Abstract
Microarray-based classifiers and associated signature genes generated from various platforms are abundantly reported in the literature; however, the utility of the classifiers and signature genes in cross-platform prediction applications remains largely uncertain. As part of the MicroArray Quality Control Phase II (MAQC-II) project, we show in this study 80–90% cross-platform prediction consistency using a large toxicogenomics data set by illustrating that: (1) the signature genes of a classifier generated from one platform can be directly applied to another platform to develop a predictive classifier; (2) a classifier developed using data generated from one platform can accurately predict samples that were profiled using a different platform. The results suggest the potential utility of using published signature genes in cross-platform applications and the possible adoption of the published classifiers for a variety of applications. The study reveals an opportunity for possible translation of biomarkers identified using microarrays to clinically validated non-array gene expression assays.
Keywords: microarray, cross-platform, gene signature, classifier, MAQC, hepatotoxicity
Introduction
The use of microarrays as molecular tools to measure transcript abundance has been pursued for more than a decade.1 The production of microarrays and the associated laboratory methods have improved and become more standardized. The maturation of this technology provided the necessary groundwork for the recent deployment of two different microarray-based diagnostic tests (that is, classifiers) related to cancer. MammaPrint uses the expression levels of 70 genes, measured using the Agilent (Santa Clara, CA, USA) platform, as a prognostic indicator to predict the risk of recurrence in lymph node-negative breast cancer patients.2 The Pathwork Tissue of Origin Test measures the transcript levels of 1500 genes, using the Affymetrix (Santa Clara, CA, USA) platform, to facilitate the diagnosis of tumors of unknown origin.3 These recent advances highlight the utility of transcript-based molecular classifiers measured by microarrays in clinical applications.
A molecular classifier is the result of a combination of signature genes (selected through machine learning to differentiate biological groups such as between disease and health) and a machine learning algorithm. The application of classifiers has been intensively analyzed in the clinic for diagnosis, prognosis and prediction of treatment outcomes. To enhance the toxicity prediction and reduce the cost in animal testing, microarray-based classifiers are often developed in the field of toxicogenomics using in vitro systems or short-term animal models to potentially replace more costly, time consuming and labor-intensive long-term animal testing. Consequently, microarray-based classifiers generated from various platforms are abundantly reported in the literature; however, the utility of the classifiers and associated signature genes in cross-platform prediction applications remains largely uncertain. The utility of these published signature genes and classifiers ultimately relies on a high degree of cross-platform transferability.
The evaluation of cross-platform microarray performance has been the focus of a large number of studies (reviewed in Yauk and Berndt4). Of the different factors that have resulted in the improved cross-platform correlations, two key areas of improvement have been in the technical aspects of producing microarray data and in the way these data are then analyzed. Technical improvements have been achieved mainly in the manufacturing of microarrays and in associated laboratory procedures.5, 6, 7, 8 Data analysis has become more comprehensive as a direct result of the development of new tools and computational approaches to facilitate analysis within and across various microarray platforms.9, 10, 11, 12, 13, 14, 15 The Food and Drug Administration(FDA)-led MicroArray Quality Control Consortium (MAQC) study16, 17, 18, 19, 20, 21 systematically evaluated both of these key areas and concluded that microarrays are capable of generating reproducible measurements within and across different laboratories as well as across different platforms if appropriate analysis procedures were applied.17, 19 This was a significant outcome as it showed that the same biological findings and insights were revealed, even when data were generated on different microarray platforms. The MAQC Consortium has now further extended their exploration of the capabilities of microarrays by establishing a set of ‘best practices' and an associated data analysis protocol for developing and validating microarray-based classifiers,22 including the presented study on cross-platform application of classifiers and signature genes.
To assess the cross-platform transferability of signature genes and classifier, we analyzed a large toxicogenomics data set that contains gene expression profiles from the target tissue (liver) generated on two different microarray platforms (Agilent and Affymetrix).23 We used a comprehensive and robust approach to evaluate whether a predictive classifier could be developed using the signature genes derived from a different microarray platform (that is, transferability of signature genes) and whether the classifier from one platform could yield an accurate prediction for the samples whose expression data were generated from another platform (that is, transferability of classifiers).
Materials and methods
Data set
The microarray-based gene expression profiling data as well as the histopathological data that were used in this study were generated previously and have been reported in detail elsewhere.23, 24 Briefly, eight different hepatotoxicants (1,2-dichlorobenzene, 1,4-dichlorobenzene, bromobenzene, diquat dibromide, galactosamine, monocrotaline, N-nitrosomorpholine and thioacetamide) were selected based on published literature regarding the differences that exist in the cell types and liver regions that are injured in response to exposure. For each compound, doses that elicited a sub-toxic (‘low'), a moderately toxic (‘medium') or a overtly toxic (‘high') response 24 h after treatment were selected. Samples were collected for gene expression profiling, clinical chemistry, hematology and histopathology at 6, 24 and 48 h after exposure. For each compound, four animals were used for each dose (including a vehicle control) and time point group, except for the diquat dibromide compound in which six animals were used for each group (because of higher degree of variability in the presence and extent of liver injury in each dose and time group). After 6, 24 or 48 h of treatment, experiments were performed according to established guidelines and an approved Animal Study Protocol was on file before initiation of the study.25
For hybridizations performed on the Affymetrix platform, RNA isolated from the liver of each individual rat was labeled and hybridized to the Rat Genome 230 2.0 Array with 31 099 probe-sets (Affymetrix) for a total of 418 hybridizations. For hybridizations performed on the Agilent platform, RNA isolated from the liver from each of the 318 treated rats was labeled and hybridized against a pooled RNA sample representing all of the individuals in the time- and compound-matched control group. The samples were hybridized to Agilent Rat Oligo Microarrays (G4130A) (22 075 probes) and a fluorophore reversal (dye-swap) was performed for a total of 636 hybridizations. The dual-color Agilent data were analyzed in two different ways in this study—ratio-based and intensity-based. The ratio-based data are the average value of the dye-swap results while the intensity-based data are the average value of Cy3 and Cy5 from the dye-swap results, corresponding only to the treated samples.
Phenotypic anchors for cross-platform analysis
For each animal in this study, a large number of endpoint measurements were collected, such as clinical chemistry, hematology and histopathology.23 Many of these parameters were relevant to the findings of liver toxicity; however, for the purposes of this study only two different endpoints were considered. First, to generate a binary classifier (the main focus of this study), the histopathological data from each of the 418 was used to identify the animals for which hepatocellular necrosis was observed (regardless of severity) and those for which it had not (summarized in Supplementary Table 1 online). Second, the response to hepatocellular injury (RHI) score24 was used as a continuous endpoint for regression analysis. All clinical data for each animal, including both binary classification and RHI values are available from Supplementary Table 2 online.
Training and test sets
The training/test set pair for each platform was generated using a hybrid approach based on chronology-based, compound-based and random splitting. Using a chronology-based approach, in which the date of hybridization is used to divide these data would be confounded by the fact that the compounds in this studied data set were profiled sequentially, which would be problematic as it is hypothesized that the molecular mechanisms underlying the toxic response to these compounds are different and also because of the lack of statistical significance associated with the limited number of compounds in this study. However, randomly splitting the data across the eight compounds could unevenly divide the samples between the training and test sets and therefore could result in inappropriately reflecting the inherent biases that may have been generated during the creation of these data. Therefore, the adopted hybrid approach was that random splitting was performed on six of the eight compounds, in which the training/test set ratio was approximately 2:1. The two remaining compounds were exclusively used for the test set. Monocrotaline was randomly selected as one of these compounds while bromobenzene was selected based on chronology. This process resulted in an approximately equal number of necrosis and non-necrosis samples in both the test and training sets. The numbers of animals with and without necrosis within each compound assigned to either the test or training set are detailed in Supplementary Table 2 online.
Probe mapping between Affymetrix and Agilent platforms
All analyses were conducted using probes that measured the same transcript on both the Affymetrix Rat Genome 230 2.0 Array and the Agilent Rat Oligonucleotide Microarray (G4130A). Three different approaches were used to identify probes associated with the same transcript.
Sequence-based mapping set (SeqMap)—a sequence-based approach to identifying common probes was generated using the approach that was also used in the MAQC-I project.19 Briefly, each probe sequence from both platforms was BLASTed against the RefSeq database. For the Affymetrix platform, each of the 11 perfect-match sequences for each probeset was independently BLASTed to the RefSeq database and the matching quality was recorded for each probe. As the 11 Affymetrix probes for each probeset usually covers several hundred bases, the criteria to be considered a perfect match to a RefSeq record was defined as at least 80% of the probes (9 out of the 11) in a probeset perfectly matched the same RefSeq record. A list of probes that perfectly matched RefSeq entries was generated for each platform and then used to find commonalities. For RefSeq entries that were perfectly matched by more than one probe from the same platform, the probe that is closest to the 3′ end was used and probes that matched more than one RefSeq entry were excluded.
RefSeq set (RefSeq)—the RefSeq database was also used in a less restrictive way to match Agilent probes with Affymetrix probesets. The probe ID of Agilent platform and the probe-set ID of Affymetrix platform were mapped to the RefSeq database available from the ArrayTrack26, 27 Library dated June 2008 that mirrored the RefSeq database in National Center for Biotechnology Information (NCBI). The common set was identified using a Venn diagram approach. In the case of multiple probes (or probesets) IDs corresponding to the same RefSeq ID for each platform, the median expression value of the probes (or probesets) was calculated and used in subsequent analyses.
Unigene set (Unigene)—for the least stringent approach for identifying matching probes across platforms, the Unigene database was used following the same method described above for the RefSeq database. The ArrayTrack Unigene database was used, which mirror the NCBI database dated 21 May 2008.
T-index for assessing the cross-platform consistency
Both transferability of signature genes and cross-platform predictivity of classifiers were evaluated using the T-index score:
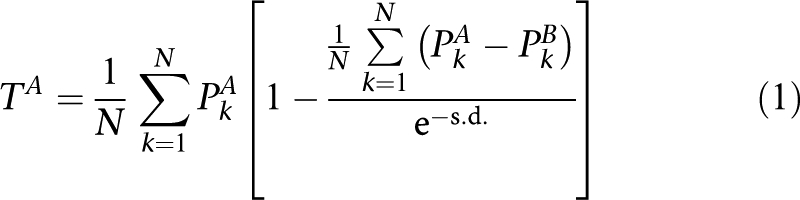 |
where TA is a measure of transferability of classifiers developed from platform A to platform B. Total of N (that is, 500) classifiers were developed. PkA and PkB are prediction accuracies for the same test set profiled by platforms A and B, respectively. s.d. is the standard deviation of (PkA−PkB). The T-index score ranges 0 to 1 with <0.5 indicating that transferability is due to chance. Larger T-index score indicates better transferability across platforms.
Results
Classifiers were generated for prediction on whether or not a given animal had evidence of hepatocellular necrosis. Both the transferability of signature genes and classifiers were assessed based on the prediction accuracy calculated using the test set (Figure 1). A T-index score was used to evaluate the degree of transferability. All of the analyses were based on the subset of transcripts that were common to both the Affymetrix and Agilent platforms. Three different methods, differing based on their level of stringency, were used to identify the common transcripts (CTs). As shown in Table 1, the number of CTs increases as the stringency of the mapping criteria decreases, thus providing an opportunity to analyze the dependency of the cross-platform transferability on the stringency that is used in identifying CTs. As the Agilent platform used a two-color design (one ‘control' and one ‘treated' sample, each labeled with a different dye, hybridized to the same microarray) in contrast to the Affymetrix platform in which a single labeled sample was hybridized to each microarray, this difference in hybridization configuration resulted in three different analysis configurations (ACs) to compare the data generated from the two different platforms (Table 1).
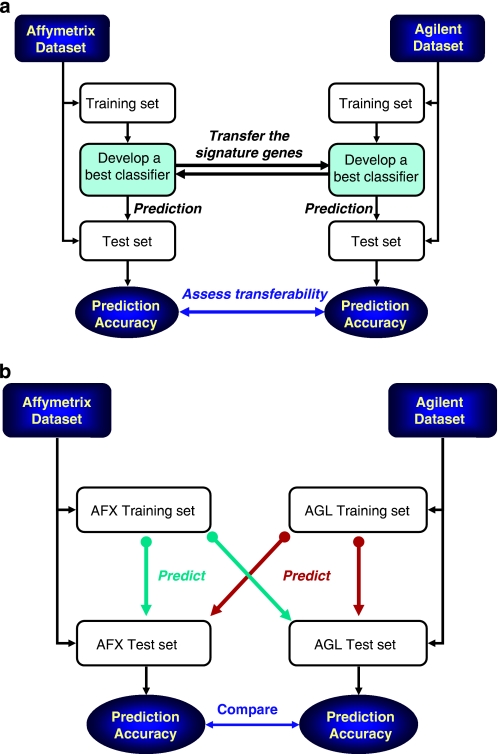
Figure 1.
Two analysis procedures for evaluation of cross-platform consistency. (a) Transferability of signature genes was assessed by first developing Affymetrix-specific classifiers using the training set data. The signature genes used by the Affymetrix classifiers were then applied to the training set data generated using the Agilent platform in order to produce Agilent-specific classifiers. Both sets of platform-specific classifiers were then used to predict their own test sets, independently. The process was repeated such that the signature genes were initially identified on the Agilent platform and then applied to the Affymetrix platform. The prediction accuracy that was achieved with both platforms was used to assess the cross-platform consistency at the level of signature genes. (b) Transferability of classifiers is to assess whether a classifier generated from one platform can accurately predict the samples profiled by another platform. Specifically, a set of classifiers using the training set data was generated for a given platform. These classifiers were then used to predict the test set for both platforms, independently. This was examined bi-directionally for both of the microarray platforms. The difference in the prediction accuracy of the test sets from the two platforms was used to evaluate cross-platform consistency at the level of classifiers.
Table 1. Description of three ACs for comparing data generated from the Affymetrix platform with data generated from the Agilent platform and three methods to identify the CT sets between two platforms.
| Abbreviation | Description |
|---|---|
| AC | |
| AC 1 | The intensity data of the Affymetrix microarrays (418 samples) compares with the averaged ratio data from the dye-swapped Agilent microarrays (318 samples). |
| AC 2 | The ratio data of the Affymetrix microarrays (318 samples) compares with the averaged ratio data from the dye-swapped Agilent microarrays (318 samples).a |
| AC 3 | The intensity data of the Affymetrix microarrays (318 samples) compares with the intensity data extracted from the Agilent platform (318 samples).b |
| CT | |
| SeqMap | The most stringent method employed a sequence-based mapping approach (4860 CTs).c |
| RefSeq | The less stringent method was to identify the probes/probesets from both platforms having the same RefSeq ID (6312 CTs). |
| Unigene | The least stringent approach was used to generate the list of Unigene-based CTs (9954 CTs). |
Abbreviations: AC, analysis configuration; CT, common transcript; MAQC, MicroArray Quality Control Phase.
The Affymetrix ratio data were calculated using its intensity data similar to the ratios produced by ‘dual' samples assayed on the Agilent platform (a treated sample compared to an average of the corresponding control samples).
In the first phase of the MAQC project, a comparative analysis between one-color and two-color data generated on the same platform revealed that the intensity value from the single channel of the two-color array exhibits similar sensitivity/specificity as the two-color ratio array data.18 Therefore, the intensity data are the average value of Cy3 and Cy5 corresponding only to the treated samples.
The method is identical to the one used in the MAQC-I project.19
The transferability analysis of both signature genes and classifiers revealed 80–90% consistency between the Affymetrix and Agilent platforms for the studied toxicogenomics data set regardless of the choice of machine learning methods, the CTs selected, and the ACs applied (the detailed results are presented below).
Transferability of signature genes
Three different algorithms, nearest centroid, K-nearest neighbor and decision forest,28, 29 were used to generate classifiers and associated signature genes for each of the three CT sets and for each of the three ACs for comparing data across the two platforms. The general analysis procedure is depicted in Figure 1a with additional details provided for each of the algorithms in Supplementary Methods. Figure 2 and Supplementary Table 3 shows the T-index scores and prediction accuracy of the classifiers that were obtained for each of the 54 permutations (two microarray platforms × three ACs × three CT sets × three algorithms). Overall, a high degree of transferability was observed with an average T-index score of 0.84. There was no significant difference in transferability regarding the directionality of the transfer (Affymetrix to Agilent or vice versa). Figure 2 clearly shows that the method for selecting the CT sets had little effect on transferability. The decision forest algorithm for classification consistently outperformed K-nearest neighbor and nearest centroid in every instance.
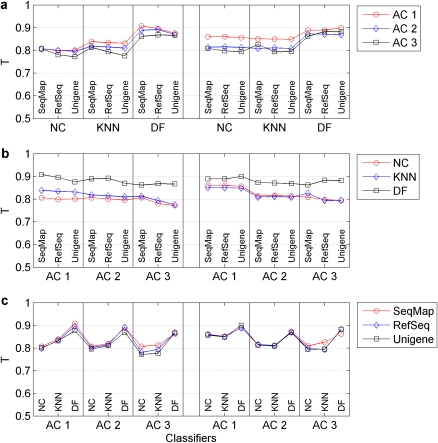
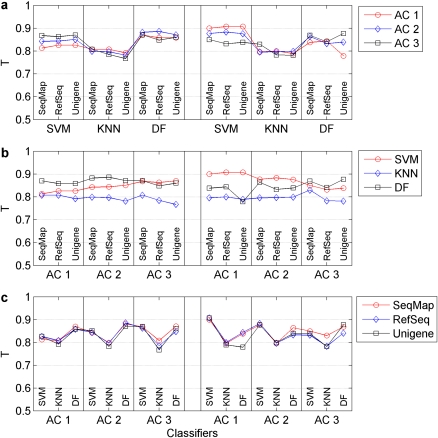
Figure 2.
Assessment of transferability of signature genes across platform using T-index scores. A total of 54 permutations were tested to assess cross-platform transferability of signature genes, which consisted of two platforms, three ACs, three approaches for generating CT lists and three classification algorithms. A T-index score was calculated for each permutation to evaluate the transferability. (a) Compares the T-index scores that were generated by the three ACs, while (b) compares the results that were achieved with the three classification algorithms. (c) Depicts the T-index scores that were obtained with the three approaches for generating CT sets across the two platforms. Both AC and methods to select the CTs had little effect on the transferability while a degree of variability was observed for different classification algorithms.
We integrated multiple different histopathological diagnoses (all related to the severity and extent of the liver injury) for each animal in this data set into a five-level of ‘RHI' score.24 We examined the animals that were misclassified based on the five-level RHI score. As depicted in Figure 3, regardless of the choice of AC, CT sets and classification algorithms used, the frequently misclassified animals usually had low RHI scores (that is, RHI=0, 1 and 2, indicating non-existent or very low levels of liver injury). The animals frequently misclassified in internal validation in one platform were also most frequently misclassified in the other platform using the transferred signature genes (Figure 4). These results suggest that if the signature genes of a classifier were generated using one platform, similar accuracy can be obtained by generating classifiers using the same signature genes within a different microarray platform. Moreover, when using the same signature genes and classification method, the platform-dependent classifiers resulted in not only similar accuracy in predicting the test set, but also the same samples are consistently misclassified, which are likely due to the biological outliers as opposed to a shortcoming in the classification approach.
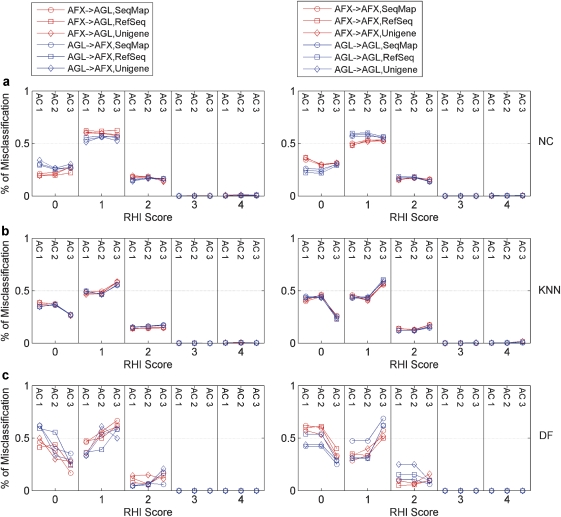
Figure 3.
Distribution of the percentage of misclassified samples across different ‘RHI' scores. Fifty-four classifiers (summarized in Figure 2 and Supplementary Table 1 online) were plotted in three panels based on the classification algorithm that was used: (a) nearest centriod (NC); (b) K-nearest neighbor (KNN); and (c) decision forest (DF). AFX → AFX (or AGL → AGL) denotes the prediction results to the test set that were obtained when the signature genes were generated using the Affymetrix (or Agilent) training set from the same platform. In contrast, AFX → AGL (or AGL → AFX) indicates that the signature genes had been identified using the training set from the opposite platform. No samples with an RHI score >2 was misclassified in any of the permutations tested. The largest misclassification rate was observed for low RHI scores (that is, RHI=0 and 1), relating to either the absence of any apparent or the presence of only minor amounts of liver injury.
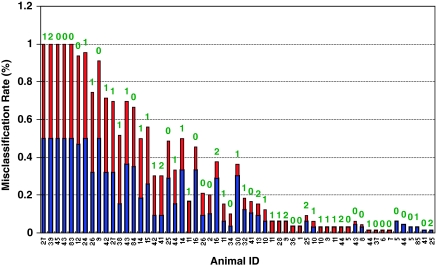
Figure 4.
The misclassification rate for each animal of the test set in the study of cross-platform transferability of signature genes. The study of cross-platform transferability of signature genes involved 54 permutations (that is, two microarray platforms × three ACs × three CT sets × three algorithms), resulting total of 108 classifiers (that is, transfer from Affymetrix to Agilent and vice versa). The misclassification rate is calculated by dividing the frequency of misclassification for each animal by the total number of classifiers (that is, 108 classifiers) for the test set. Each bar is divided into two colors; the blue is associated with the misclassification for the classifiers selected by the cross-validation while the red is for the classifiers using the transferred signature genes. The label above the bar is the RHI score. All the misclassified animals had the RHI scores of 0, 1 or 2, and no animals with RHI=3 and 4 were misclassified. the samples (that is, animals) misclassified by the cross-validation driven classifiers in one platform likely occurred in another platform using the transferred signature genes, indicating the performance of the classifiers was not affected by the choice of the signature genes as long as they were validated in any platform. Animals 27, 39, 45, 43 and 83 were misclassified by all the classifiers.
The results thus far have shown that cross-platform transferability can be achieved using transcript level data. However, it has been well documented that cross-platform reproducibility of microarray data are even greater when the data are examined at the biological process/pathway level.5 To analyze this, we examined the transferability of data after the transcript data had been mapped to 352 canonical pathways using GeneGo's MetaCore application30, 31 and the analysis procedure detailed in Supplementary Methods. Supplementary Table 4 summarizes the pathway-based classifier results. Consistent with previous reports regarding data reproducibility, improved cross-platform transferability was observed using pathway level data as compared with transcript level data. This is evidenced by the overall higher T-index scores in Supplementary Table 4 as compared with Supplementary Table 3. Similar to the transcript level analyses, neither the AC nor the method for choosing CTs significantly influenced the results.
Although good results were obtained for developing molecular signatures for a binary classification system (that is, the presence or absence of necrosis), we further examined the transferability of signature genes across platforms when using a continuous variable (that is, endpoint). The RHI score was selected as the endpoint and cross-platform transferability was evaluated using three different algorithms (that is, general linear model, partial least square and partition tree). The analysis procedure is detailed in Supplementary Methods and the results from 43 model settings from three algorithms based on AC 3 and the CT set of SeqMap were summarized in Supplementary Figures 1 and 2. As depicted in Supplementary Figure 1, the training models developed on the training set of both platforms had the comparable root mean square error, regardless of whether the signature genes were derived from the internal validation conducted within the same platform or were transferred from the other platform. Moreover, these training models yielded similar prediction accuracy to their respective test sets. The Pearson's correlation of prediction between the training models of two platforms, one using signature genes derived from the internal validation while the other using the transferred signature genes, were >0.97, which is slightly higher than the correlation (0.94–0.96) values that were generated with each respective test set (Supplementary Figure 2). These results show a high cross-platform transferability of signature genes when using a continuous endpoint variable.
Transferability of classifiers
The cross-platform transferability of classifier was analyzed as outlined in Figure 1b. The classifiers were generated using the training set from a given platform and then used to predict the test sets from gene expression data that was generated by both platforms. The prediction accuracies for the test set of both platforms were compared using the T-index score to determine the cross-platform predictivity. K-nearest neighbor, decision forest and support vector machine were applied to each of three ACs for all the three CT sets. The analysis procedures for three distinct classification algorithms are summarized in Supplementary Methods.
As with the evaluation of the transferability study of signature genes, a total of 54 different classifiers were generated with the cross-platform batch correction for ACs 1 and 3, but not for AC 2. Supplementary Table 5 online summarizes the T-index scores of these classifiers and illustrates the high prediction accuracy that was observed for both platforms with an average T-index score of 0.84. Figure 5 clearly shows that there was little difference among the prediction accuracy regardless of which of the three ACs to compare data across the platforms and which of the methods for selecting the CT sets were used. A slight variation among classification algorithms used was observed for cross-platform transferability analysis.
Figure 5.
Assessment of transferability of classifiers across platform using T-index scores. A total of 54 permutations were tested to assess cross-platform predictivity of classifiers, which consisted of two platforms, three ACs, three approaches for generating CT sets and three classification algorithms (that is, support vector machine (SVM), KNN and DF). AFX → AGL (or AGL → AFX) denotes that the classifiers were generated from the Affymetrix (or Agilent) platform and then used to predict the test sets that profiled by the opposite platform. A T-index score was calculated for each permutation to evaluate the cross-platform predictivity based on a comparative analysis of prediction results obtained from both of the test sets that each set of classifiers was used to predict. (a) Compares the T-index scores that were generated by the three ACs, while (b) compares the results that were achieved with the three classification algorithms. (c) Depicts the T-index scores that were obtained with the three approaches for generating CT lists across the two platforms. The results indicate that the cross-platform predictivity was independent of the AC and the method for identifying the CTs, but varied slightly with the classification algorithms.
Cross-platform batch correction is necessary to use a classifier from one platform to predict the samples profiled by the other platform because the scale in measurement for the absolute transcript abundance is different between platforms. As depicted in Figure 6 and summarized in Supplementary Table 6, when using intensity values generated by at least one of the platforms (ACs 1 and 3), the predictivity was low unless a cross-platform batch correction was performed. In the case of AC 3 (the comparison of intensity-based data generated by both platforms), a simple data transformation such as the mean centering method is sufficient to correct the batch effect (Supplementary Table 6). However, when classifiers from the Affymetrix intensity data were used to predict the ratio data generated by samples profiled on the Agilent platform (that is, AC 1), both data transformation and scaling was required. Not surprisingly, when ratio-based data were compared (AC 2), no cross-platform batch correction was required.
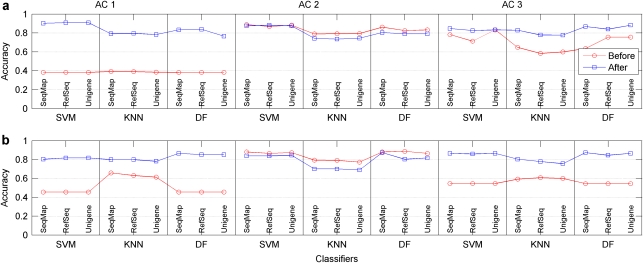
Figure 6.
Assessment of cross-platform batch correction on the transferability of classifiers across platform. The effect of cross-platform batch correction was evaluated for each of the 54 permutations before and after the batch correction was applied to the data. (a) Compares the prediction accuracy that was obtained by generating classifiers using data from the Agilent platform and then predicting the test set data from the Affymetrix platform. (b) Depicts the results from the reverse approach in which the classifiers were generated using data from the Affymetrix platform and then used to predict the test set data from the Agilent platform. The results showed that cross-platform batch correction was necessary for ACs 1 and 3, but not required when the ratio data was used (AC 2).
Discussion
There is great interest in using classifiers (that is, molecular signatures) in clinical applications, predictive toxicology and risk assessment. With the variety of different microarray platforms that are commercially available, natural questions arise regarding (1) can signature genes identified from one platform be directly used by another platform to generate a predictive classifier, thereby suggesting the potential utility of using published signature genes in cross-platform applications? and (2) will a classifier developed using one platform yield an accurate prediction for samples whose gene expression data were generated using a different platform, which would suggest the possible adoption of the published classifiers for a variety of applications? Using data from a large toxicogenomics study that were generated using both the Agilent and Affymetrix microarray platforms, the results presented here describe a comprehensive evaluation of these specific questions, which revealed 80–90% cross-platform consistency regardless of the choice of machine learning methods, the CTs selected, and the ACs applied.
There are many parameters that can be varied when comparing data across microarray platforms, each of which could theoretically affect the conclusions that are drawn. For example, the distinct experimental designs associated with the one-color Affymetrix platform and the two-color Agilent platform resulted in three different ways to compare the data between the two platforms (Table 1). In addition, the method used to identify the common list of transcripts that were measured by both platforms (Table 1) and also the algorithm used for classification of the samples could affect the cross-platform comparison results. Furthermore, the cross-platform consistency can be evaluated either at the level of individual transcripts or at the level of biological pathways. Finally, the cross-platform consistency can be assessed in either the binary classification or by regression analysis. Given their combinatorial nature, it is not feasible to analyze all possible factors that may influence microarray results. Therefore, in this study we focused on evaluating factors that we believe are most important and most likely to affect the interpretation of cross-platform consistency results.
We examined a number of the key factors that influence the cross-platform transferability of signature genes. The choice of AC or CT determination generally had minimum effect on the cross-platform consistency, while different classification methods varied slightly in regards to the accuracy that each was capable of achieving. Cumulatively, the results show that the high degree of cross-platform transferability of signature genes for both binary classification and continuous endpoint prediction.
In terms of cross-platform transferability of classifiers, when ratio-based data were calculated from the one-color Affymetrix platform and compared with the ratios that were generated directly by the two-color Agilent platform (AC 2), no cross-platform batch correction was needed to achieve high cross-platform predictivity. However, when the intensity data were extracted from the two-color Agilent platform and compared with the intensity data generated directly by the one-color Affymetrix platform (AC 3), a simple mean centering data transformation was necessary to achieve the same level of cross-platform predictivity. Finally, a more complicated batch correction was required when the intensity data from the Affymetrix platform compared with the ratio data from the Agilent platform (AC 1). This emphasizes the importance of ensuring data equivalency from different platforms before making cross-platform analysis.
There were unexpected phenotypic results within the context of this multiple compound study. For example, on histological examination of the liver sections from each of the 418 animals, it was observed that two of the control animals had minimal amounts of liver necrosis. The cause was idiopathic because these animals had not been exposed to any toxicants, but as the damage was present, these animals were counted in the ‘necrosis' group in this study. In addition, in spite of the fact that an inbred strain of animals was used, there was considerable variability across animals in the phenotypic response to some of the toxicants at particular dose and time intervals. When one considers this variability, it was not surprising that several animals were consistently misclassified regardless of the methods used to generate the classifiers with or without using the transferred signature genes. However, in order to better understand how well the classifiers and signatures genes performed, the misclassified animals were analyzed further. In most of the permutations tested, the majority of the misclassified animals either had minimal amount of necrosis (that is, predicted as false negatives) or none at all (that is, predicted as false positives) (Figure 4). From a technical standpoint, this result was not unexpected. Two separate cross-sections of the left lobe of the liver were harvested at necropsy. One section was used for histopathology and the other section was used for gene expression profiling. The different levels of hepatocellular necrosis (minimal, mild, moderate and marked (severe)) were based on the amount of necrosis that was observable in the section that was used for histopathology.23 At the more pronounced levels of necrosis, one would expect that the damage would not be localized in specific regions of the liver, but might rather present throughout. This is consistent with the observations that samples with moderate or marked necrosis were not misclassified in this study. However, with less pronounced levels of the liver damage, the injury may not be homogenous throughout the liver, but rather localized. This could explain why the observation of necrosis was made in one section of the tissue, but the damage was not present in the section that was used for gene expression profiling, hence leading to the misclassification as a result of a sampling issue. The second most common set of misclassifications was when an animal had no observable necrosis but classified to have necrosis. From a technical perspective, this could simply be the reverse of the sampling issue detailed above. However, from a biological perspective, this observation was also expected. Within the context of a given dose group, some animals did not have observable liver necrosis at the 6-h time point (and therefore would be included in the ‘non-necrosis' group), yet all of the animals at the next time point exposed to the same dose had notable necrosis. We considered these animals to be representative of ‘incipient toxicity', in which the liver responds to the toxic insult at the level of transcription, but has not yet manifested in the cellular phenotype.32 This could potentially explain why the necrotic signal was detected at the molecular (transcript) level and hence the ‘necrosis' calls by the classifier, but why this was not yet observable at the cellular (histopathological) level.
There are several additional interesting observations that can be made when these results are examined in aggregate. First, there is no significant difference in the degree of transferability for both signature genes and classifiers when generated on the Agilent platform and transferred to the Affymetrix platform as compared with these in the reverse order, suggesting the comparability of data derived from the Affymetrix and Agilent microarray platforms and thus further supporting the findings of the MAQC-I project regarding the reproducibility of data generated on different microarray platforms. Second, the T-index scores for the transferability of signature genes were statistically comparable (P-value <0.05) to the scores assessing the cross-platform predictivity of classifiers, suggesting a positive correlation between signature genes and classifiers in terms of cross-platform transferability. Finally, the gene expression profiles derived from either platform performed well at separating samples in a principal component analysis based on the presence or absence of necrosis and also based on the severity of the necrosis (that is, RHI) (Supplementary Figure 3). Undoubtedly, there is a strong transcriptional response to necrosis occurring in the liver that was observed in this study and therefore the cross-platform consistency of biological responses evoked by weaker biological stimuli should be the subject of further investigation.
In summary, the positive findings of cross-platform transferability in signature gene and classifiers not only open potential opportunities and applications for the existing and future signature genes and classifiers for cross-platform applications, but also suggest the possibility of identifying clinically relevant biomarkers based on data generated using expression microarrays, and then assaying these biomarkers using a lower-density platform (for example, quantitative-PCR) in clinical and risk assessment applications.
Acknowledgments
Dr Xiaohui Fan thanks the support by Chinese Key Technologies R&D Program (No.2005CB23402), the National Science Foundation of China (No. 30801556) and Research Fund for the Doctoral Program of Higher Education of China (No. 20090101110126) for participating in the MAQC project at National Center for Toxicological Research of the US Food and Drug Administration. The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Glossary
- AC
analysis configuration
- CT
common transcript
- DF
decision forest
- GO
gene ontology
- KNN
K-nearest neighbor
- MAQC
MicroArray Quality Control
- NC
nearest centriod
- RHI
response to hepatocellular injury
- SVM
support vector machines
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Dumur CI, Lyons-Weiler M, Sciulli C, Garrett CT, Schrijver I, Holley TK, et al. Interlaboratory performance of a microarray-based gene expression test to determine tissue of origin in poorly differentiated and undifferentiated cancers. J Mol Diagn. 2008;10:67–77. doi: 10.2353/jmoldx.2008.070099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Berndt ML. Review of the literature examining the correlation among DNA microarray technologies. Environ Mol Mutagen. 2007;48:380–394. doi: 10.1002/em.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Kuo WP, Liu F, Trimarchi J, Punzo C, Lombardi M, Sarang J, et al. A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat Biotechnol. 2006;24:832–840. doi: 10.1038/nbt1217. [DOI] [PubMed] [Google Scholar]
- Yauk CL, Berndt ML, Williams A, Douglas GR. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32:e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KJ, Dumur CI, Taylor GS, Chaplin MD, Guiseppi-Elie A, Grant G, et al. Application of a correlation correction factor in a microarray cross-platform reproducibility study. BMC Bioinformatics. 2007;8:447. doi: 10.1186/1471-2105-8-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Becker KG, Cho-Chung YS, Nesterova M, Watkins T, Wood W, et al. A rapid method for microarray cross platform comparisons using gene expression signatures. Mol Cell Probes. 2007;21:35–46. doi: 10.1016/j.mcp.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Culhane AC, Perriere G, Higgins DG. Cross-platform comparison and visualisation of gene expression data using co-inertia analysis. BMC Bioinformatics. 2003;4:59. doi: 10.1186/1471-2105-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Luthi-Carter R, Delorenzi M. Cross-species and cross-platform gene expression studies with the Bioconductor-compliant R package ‘annotationTools'. BMC Bioinformatics. 2008;9:26. doi: 10.1186/1471-2105-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Kamath K, Zhang KY, Pulapura S, Achar A, Nunez-Iglesias J, et al. Integrative array analyzer: a software package for analysis of cross-platform and cross-species microarray data. Bioinformatics. 2006;22:1665–1667. doi: 10.1093/bioinformatics/btl163. [DOI] [PubMed] [Google Scholar]
- Severgnini M, Bicciato S, Mangano E, Scarlatti F, Mezzelani A, Mattioli M, et al. Strategies for comparing gene expression profiles from different microarray platforms: application to a case-control experiment. Anal Biochem. 2006;353:43–56. doi: 10.1016/j.ab.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Shabalin AA, Tjelmeland H, Fan C, Perou CM, Nobel AB. Merging two gene-expression studies via cross-platform normalization. Bioinformatics. 2008;24:1154–1160. doi: 10.1093/bioinformatics/btn083. [DOI] [PubMed] [Google Scholar]
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, et al. Performance comparison of one-color and two-color platforms within the Microarray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy R, Fulmer-Smentek S, Jensen RV, Jones WD, Wolber PK, Johnson CD, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Lucas AB, Shippy R, Fan X, Fang H, Hong H, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat Biotechnol. 2006;24:1132–1139. doi: 10.1038/nbt1237. [DOI] [PubMed] [Google Scholar]
- The MicroArray Quality Control (MAQC) Consortium MAQC-II Project: a comprehensive survey of common practices for the development and validation of microarray-based predictive models Nat Biotechnol 2010. accepted). [DOI] [PMC free article] [PubMed]
- Lobenhofer EK, Auman JT, Blackshear PE, Boorman GA, Bushel PR, Cunningham ML, et al. Gene expression response in target organ and whole blood varies as a function of target organ injury phenotype. Genome Biol. 2008;9:R100. doi: 10.1186/gb-2008-9-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobenhofer EK, Boorman GA, Phillips KL, Heinloth AN, Malarkey DE, Blackshear PE, et al. Application of visualization tools to the analysis of histopathological data enhances biological insight and interpretation. Toxicol Pathol. 2006;34:921–928. doi: 10.1080/01926230601072319. [DOI] [PubMed] [Google Scholar]
- National Research Council Guide for The Care and Use of Laboratory AnimalsNational Academy Press: Washington, 1996.
- Tong W, Cao X, Harris S, Sun H, Fang H, Fuscoe J, et al. ArrayTrack--supporting toxicogenomic research at the U.SFood and Drug Administration National Center for Toxicological Research. Environ Health Perspect 20031111819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Harris S, Cao X, Fang H, Shi L, Sun H, et al. Development of public toxicogenomics software for microarray data management and analysis. Mutat Res. 2004;549:241–253. doi: 10.1016/j.mrfmmm.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Tong W, Xie Q, Hong H, Shi L, Fang H, Perkins R, et al. Using decision forest to classify prostate cancer samples on the basis of SELDI-TOF MS data: assessing chance correlation and prediction confidence. Environ Health Perspect. 2004;112:1622–1627. doi: 10.1289/txg.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Hong H, Fang H, Xie Q, Perkins R. Decision forest: combining the predictions of multiple independent decision tree models. J Chem Inf Comput Sci. 2003;43:525–531. doi: 10.1021/ci020058s. [DOI] [PubMed] [Google Scholar]
- Ekins S, Bugrim A, Brovold L, Kirillov E, Nikolsky Y, Rakhmatulin E, et al. Algorithms for network analysis in systems-ADME/Tox using the MetaCore and MetaDrug platforms. Xenobiotica. 2006;36:877–901. doi: 10.1080/00498250600861660. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Heinloth AN, Irwin RD, Boorman GA, Nettesheim P, Fannin RD, Sieber SO, et al. Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci. 2004;80:193–202. doi: 10.1093/toxsci/kfh145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.