in the training set data, classification performance differed most prominently with respect to the clinical endpoint and with respect to the applied classification algorithm. In contrast, equivalent overall performance metrics resulted from one-color and two-color data measurements, thereby substantiating that this factor does not significantly influence the classification performance. As an exception, a slight tendency towards higher prediction performance with one-color data was noted for classifiers built on PLS and LR, for which this effect was detectable for all endpoints (Figure 3, Supplementary Figure 2). Apart from that, we also observed marginally higher performance values for models built on PT and one-color data for the negative endpoint M. However, as illustrated in Supplementary Figure 1, these effects were clearly restricted to these models and the combination of specific models with certain endpoints, respectively. Thus, the putative higher prediction performance of the one-color data was not a general finding but rather a specific effect occurring only with these algorithms, or with a combination of specific algorithms and the chosen feature selection method.
Figure 3.
Comparison of classification results from one-color vs two-color data on an independent validation set. A set of 9986 common probes and 244 samples was used to train the classifiers. Eight classification methods, discriminant analysis, general linear model selection, logistic regression, partial least square (PLS), partition tree, radial basis machine, prediction analysis of microarrays (PAM) and support vector machines plus recursive feature elimination (SVM+RFE) were selected for prediction. MCC (a) and AUC (b) of the prediction results using one-color and two-color data are plotted against each other. Endpoints are coded by color: OS_MO (endpoint J), red; EFS_MO (endpoint K), green; patient's sex (endpoint L), blue; random classes (endpoint M), orange.
Overlap of resulting gene signatures
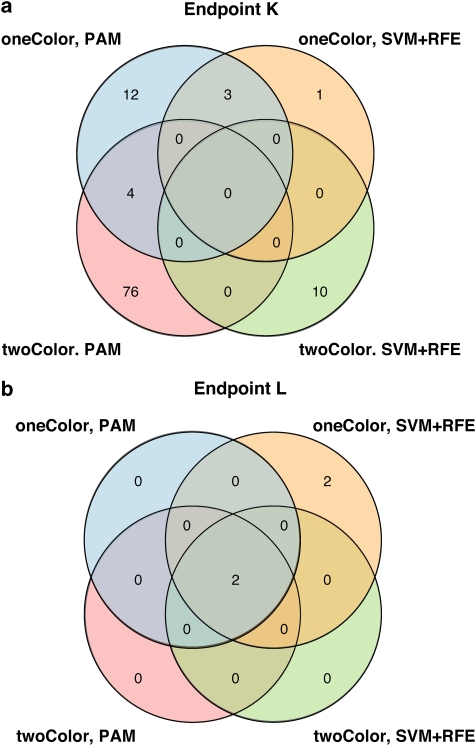
Finally, models that were built on either PAM or SVM-RFE were also compared with respect to their resulting gene signatures. As a general observation, classifiers derived from one-color data tended to include fewer features than those from two-color data in our study (Supplementary Table S5). Apart from that, it was noted that features selected in training from one- or two-color data showed only little overlap (Figure 4a) for the clinical endpoints. Interestingly, we found that the degree of overlap decreased with increasing difficulty of the predicted endpoint. Hence, for the positive endpoint L (patients' sex), three of four approaches concurrently selected a total of two identical features (Figure 4b). Moreover, the four features selected by the remaining approach also comprised these two features (overlap 50%, Figure 4b). Intriguingly, the features recurrently selected for classification of this endpoint, X (inactive)-specific transcript (XIST) and ribosomal protein S4, Y-linked 1 (RPS4Y1), are known to be expressed in a highly sex-specific manner. XIST is a non-coding RNA involved in silencing of the second X chromosome in female cells,17 and RPS4Y1 is a ribosomal protein encoded by Y chromosome. Thus, with respect to this endpoint, both systems not only performed equivalently well with only a very small number of features, but also demonstrated similar potential to identify features with mechanistic relevance, as indicated by the biological function of the selected candidates.
Figure 4.
Overlap between features recurrently selected for classification by either PAM or RFE. (a) Features selected for classification of EFS_MO (endpoint K). (b) Features selected for prediction of patient's sex (endpoint L).
Discussion
In this study, we conducted a systematic comparison of the classification performance resulting from one- and two-color gene-expression measurements of a considerable number of 478 clinical samples. From these, a large number of classification models was generated to predict four endpoints defined for the MAQC-II study and performance was assessed by the metrics MCC, AUC, RMSE and accuracy both for internal and external validation. Cumulatively, our analysis indicates a largely equivalent overall performance of both platforms, thereby suggesting that the choice of a one-color or of a two-color platform does not need to be a primary factor in decisions regarding experimental microarray design in classification studies.
To allow for warranted conclusions from our calculations, gene expression profiling was performed using the one- and two-color platform from the same manufacturer, Agilent Technologies. Furthermore, since two different microarray designs (2 × 11K vs 4 × 44K) had to be used for this study, predictive models were built on a set of roughly 10 000 probes common to both designs. Although every effort was made to conduct the analyses comprised in this study as similar as possible, some minor differences in the experimental procedures were inevitable. First, because of a change to a higher density microarray design (4 × 44k) for the one-color profiles, the protocols differed slightly, mainly with respect to the hybridization temperature (60 °C for two-color, 65 °C for one-color, see Supplementary Table 3). Second, two-color samples were measured as dye-swap replicates yielding two profiles per patient, whereas single-color arrays where performed without replications. A third factor was the standardization of the two-color data relative to a signal from a reference channel, which is intrinsic to the method. Despite these differences, classification performance was observed to be highly comparable as measured by repeated internal 5 × cross validation, thereby underlining the validity of our approach. A marginally higher robustness of classifiers trained from two-color data was suspected based on the lower variance of performance measures observed with two-color measurements. However, considering the fact that two-color data were generated as dye-flipped replicates (resulting in two measurements per patient), this observation can be explained by the higher degree of replication in the two-color data set. Alternatively, measurement against an internal reference on the same chip could have resulted in a stabilization of the two-color data.
Similarly, predictive performance on an independent validation also was comparable between one-color and two-color measurements. However, a slightly higher performance of classifiers trained on one-color data was observable for three of the eight classification algorithms used, namely PLS, LR and for endpoint M also PT. This finding could point to a tendency of overfitting of these classifiers when trained on two-color data. However, it should be noted that a higher classification performance of the negative endpoint M (as observed for PT built on one-color data) rather indicates a limited suitability of that classifier. Alternatively, the observation of higher performance of some models with one-color data might be related to the feature selection algorithms used in combination with these algorithms. As the number of features selected for classification was generally lower for classifiers generated from one-color data, it can be hypothesized that one-color gene-expression measurements were less noisy and more stable as compared to log-ratio data from two color arrays. However, since this effect was neither visible with other classification algorithms nor in the internal validation by cross-validation, we conclude that the observed differences in performance do not indicate a general effect that results from the choice of the platform.
With respect to the varying degrees of overlap observed for the selected signature genes, our finding that the potential overlap of features decreases with increasing difficulty of the predicted endpoint is also in line with results of the MAQC-II main study, in which it was shown that feature list stability is inversely correlated with endpoint difficulty.1 However, the higher degree of replication underlying the two-color data set clearly influenced feature selection in our study, thereby making it difficult to draw definite conclusions from this analysis. In addition, overlapping genes do not appear to be a true indicator of performance, since it has been shown clearly that differing sets of genes may be derived from a single data set and perform equally well in terms of classification.18 Yet, the fact that both platforms identified features with biological relevance with high overlap for an easily predictable endpoint (L) appears to further underscore the equivalent potential of both approaches.
Neuroblastoma was used in this study as a model system to address our primary question, the difference or similarity in performance of classifiers trained on one-color or two-color microarray-based gene expression measurements. While similar studies on other tumor entities have not been performed so far, it is reasonable to believe that the results can be extrapolated to other entities with similar approaches, that is, prediction of a binary clinical endpoint based on high-dimensional gene expression measurements, provided that microarray-based classification allows at all for such prediction.
Acknowledgments
This study was supported by grants 01GS0895 (MF) and 01GS0896 (BB) from the German Federal Ministry of Research and Education (BMBF) through the National Genome Research Network. BB was further supported by a grant from the European Commission (LSHC-CT-2006-037260). Furthermore, this study was supported by the Deutsche Krebshilfe (grant 50-2719-Fi1 to MF) and by the Auerbach Stiftung.
Glossary
- AUC
area under the receiver operating-characteristics curve
- DAT
data analysis team
- DEG
differentially expressed genes
- LR
logistic regression
- MAQC
microarray quality control
- MAQC-I
microarray quality control phase I on technical performance
- MAQC-II
microarray quality control phase II on predictive modeling
- MCC
Matthews correlation coefficient
- NB
neuroblastoma
- PAM
prediction analysis of microarrays
- PLS
partial least square
- PT
partition tree
- RBWG
regulatory biostatistics working group of MAQC-II
- RBM
radial basis machine
- RFE
recursive feature elimination
- RMSE
root mean-squared error
- SVM
support vector machines
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Disclaimer
The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Supplementary Material
References
- The MicroArray Quality Control (MAQC) Consortium Shi L, Campbell G, Jones W, Walker SJ, Campagne F, Pusztai L, et al. The MAQC-II Project: a comprehensive study of common practices for the development and validation of microarray-based predictive models Nat Biotechnol(submitted). [DOI] [PMC free article] [PubMed]
- Chen Y, Dougherty ER, Bittner ML. Ratio-based decisions and the quantitative analysis of cDNA microarray images. J Biomed Optics. 1997;2:364–374. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21 (1 Suppl:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Shi L, Tong W, Su Z, Han T, Han J, Puri RK, et al. Microarray scanner calibration curves: characteristics and implications. BMC Bioinformatics. 2005;6 (Suppl 2:S11. doi: 10.1186/1471-2105-6-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32 (Suppl:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Peixoto BR, Vencio RZ, Egidio CM, Mota-Vieira L, Verjovski-Almeida S, Reis EM. Evaluation of reference-based two-color methods for measurement of gene expression ratios using spotted cDNA microarrays. BMC Genomics. 2006;7:35. doi: 10.1186/1471-2164-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat Methods. 2005;2:337–344. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- Smyth GK.Limma: linear models for microarray dataIn: Gentleman R, Carey V, Dudoit S, Irizarry RA, Huber W (eds)Bioinformatics and Computational Biology Solutions using R and Bioconductor Springer: New York; 2005397–420. [Google Scholar]
- Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognition. 1997;30:1145–1159. [Google Scholar]
- Hand DJ, Till RJ. A simple generalization of the area under the ROC curve for multiple class classification problems. Machine Learning. 2001;45:171–186. [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46:389–422. [Google Scholar]
- Mathews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Boumil RM, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet. 2001;10:2225–2232. doi: 10.1093/hmg/10.20.2225. [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set. Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.