Abstract
Gene expression signatures of toxicity and clinical response benefit both safety assessment and clinical practice; however, difficulties in connecting signature genes with the predicted end points have limited their application. The Microarray Quality Control Consortium II (MAQCII) project generated 262 signatures for ten clinical and three toxicological end points from six gene expression data sets, an unprecedented collection of diverse signatures that has permitted a wide-ranging analysis on the nature of such predictive models. A comprehensive analysis of the genes of these signatures and their nonredundant unions using ontology enrichment, biological network building and interactome connectivity analyses demonstrated the link between gene signatures and the biological basis of their predictive power. Different signatures for a given end point were more similar at the level of biological properties and transcriptional control than at the gene level. Signatures tended to be enriched in function and pathway in an end point and model-specific manner, and showed a topological bias for incoming interactions. Importantly, the level of biological similarity between different signatures for a given end point correlated positively with the accuracy of the signature predictions. These findings will aid the understanding, and application of predictive genomic signatures, and support their broader application in predictive medicine.
Keywords: genomic signatures, enrichment analysis, network reconstruction, biological pathways, interactome, MAQCII
Introduction
The analysis of high-content omics data can be roughly divided into two main approaches: statistical analysis and functional (pathway or systems) analyses. Statistical analysis reduces thousands of data points (for example, genome-wide gene expression values) to a relatively short list of genes. These genes represent a multivariant descriptor of the studied condition (end point), and may be further refined to distinguish between or predict phenotypic outcomes (a ‘gene signature'). Over the last decade, myriads of gene signatures have been reported to predict metastases in breast cancer,1, 2 classify cancer subtypes3, 4 or predict drug response and toxicity.5, 6, 7, 8, 9
Functional analysis (FA) of high-content molecular data aims to reveal the underlying biology driving the observed changes by identifying key pathways and mechanisms associated with the studied phenotype (reviewed in Nikolsky and Bryant10). Early FA involved mapping of expressed genes onto pathway maps such as Kyoto Encyclopedia of Genes and Genomes (KEGG)11 or functional ontologies such as gene ontology (GO).12 FA has more recently expanded into three main approaches: enrichment in biological ontologies,13 biological network reconstruction, and interactome analysis.14, 15 The power of FA has been demonstrated in studies of common diseases,16, 17, 18, 19, 20 and in toxicity and drug response analyses.21, 22
FA usually does not work well on gene signatures, making it difficult to understand the underlying biology of classifying gene sets, and thereby understand the link between the signature genes and the phenotype being studied. This is mainly because of the size of the signatures, which are typically small—from several to a few dozen genes. FA methods operate best on large, preferably whole genome, data sets nonstringently filtered by fold change, P-value or FDR thresholds,23, 24 or not limited in size at all (gene set enrichment analysis13). Signature generation often uses advanced mathematical models (comprehensively summarized and tested in Microarray Quality Control Consortium II (MAQCII)25) to achieve the best predictive performance using the smallest number of genes, irrespective of the biological function of those genes. The quality of the resulting lists of biomarkers, in terms of predictivity and stability, can be assessed by a wide spectrum of techniques (see Boulesteix and Slawski26 for a recent review), ranging from set theory to algebraic computational biology methods.27 It is challenging to explain the mechanism, form networks or represent specific biological processes from small signatures. Not surprisingly, there have been few attempts to analyze and compare gene signatures in a functional context. A recent study28 compared six breast cancer signatures by enrichment analysis (EA) in GO processes, and BioCarta and KEGG pathways, showing that the signatures, although similar in predictive performance, shared very few genes.
The MAQCII experimental design created a unique opportunity to conduct the first comprehensive FA study on statistically generated predictive gene signatures. The data are both massive and diverse, with six whole-genome gene expression data sets representing 13 phenotypic end points, 3 for chemically induced pathology (carcinogenesis or organ damage), and 10 for 3 different types of cancer.29, 30, 31, 32, 33 We conducted a meta-analysis across 262 distinct signatures, generated by 33 MAQCII data analysis teams for each end point. We applied FA to assess the biological and topological composition of individual and merged (union) signatures, and to investigate biological consistency and interconnectivity between different gene signatures. The diverse collection of signatures provided large enough union data sets for FA, in which many functional dependencies and correlations became apparent in cross-signature comparisons. In this study, we report the results of this analysis, along with two novel analytical techniques for gene list comparison.
Materials and methods
Data sets, end points and analysis teams
A detailed description of the MAQC phase II (MAQCII) project is given in Shi and colleagues.45 A brief summary of the data sets used, end points classified and analysis teams participating is given in Tables 1 and 2.
Table 1. MAQCII data sets and classification end points.
| Date set provider | End point code | End point description | Number of samples | Positives | Negatives |
|---|---|---|---|---|---|
| Hamner Institutes33 | A | Chemical tumorigenesis in mouse lung | 70 | 26 | 44 |
| Entelos29 | B | Chemical nongenotoxic carcinogenesis in rat liver | 216 | 73 | 143 |
| NIEHS46 | C | Necrosis in rat liver | 214 | 79 | 135 |
| MD Anderson Cancer Center30 | D | Clinical breast cancer treatment response | 130 | 33 | 97 |
| E | Breast cancer estrogen receptor status | 130 | 80 | 50 | |
| University of Arkansas32 | F | Overall survival milestone outcome in multiple myeloma | 340 | 51 | 289 |
| G | Event-free survival in multiple myeloma | 340 | 84 | 256 | |
| H | Control parameter S1 (gender) | 340 | 194 | 146 | |
| I | Control parameter R1 (random) | 340 | 200 | 140 | |
| University of Cologne31 | J | Overall survival milestone outcome in neuroblastoma | 238 | 22 | 216 |
| K | Event-free survival milestone outcome in neuroblastoma | 239 | 49 | 190 | |
| L | Control parameter S (gender) | 246 | 145 | 101 | |
| M | Control parameter R (random) | 246 | 145 | 101 |
Table 2. Analysis teams25.
| Organization abbreviation | Organization name |
|---|---|
| ABT | Abbott Laboratories |
| Almac | Almac Diagnostics, UK |
| CAS | Chinese Academy of Sciences, China |
| CBC | CapitalBio Corporation, China |
| CDRH | Center for Devices and Radiological Health, FDA |
| CDRH2 | Center for Devices and Radiological Health, FDA |
| CIPF | Centro de Investigacion Principe Felipe, Spain |
| Cornell | Weill Medical College of Cornell University |
| Cornell2 | Cornell University |
| DKFZ | German Cancer Research Center, Germany |
| EPA | US Environmental Protection Agency |
| FBK | Fondazione Bruno Kessler, Italy |
| GeneGo | GeneGo Inc. |
| GHI | Golden Helix Inc. |
| GT | Georgia Institute of Technology, Emory University |
| JHSPH | Johns Hopkins Bloomberg School of Public Health |
| KU | University of Kansas |
| Ligand | Ligand Pharmaceuticals |
| NCTR | National Center for Toxicological Research, FDA |
| NIEHS | National Institute of Environmental Health Sciences |
| NWU | Northwestern University |
| Princeton | Princeton University |
| Roche | Roche Palo Alto LLC |
| SAI | Systems Analytics Inc. |
| SAS | SAS Institute Inc. |
| SDSU | South Dakota State University |
| SIB | Swiss Institute of Bioinformatics, Switzerland |
| SA | SuperArray Bioscience Corporation |
| Tsinghua | Tsinghua University, China |
| UAMS | University of Arkansas for Medical Sciences |
| UCLA | Cedars-Sinai Medical Center of UCLA |
| UIUC | University of Illinois at Urbana-Champaign |
| UML | University of Massachusetts Lowell |
| USM | University of Southern Mississippi |
| ZJU | Zhejiang University, China |
| GSK | GlaxoSmithKline |
Evaluation of network topology
Topology analysis gives information about how tightly the GeneGo network nodes from the explored data set are connected.
Degree is the average number of links (interactions) connected to a node (protein). As the GeneGo database of biological interactions includes directionality of effect, the nodes may be characterized by IN and OUT degree, giving the average number of outgoing and incoming interactions.
The clustering coefficient captures the degree connectivity between a node's neighbors. It is defined as: 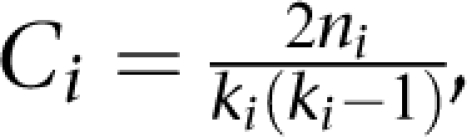
, where ni is the number of links among the ki neighbors of node i. As ki(ki–1)/2 is the maximum number of such links, the clustering coefficient is a number between 0 and 1. The average clustering coefficient is obtained by averaging over the clustering coefficient of individual nodes. A network with a high clustering coefficient is characterized by highly connected subgraphs.
Enrichment by protein classes
All signatures were analyzed for relative enrichment with certain protein classes. The results were ranked by a P-value. The P-values were calculated using the basic formula for a hypergeometric distribution (listed below), where r is the number of objects of particular protein class from the set of interest (signatures); R is the number of objects in the set of interest; n is the number of objects of particular protein class in the whole GeneGo global network; N is the number of objects in the GeneGo global network.
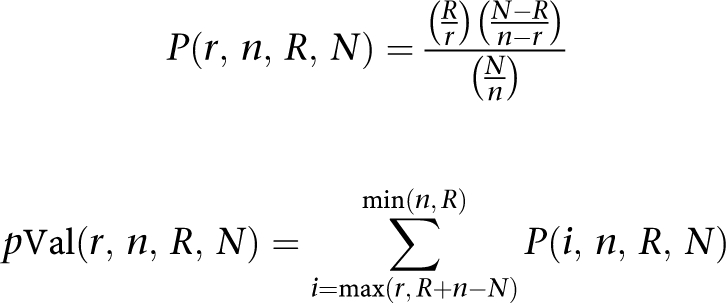 |
EA in functional ontologies
For FA, we used a number of public and proprietary functional ontologies in MetaCore v6.0 (http://www.genego.com). MetaCore includes the public ontologies GO biological processes (GO Processes), GO molecular function (GO Molecular Function) and GO cellular component (GO Localization), and the proprietary ontologies GeneGo Pathway Maps, GeneGo Biological Processes, GeneGo Disease Biomarker Networks, GeneGo Drug Target Networks, GeneGo Toxicity Networks, GeneGo Metabolic Networks, GeneGo Diseases (by Biomarkers) and GeneGo Metabolic Networks (Endogenous).
All signatures were analyzed for relative enrichment with certain categories from several functional ontologies, including GO and GeneGo cellular processes, canonical pathway maps, diseases or molecular functions. The results were ranked by P-value for a hypergeometric distribution where the P-value essentially represents the probability of particular mapping arising by chance, given the numbers of genes in the set of all genes on maps/processes/diseases/molecular functions, genes on a particular map/process/disease/molecular function and genes in the analyzed experiment. Significance cutoff was selected at P<0.05.
GeneGo canonical pathway maps comprise several hundred pictorial representations of human and rodent signaling and metabolic pathways. GeneGo cellular processes is a proprietary ontology of biological processes based on functionally interacting groups of genes. The GeneGo disease ontology contains more than 8000 genes with their known links to over 500 human diseases.
Relative connectivity of proteins inside the data set (intraconnectivity), and between the set and the global interactome
All signature genes were associated with their proteins, and all protein lists were screened for the number of interactions with the global interactome (GeneGo global network (interconnections)) and within the individual protein lists (intraconnections). All proteins were divided onto seven different functions (protein target classes): transcription factors (TFs), receptors, ligands, kinases, proteases, phosphatases and metabolic enzymes. The expected number of interactions for a given protein with (for interconnected)/ within (for intraconnected) the protein list is determined as a fraction of the total number of its interactions in the GeneGo global network proportional to the size of the protein list. If the number of interactions with (for interconnected)/within (for intraconnected) the protein list is larger than expected, the protein is considered overconnected; if the number of interactions is lower than expected, the protein is considered underconnected. The degree of over- and underconnectivity can be evaluated by z-score and P-value. The z-score signifies the difference between the obtained number of proteins and the expected average number of proteins corresponding to genes expressed in units of standard dispersion. P-values were calculated using the basic formula for a hypergeometric distribution listed above, where r=number of proteins derived from current protein list that have interactions with given protein; R=total number of proteins in the GeneGo global network that have interactions with given protein; n=total number of proteins in given protein list; and N=total number of proteins in the GeneGo global network.
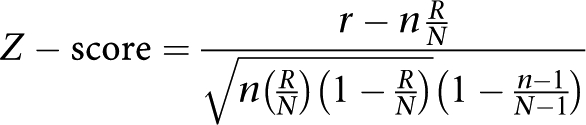 |
Upstream and downstream targets
All signatures were analyzed for the most frequent upstream targets that have outgoing links to genes in signatures, and downstream targets, which have incoming links from genes in signatures. The results were ranked by P-value. The P-values were calculated using a binomial distribution.
Calculating significance for topological properties of end points and signatures
Topological properties were calculated from the global protein interaction network database of MetaCore v6.0. The pool of genes present on the microarrays for each end point was randomly sampled 10 000 times, and multiple random sets of sizes corresponding to end points (union of signatures) were generated. Topological features such as largest direct interaction (DI) cluster size and average shortest paths were calculated for the random sets and compared to the topological properties of the corresponding end points. The average shortest path of a set was calculated as an average over the shortest paths between all possible gene pairs from the set in the global interaction network.
To assign significance to the sizes of the DI networks of end points, we evaluated the relative frequency of instances for which the random DI networks were larger than or equal to the DI network of the end points, and P-values were calculated. Similarly, the average shortest paths of end points was compared to the average shortest paths of corresponding random sets, and we evaluated the number of instances for which the random sets had smaller or equal average shortest paths, and P-values were calculated.
Similarly, we calculated the significance of the topological properties for signatures by randomly sampling the microarrays and generating random sets of the sizes corresponding to the sizes of the signatures.
Calculating significance for the overlap of signatures and end points
To assign a significance level to the overlap of signatures and end points, we generated 10 000 random samples of the same sizes as the signatures and end points from the pools of genes present on the microarrays. Next, we calculated the overlaps between the gene sets of signatures and end points and estimated P-values as relative frequency of instances in which the random sets had higher or equal overlap than the genes from the corresponding signatures and the end points.
Calculating network distance between sets of genes
The network distance between two sets of genes (A and B) was calculated as the average shortest path between all genes from A to B. This distance was normalized with the sizes of A and B. The distance can be written as:
〈SAB〉 is the average undirected shortest path between all genes from A to B. 〈sA〉 and 〈sB〉 are the average shortest paths between gene pairs within A and B.
κ Statistics
κ Statistics is a statistical measure of interrater agreement. The input for κ involves a couple of raters or learners, which classify a set of objects into categories. In this study, we used it to compare different signatures for their congruency. Similar to the design in Huang et al.,34 we consider each team as a learner or rater and each object (probe) in the union of signatures' probes is categorized by each team as 1 (selected) or 0 (unselected). Using this 0/1 matrix (object × learner) as input, we used the Cohen's κ function {concord} package in R35 to derive κ values, z-scores and P-values for congruency. Instead of using Cohen's κ,36 we used Siegel and Castellan's κ (1988), by assuming pooled classification proportions and an adjustment for bias, in which the different methods systematically differ in their categorization.37 Such calculations are also provided by the R package. The congruency can be between two raters (teams) as pairwise or on all teams as overall congruency. Our analysis is not only on signature genes, but also on a set of pathways (arrived at by EA) for each team, which calculates the congruency at the biological pathway level.
Pairwise trees at signature, pathway and network levels, and their comparisons
κ Statistics (z-scores) were calculated for signature congruency based on three parameters: feature intersection, EA (disease biomarkers) and network distance. The pairwise matrix was calculated and the cluster trees were plotted for feature intersection, pathway (disease) and network distance, respectively. The hierarchical tree was plotted using the hclust function of R35 and ‘complete linkage' as the agglomeration method.38 As z-scores can be negative and the higher z-scores correspond to shorter ‘distance', the ‘similarity' used a fixed positive number minus each z-score, while keeping the relative ‘distance' as the input for plotting the trees.
A general comparison of two trees is a matter of graph similarity. In this study, we introduced a simple but efficient approach for our hierarchical trees. Assuming that we are comparing signature tree with disease pathway tree for end point A: starting with ‘distance matrix', for each team, we selected its five nearest neighbors (if we set neighborhood=5) from the signature distance matrix and from the disease pathway distance matrix, respectively. Then for each of the 34 teams, we compared their 5 nearest neighbors as overlap ratios. The average of 34 such overlap ratios is considered as a statistic for similarity between signature tree and disease tree for end point A. Comparisons for other end points were calculated using the same approach.
Results
We analyzed all gene signatures produced by 33 teams for 13 end points (labeled A–M), a total of 262 gene lists between 3 and 200 probes in size. The number of signatures for different end points varied from 17 to 27, as not every team submitted signatures for all end points. In addition to analyzing individual signatures, we created nonredundant unions for each end point, comprising all the genes represented in one or more of the individual signatures for that end point. Signature unions varied in size from 92 (end point L) to 659 genes (F) (Supplementary Table 1).
Gene signatures for different end points vary in their composition by protein function
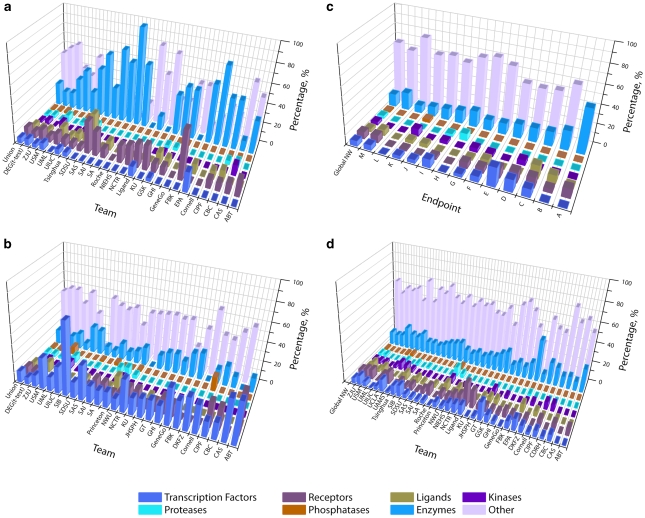
We divided genes for each signature into eight broadly defined encoded protein functions: TFs, receptors, ligands, kinases, proteases, phosphatases, enzymes and ‘other' (for example, unspecified binding proteins) (Supplementary File 1). We considered as controls the overall distribution of protein function in the MetaCore database (about 20 000 human proteins) and in the sets of differentially expressed genes (DEGs) for each end point. Signatures varied significantly in protein functions in an end point and model-dependent manner (Figure 1). Signatures C, D, E and I were enriched in TFs; A in metabolic enzymes; C, D and M: receptors; and H: proteases. The protein function enrichment pattern in signatures tended to reflect the biology of the corresponding phenotype. Signatures for end point A (lung tumorigenicity) featured a high fraction of xenobiotic-metabolizing enzymes. In contrast, signatures for end point E (breast cancer) were highly enriched with TFs, reflecting intensive transcriptional activity in invasive breast tumors.
Figure 1.
Protein function enrichment in signatures and unions for all end points. (a) Protein class distribution for end point A, individual signatures. (b) Protein class distribution for end point E, individual signatures. (c) Protein class distribution for union signatures for all 13 end points. (d) Protein class distribution for all end points; all signatures.
Signature network topology is end point dependent
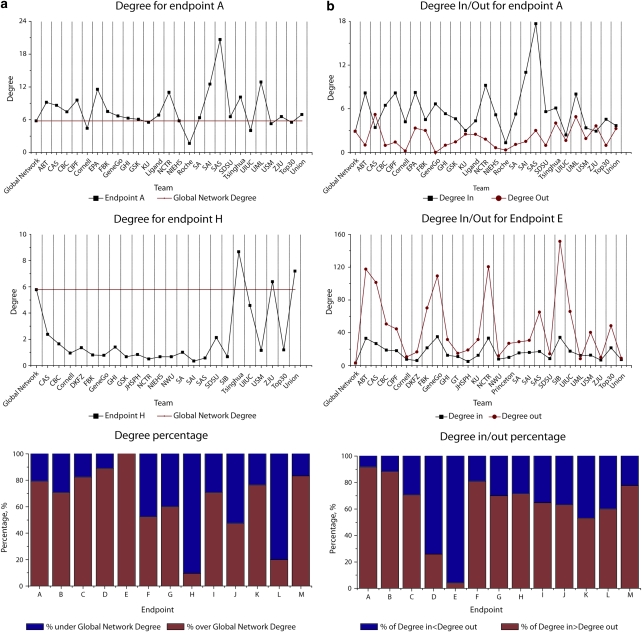
We evaluated the interconnectivity of signatures and their unions, relative to the general connectivity of the human interactome (about 300 000 experimentally validated human protein interactions recorded in MetaCore v6.0). Specifically, we calculated the relative enrichment of signature genes with IN and OUT hubs (proteins with the highest number of incoming (upstream) and outgoing (downstream) interactions, correspondingly)39 (Figure 2; Supplementary File 3). Signature genes generally encoded proteins with more interactions than the average human protein. In all, 10 out of the 13 union signatures were enriched in hubs (highly connected proteins with many interactions). The degree of enrichment with IN or OUT interactions reflected the observed protein function distribution: a high degree of IN was observed with a high fraction of ‘effector' proteins such as core metabolic enzymes (end points A, B and C, which represent pathological responses to xenobiotic exposures), whereas a high degree of OUT is indicative of an enrichment in TFs (end point E). Union signatures for 11 of the 13 end points were enriched with IN interactions compared with the global interactome (interactions driven by an upstream protein, such as transcriptional regulation or phosphorylation).
Figure 2.
Network topology enrichment for signatures and unions for all end points. (a) Enrichment in ‘degree'—average number of interactions per gene in signatures for end points A, E and median signatures for all end points. (b) Distribution of degrees IN and degrees OUT for end points A and E and for all end points.
Signature genes are interconnected, but connectivity does not correlate with signature performance
An important measure of functional connectivity between genes is the ability of their protein products to form DI networks.40 The size of the DI network and the probability (P-value) of assembling a DI network with the same number of nodes from a random set of genes serves as a quantitative measure of interconnectivity (see Materials and methods for details). We built DI networks for all 262 signatures and 13 unions (Supplementary Table 2; Supplementary File 4). The ability of signature gene lists to generate DI networks was dependent on the phenotype and the statistical method used to derive the signature. No significant DI networks were assembled from any signature for end points A, B, H and L. Conversely, between 45 and 85% of signatures assembled into statistically significant DI networks for end points C, D, E and K. Signatures generated by certain analysis groups (likely reflecting the methodology used) formed DI networks more consistently than other teams' signatures. Over 40% of signatures derived by the SAI, Tsinghua and ZJU teams formed networks. Signatures from other teams, however, did not assemble into DI networks regardless of the end point (DKFZ, GT, JHSPH).
Signature unions for 10 of the 13 end points formed statistically significant DI networks (Supplementary File 4). No networks were formed by the unions of end points B, H and L. Altogether, 22 out of 33 teams generated signatures capable of forming statistically significant DI networks, indicating that the genes comprising most signatures were interconnected to a significantly higher degree than expected for a random gene list of the same size. Therefore, we conclude that the predictive models used tended to select interconnecting genes. We saw no correlation between signature performance and interconnectivity between its genes, however, so there does not appear to be any relationship between the biological interconnectedness of signature genes and their predictive power.
EA of gene signatures and unions
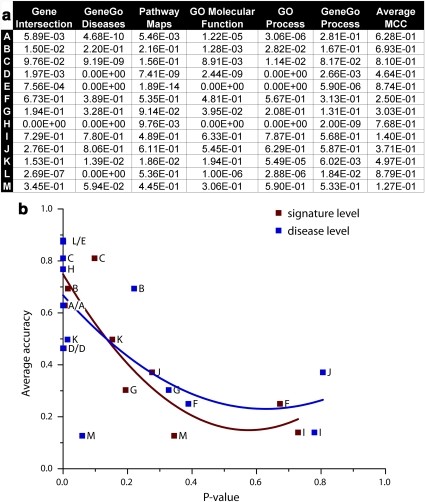
EA is a standard FA method to highlight and rank biologically relevant pathways, processes, disease markers or other functionality in a gene list. Relative enrichment can be evaluated by hypergeometric distribution P-value,41 gene set enrichment analysis score13 and other metrics. We subjected all 262 signatures, signature unions and sets of DEGs (as defined by t-test) for each end point to EA across five ontologies in MetaCore: canonical pathway maps, GeneGo process networks, GeneGo disease biomarkers, GO cellular processes and GO molecular functions (Supplementary File 5, Supplementary Archive 2). We also tested consistency in ontology distributions between signature unions, all DEGs and end point phenotypes. The enrichment pattern for both unions and DEGs was highly end point dependent, and, in most cases, matched the biology of the end point phenotype. For example, unions A and C (xenobiotic-induced lung tumor and liver necrosis, respectively) were highly enriched in pathways for oxidative stress response, glutathione metabolism, and drug-metabolizing enzymes and their transcriptional regulation by NRF2 and CAR. Unions D and E (breast cancer end points) featured enrichment in breast cancer biomarkers and pathways characteristic of invasive carcinogenesis, such as cell adhesion, PLAU signaling, estrogen receptor signaling and apoptosis. Unions F and G (multiple myeloma) and J and K (glioblastoma) were enriched in cell-cycle pathways, and checkpoint proteins typical of early cancer development. Interestingly, the average model performance for an end point (evaluated by the Matthew's correlation coefficient, MCC) was strongest when the union's enrichment in the disease biomarkers ontology best reflected the phenotype of the end point. MCC for end point C was the highest among the three xenobiotic-induced pathology end points. The signature union for C (overall necrosis score) was enriched for ‘drug toxicity' (P<E−17); ‘drug toxicity' was less highly enriched (P-value in the E−6 range) for A and B (carcinogenicity), which were also less accurately classified by signatures. In the breast cancer data set, signatures for end point E (estrogen receptor status) had a higher MCC than those for D (treatment response), and a very strong enrichment for breast cancer-related genes (P<E−16) was seen in the E signature union. For end point A, the enrichment pattern of DEGs was more consistent with the end point phenotype (chemically induced lung cancer in mice) than that of the signature union. The signature union for A showed two clear trends: general drug-induced toxicity and specific lung cancer responses. Lung cancer-related pathways and biomarkers were also enriched in DEGs for end point C (liver necrosis) (P<E−8 for lung neoplasms, compared with P<E−4 for end point A signature union genes). DEGs for end point A were enriched with carcinogenesis pathways and processes, whereas signature union genes for A were enriched with drug response and oxidative stress pathways (Supplementary File 5). MCC performance for A was also the worst among the three xenobiotic response data sets.
Ontology enrichment of outlier unions
FA revealed four outlier signature unions, those for end points H, I, L and M. These unions showed dramatic discordance with the corresponding data sets, and clear differences to other unions, but for very different reasons.
End points I and M were ‘dummy' end points, defined by randomly generated label sets and introduced as a negative control for model performance. In general, genes in signatures for I and M had a model-dependent functional bias similar to other end points. Thus, I and M signatures were enriched in hub genes, similar to others. Unions for I and M formed statistically significant DI networks, and contained common housekeeping genes, as seen for other signatures. Moreover, P-value distributions in ontology EA for I and M union signatures was characteristic of functionally cohesive data sets. These data suggests a systematic topology bias toward highly connected genes implicated in most models. However, ontology enrichment was in striking dissonance with the expected functionality of the original data sets. For the disease biomarker ontology, both I and M were highly enriched in genes associated with cardiac and blood vessel diseases (P<10−8), highly discordant with the cancer-related phenotype of the data sets, multiple myeloma and neuroblastoma, correspondingly. Similarly, distributions in other ontologies were enriched with pathways and processes also inconsistent with the cancer source of the data sets, but consistent with cardiovascular development (Supplementary File 5).
End points H and L were composed of gender class labels as a positive control for model performance. These also were clear outliers in most of our analyses. The results of EA on union signatures for H and L was inconsistent with the expected cancer phenotype. The P-values for all ontologies (with the exception of disease biomarkers) were several orders of magnitude higher than that for other unions (except the dummy end point, J). Moreover, the enriched diseases were linked to male and female reproductive systems, rather than cancer, including MeSH terms (http://www.nlm.nih.gov/mesh) for urogenital diseases, gonadal dysgenesis and sex-differentiation disorders. Enriched GeneGo and GO processes for H and L included male sex differentiation and androgen receptor signaling, and reproductive categories such as spermatogenesis (Supplementary File 5). Signatures for both H and L were enriched in underconnected genes, in stark contrast to the other signatures, which were enriched in highly connected hubs. Interestingly, signatures for these two end points were most similar in gene content in a pairwise comparison of all signature unions (Supplementary File 6), and these were the only end points for which the unions failed to form statistically significant DI networks. The functional composition of signatures for H and L, although they differ from those of the other end points, reflect their corresponding class label (gender), and indicate that classification for this phenotype is made on the basis of genes involved in gender-specific processes. The consistency in composition of signature unions for H and L underlines the robustness of the models in selecting end point-relevant genes from very different data sets.
In addition to highlighting the biological relevance of gene signatures, the ability of our functional analyses to clearly differentiate the positive- and negative-control label sets from the other end points, and to reveal the sex-related nature of genes that classify gender from different data sets, demonstrates the validity of the approach in investigating the biological basis of classifier gene signatures for other phenotypes.
Signature similarity
A key issue in disease biology is the heterogeneity of human samples and the problem of subclustering of samples in a study (cohort) in a clinically relevant way to allow phenotypic anchoring of gene expression data to well-defined clinical classes. Traditionally, expression data sets are clustered based on sharing DEGs between samples.4 However, recent data suggests that shared functionality (differentially affected pathways, subnetworks) may represent a more robust classifier than gene expression alone.17, 40 We addressed this issue by investigating the similarity (congruency) between different signatures for the same end point at three levels. First, the degree of overlap in gene content was measured. Second, congruency at the level of gene functionality (biological pathways, processes and disease associations) was investigated. Third, we measured topological congruency by the average distance between the signature gene sets within the global interactome.
Similarity based on gene content
We calculated pairwise intersections (overlaps) of gene content between signatures and evaluated their significance by P-value. Signatures formed statistically significant intersections in an end point-dependent manner, ranging from 38% of pairs (end point J) to 100% (end point H) (Supplementary File 6). Importantly, some genes were consistently repeated between signatures and unions. Common genes between unions included housekeeping genes, such as the 60S and 40S ribosomal subunit genes (10 and 9 unions, respectively), elongation factor 1-α (7 unions) and ribonucleotide reductase (5 unions) (Supplementary Table 3). Altogether, 9 of the 39 most common genes encoded immunoglobulins of different A, G, E and M complexes, particularly in breast cancer and multiple myeloma signature unions. The most common genes were generally enriched with IN interactions (10.5–24.5 interactions per protein on average).
Pairwise similarity between signatures was calculated by κ values, z-score and P-value metrics, taking into account both unique and common features (probes or probe sets) between signatures (Supplementary File 7). κ Values (value, z-score and P-value) were calculated for each pair of signatures for each phenotype. The higher the z-score, the lower the P-value and the higher the congruency. Degree of congruency varied between teams. For instance, the GSK team's signatures for A were similar to signatures from SDSU (P=0) and GHI (P=0), but dissimilar to NCTR (P=0.52) or CAS (P=0.41) signatures. Some signatures, for instance, those from the ZJU team, were dissimilar to all others (minimum pairwise P-values for ZJU=0.77). Gene content-based congruency was also end point specific (Supplementary File 8).
Similarity based on ontology enrichment
A pattern of distribution in EA can be used as a measure of similarity between gene lists or experiments. We evaluated congruency between all signatures within and across end points using κ statistics for the enrichment of all 262 signatures across 5 ontologies (see Materials and methods). For instance, for each end point, the composition of units of the disease ontology with P<0.05 were compared according to pairwise.path.z-score and pairwise.path.P-value (Supplementary File 9). Similarity between signatures is shown in a hierarchical tree (Supplementary File 8). Pairwise congruency (P-values) at the pathway level varied significantly in an end-point-dependent manner (Supplementary File 8).
Similarity based on network distance
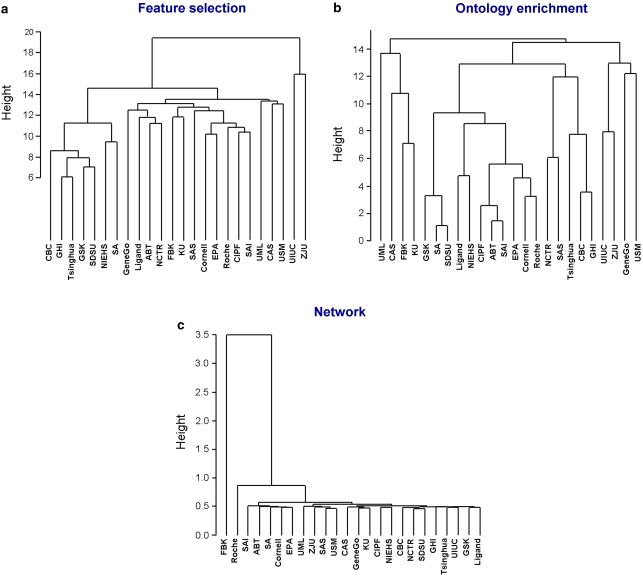
Similarity between gene sets can also be assessed based on the network distance between them. The topological distances between all signature pairs were calculated as the ratio of the average shortest path between proteins belonging to different signatures to the average shortest paths between proteins from the same signature. Hierarchical clustering38 was applied to generate signature trees. Signatures characterized by proximity on the global interaction networks were grouped together (for example SAI, ABT, SA, Cornell, EPA in Figure 3), whereas other signatures (FBK in Figure 3) were characterized by large network distances from all other signatures.
Figure 3.
Signature similarity for end point A based on (a) feature content; (b) ontology enrichment; (c) network closeness. Using the κ statistics, we generated hierarchical trees using z-scores as a measurement of distance. Trees for the 12 other end points are given in Supplementary File 9.
The hierarchical trees generated from gene content, ontology enrichment (disease biomarkers) or network distance measure relative pairwise closeness of the signatures. Trees for the same end point are therefore directly comparable (Supplementary Table 4). t-Test analysis showed that different signatures for the same end point are more similar when compared by gene content and ontology enrichment, than when compared by network similarity (Supplementary Table 3).
We further evaluated whether gene content or ontology enrichment is more relevant as a measure of similarity. Each signature was pairwise compared with every other signature by gene content and ontology using κ P-value (Supplementary Table 5). At a cutoff of P=0.05, we calculated pairwise changes from nonsignificance at the gene level to significance at the enrichment level, and changes from significance by genes to nonsignificance by enrichment. For signatures from the ABT team, the values were 15 and 0, meaning that signatures from 15 other teams became significantly more congruent to those from ABT when gene overlap was substituted with ontology overlap. No teams' signatures became more significant in the other direction. The difference for ABT is 15 (15−0), compared to the null hypothesis of zero. We repeated the above process for each team for every end point (Supplementary Table 6). For end points A, C, D, E, F, Land M, the GeneGo Disease Biomarkers ontology produced significantly higher pairwise congruency than with gene content. For end points H and K, the P-values were nonsignificant following multiple test adjustment. For end points B, G, I and J, P-values were higher than 0.2 (not significant). The P-value for the same t-test across all 13 end points simultaneously was 5.3E−19, confirming that congruency at the level of functional ontologies was significantly higher than by gene content.
Correlation between signature similarity and model performance
We evaluated the correlation between signature similarity (measured as κ at the level of feature intersection and ontology congruency, Figure 4a) to model performance (internal validation on the training sets and external validation). The congruence value was compared to the average model performance for an end point (Figure 4b, Supplementary Table 7). Overall, we saw a strong positive correlation (P<0.001) between signature congruency and model performance, suggesting that certain phenotypes have a stronger characteristic transcriptional fingerprint than others, and are therefore more amenable to addressing with the gene signature approach.
Figure 4.
Correlation between signature similarity and model performance. (a) Correlation between signature intersection and ontological enrichment similarity (five ontologies) for 13 end points. (b) Correlation between signature similarity and model performance. The similarity was calculated by κ statistics as P-values among all submitted gene lists for each end point. MCC was used to evaluate model performance.
Transcriptional regulators of signature genes, and genes regulated by signature genes, are end point related
Proteins functionally important for a particular phenotype have many interactions with proteins encoded by genes differentially expressed in the phenotype.16 This is logical, as proteins function in physically connected groups (complexes, pathways, network modules) and functionally related genes are co-regulated by TFs. To identify the ‘most relevant' (that is, most connected) proteins for each end point, we evaluated (1) for each protein from a given signature, relative connectivity between it and other proteins from the signature (intraconnectivity); (2) for each protein from a given signature, relative connectivity between it and all proteins from the human proteome (defined by the MetaCore database of 20 000 human proteins and 300 000 directional protein interactions20 (Figure 5, see Materials and methods). Proteins were considered overconnected when the number of observed interactions exceeded the number of expected interactions42 (details in Materials and methods). In general, connectivity between genes within individual signatures (intraconnectivity) was low (Supplementary File 10). However, several TFs were overconnected with the genes from multiple signatures for end points C, D, E, F, G, J and K (Supplementary File 10). For instance, ATF3 regulates (physically binds to the promoters of) genes from five signatures. ESR1 regulates genes from 7 and 15 signatures for end points D and E, correspondingly.
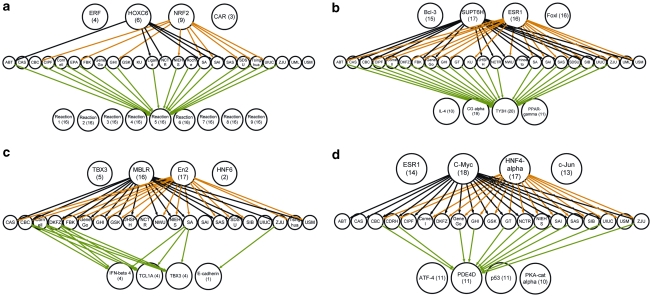
Figure 5.
Interactome analysis of signature gene lists. The most frequent upstream and downstream genes for each signature generated for end points A (a), E (b), H (c) and J (d).
Considering all interaction mechanisms, we found that signature genes were not overconnected with the global proteome, with the exception of end points A, F and K. However, signatures for all end points featured a disproportionably large fraction of targets of particular TFs (defined as overconnected by transcription regulation interactions) (Supplementary File 10). Most signature genes were regulated by very few upstream TFs in a highly end-point-specific manner (Supplementary File 10). For instance, 9 out of 24 signatures for end point A were regulated by NRF2 (a key TF in controlling cellular oxidative stress response). Of 24, 17 signatures for E included direct targets of forkhead family TF HNF3-α (epithelial transcription) and of 24, 16 included direct targets of ESR1. Of 21, 15 signatures for H were co-regulated by both En2 (developmental homeo-domain-containing TF) and MBLR (developmental polycomb group TF) (Supplementary File 10). Downstream signaling from signature genes also was highly clustered. Signature genes regulated a limited number of downstream genes and reactions (Supplementary File 11). Out of 24, 16 signatures for end point A contained drug-metabolizing enzymes; out of 24, 20 signatures for E contained genes regulating tyrosine 3-monooxygenase TY3H; 19 contained genes regulating CG-α and 11 contained genes regulating interleukin-4. In end point H, downstream signaling is seen with only 4 out of 20 signatures (those from CBC, Cornell, DKFZ and SA), which regulate IFN-β, TCL1A and TBX3.
Discussion
Our observations suggest that gene signatures derived statistically and by machine-learning algorithms do not independently make biological sense in the context of functional biological units such as pathways. Instead, functional correlations manifest only at the level of genotype–phenotype association and protein interaction, where signatures are logically distributed across ontologies of cellular processes, pathways and biomarkers, and physically connected into significant networks. Correlation through ontology enrichment was particularly pronounced for the nonredundant gene unions of all signatures for a given phenotype (lower distribution P-values for unions than for individual signatures). This suggests that different statistical models selected different subsets of genes from the same pathways and processes, and highlights the redundancy of molecular signatures where variable selection can lead to many quantitative solutions of equal reliability in terms of prediction rates.43, 44 Biological pathways are partly reconstructed from individual signatures in signature unions.
Signatures selected by different methods share certain features across all end points. Most signatures and all unions were enriched in hubs (the 25% highest-connected human proteins). Moreover, signature genes predominantly encode IN proteins, with most of their interactions being upstream. A high ratio of IN to OUT interactions is typical of effector proteins such as metabolic enzymes, cytoskeleton and cell matrix proteins—housekeeping genes encoding homeostatic functions. Genes encoding effector proteins are more likely to show condition-related changes in expression levels than transiently expressed regulatory genes, contributing to a higher probability of selection in predictive models.
Phenotype (end point) dependency of gene signatures is evident from multiple analyses. In addition to a high fraction of OUT interactions, end points D and E (breast cancer phenotypes) featured the largest number of statistically significant DI networks, both within their individual signatures, and within their signature unions. The number of intersignature links was also the highest for the breast cancer end points D and E. Breast cancer is a complex and heterogeneous disease with many different subtypes, involving hundreds of pathways and processes. Models applied by different teams likely selected genes responsible for different, yet related processes of carcinogenesis. Interestingly, some signatures were enriched in regulatory genes and some in regulated genes within the highly connected, relatively small unions for end points D and E.
Some genes were consistently represented in multiple signatures across different end points. A total of 38 genes were selected in at least 4 out of 13 union signatures. Two genes, encoding 60S and 40S rRNA, were selected in 10 and 9 unions, respectively. eEF1A1 (elongation factor 1) was chosen in seven unions, including both xenobiotic-induced pathology and cancer end points. A number of DNA exchange and protein biosynthesis genes made the ‘top 38', including CENP-A (histone H3-like centromeric protein A), ribonucleotide reductase, Holliday junction recognition protein and POLE2 (DNA polymerase-ɛ subunit 2). This suggests that the core cellular functions are profoundly altered in both drug response and homeostasis-changing diseases such as cancer. A large number of genes encoding immunoglobulins appeared in signatures for multiple end points (11 out of 38 top genes), partly due to their high relevance for breast cancer and multiple myeloma end points.
Although housekeeping genes are highly represented in multiple unions, individual unions are also enriched in tissue-specific genes, evident from EA in disease biomarkers (Supplementary File 6; Supplementary File 14). With the exception of the ‘dummy' data sets I and M, unions are enriched with disease-associated genes in a manner highly consistent with the end points (P-value range E−6 to E−17 for different end points; Supplementary File 14). It was recently shown that tissue-specific genes are twice as likely as housekeeping genes to be associated with diseases.45
Signatures for the same end point showed both similarity (or congruency—shared gene content and enriched ontologies) and synergy (interconnectivity). We compared congruency by two statistical approaches at both the gene content and the functional level (disease biomarkers, canonical pathways, GO and GeneGo cellular processes). Congruency at the functional level was consistently higher than at the feature (gene) level for all end points. A logical explanation for this observation is that different statistical models may select different sets of genes from the same biological categories. Higher pathway congruency supports the assumption of common underlying biological mechanisms for each end point. Signature congruency is in agreement with the observation of ‘synergy' between signatures in ontology EA, evidenced by lower P-values for unions compared with the individual signatures for the same end point42 (data not shown). These two observations suggest that FA procedures are robust and efficient tools for the measurement of similarity between data sets and gene lists. The pathway/ontology congruency technique we developed will be useful in such critical applications as patient cohort stratification and clustering of clinical samples in biomarker discovery, with important applications in personalized medicine for precise diagnosis and treatment choice.
A relationship between signature congruency at the gene level (gene list stability) and average prediction performance was noted,25 and we further observed a correlation between signature congruency at the functional level and model performance on both training and validation sets. In general, the more congruent the signatures, the better the average prediction performance. This is an unexpected and important finding. The better performing sets of predictor genes appear to be biologically related, and are more reflective of the phenotype. The congruency of different gene classifiers for a given phenotype or outcome may therefore also be a useful indicator of the amenability of the end point to accurate prediction using the signature approach.
For every end point, different signatures were co-regulated by surprisingly few significantly overconnected TFs. Signature genes also had a small number of interactions with overconnected downstream genes and reactions. Typically, the common regulators and regulated genes do not belong to the signatures they regulate. The common regulators and regulated genes are likely to be important in the biology of the phenotype and can potentially be used as conditional biomarkers.
Overall, our analysis suggests that although gene signatures, when analyzed individually, are difficult to link to the predicted phenotype (end point), the genes identified by various methodologies do have biological relevance. The union of signature genes for each end point provided a better indication of the underlying biology of the phenotype studied. Signatures for a given end point were also congruent at the level of protein functionality and interconnection. Phenotypes that showed a greater biological homogeneity, evidenced by higher functional overlap between different signatures for the end point, were likely to generate better-performing gene signatures overall.
When generating predictive signatures for critical applications, the use of multiple statistical feature selection approaches, several different classification algorithms or a recursive feature elimination approach (or a combination of these approaches) may be employed to generated multiple, independent classifying gene sets to predict the same outcome. A comparison of these signatures at the feature level and at the functional, biological level (biological processes, diseases and interactions), would give confidence both that the final choice of predictive model is likely to perform robustly in prospective application, and that the classification is biologically sound. The approaches presented here, alongside the wealth of information on best practices for signature generation and performance assessment resulting from the MAQCII project, will enhance the development, analysis and appropriate use of gene signatures, and their application to compound safety assessment, disease diagnosis and personalized medicine.
Glossary
- DEGs
differentially expressed genes
- DI
direct interactions (network)
- FA
functional analysis
- GO
gene ontology
- GSEA
gene set enrichment analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAQCII
Microarray Quality Control Consortium II
- TF
transcription factor
- TR
transcriptional regulation (an interaction mechanism).
Dr Russell Thomas' research was supported by the American Chemistry Council's Long-Range Research Initiative and a contract by Honeywell Inc. All other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoi H, Potti A, Delorenzi M, Mauriac L, Campone M, Tubiana-Hulin M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–1078. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- Eun JW, Ryu SY, Noh JH, Lee MJ, Jang JJ, Ryu JC, et al. Discriminating the molecular basis of hepatotoxicity using the large-scale characteristic molecular signatures of toxicants by expression profiling analysis. Toxicology. 2008;249:176–183. doi: 10.1016/j.tox.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Huang Y, Penchala S, Pham AN, Wang J. Genetic variations and gene expression of transporters in drug disposition and response. Expert Opin Drug Metab Toxicol. 2008;4:237–254. doi: 10.1517/17425255.4.3.237. [DOI] [PubMed] [Google Scholar]
- Natsoulis G, El Ghaoui L, Lanckriet GR, Tolley AM, Leroy F, Dunlea S, et al. Classification of a large microarray data set: algorithm comparison and analysis of drug signatures. Genome Res. 2005;15:724–736. doi: 10.1101/gr.2807605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troester MA, Hoadley KA, Parker JS, Perou CM. Prediction of toxicant-specific gene expression signatures after chemotherapeutic treatment of breast cell lines. Environ Health Perspect. 2004;112:1607–1613. doi: 10.1289/txg.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolsky Y, Bryant J.Protein Networks and Pathway Analysisvol. 563.Human Press: Clifton, NJ; 2009345p. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14 (Spec No. 2):R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- Vidal M. Interactome modeling. FEBS Lett. 2005;579:1834–1838. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Fischer HP. Towards quantitative biology: integration of biological information to elucidate disease pathways and to guide drug discovery. Biotechnol Annu Rev. 2005;11:1–68. doi: 10.1016/S1387-2656(05)11001-1. [DOI] [PubMed] [Google Scholar]
- Kiechle FL, Zhang X, Holland-Staley CA. The -omics era and its impact. Arch Pathol Lab Med. 2004;128:1337–1345. doi: 10.5858/2004-128-1337-TOEAII. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Wang SJ, Tsai CA, Lin CJ. Selection of differentially expressed genes in microarray data analysis. Pharmacogenomics J. 2007;7:212–220. doi: 10.1038/sj.tpj.6500412. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Schaffer J, Boldrick J. Multiple hypothesis testing in microarray experiments. Stat Sci. 2003;18:71–103. [Google Scholar]
- Consortium TMQC The MAQC-II Project: a comprehensive study of common practices for the development and validation of microarray-based predictive models Nat Biotechnol(in press). [DOI] [PMC free article] [PubMed]
- Boulesteix AL, Slawski M. Stability and aggregation of ranked gene lists. Brief Bioinform. 2009;10:556–568. doi: 10.1093/bib/bbp034. [DOI] [PubMed] [Google Scholar]
- Barla A, Jurman G, Riccadonna S, Merler S, Chierici M, Furlanello C. Machine learning methods for predictive proteomics. Brief Bioinform. 2008;9:119–128. doi: 10.1093/bib/bbn008. [DOI] [PubMed] [Google Scholar]
- Shen R, Chinnaiyan AM, Ghosh D. Pathway analysis reveals functional convergence of gene expression profiles in breast cancer. BMC Med Genomics. 2008;1:28. doi: 10.1186/1755-8794-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci. 2007;99:90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Pluta L, Yang L, Halsey TA. Application of genomic biomarkers to predict increased lung tumor incidence in 2-year rodent cancer bioassays. Toxicol Sci. 2007;97:55–64. doi: 10.1093/toxsci/kfm023. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman RR. A language for data analysis and graphics. J Comput Graph Statist. 1996;5:299–314. [Google Scholar]
- Cohen J. A coefficient of agreement for nominal scales. Edu Psychol Measure. 1960;20:37–46. [Google Scholar]
- Siegel S, Castellan NJ. Nonparametric Statistics for the Behavioral Sciences. McGraw-Hill: New York; 1988. [Google Scholar]
- Murtagh F.Multidimensional Clustering Algorithmsvol. 4.Physica-Verlag: Wien-Würzburg; 1985 [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J Roy Stat Soc. 1922;85:87–94. [Google Scholar]
- Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V, et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 2008;68:9532–9540. doi: 10.1158/0008-5472.CAN-08-3082. [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G, Pearson CI, Gollub J, Eynon PB, Ferng J, Nair R, et al. The liver pharmacological and xenobiotic gene response repertoire. Mol Syst Biol. 2008;4:175. doi: 10.1038/msb.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezso Z, Nikolsky Y, Sviridov E, Shi W, Serebriyskaya T, Dosymbekov D, et al. A comprehensive functional analysis of tissue specificity of human gene expression. BMC Biol. 2008;6:49. doi: 10.1186/1741-7007-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobenhofer EK, Auman JT, Blackshear PE, Boorman GA, Bushel PR, Cunningham ML, et al. Gene expression response in target organ and whole blood varies as a function of target organ injury phenotype. Genome Biol. 2008;9:R100. doi: 10.1186/gb-2008-9-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.