Abstract
GW182-family proteins are essential for microRNA-mediated translational repression and deadenylation in animal cells. Here we show that a conserved motif in the human GW182 paralog TNRC6C interacts with the C-terminal domain of polyadenylate binding protein 1 (PABC) and present the crystal structure of the complex. Mutations at the complex interface impair mRNA deadenylation in mammalian cell extracts, suggesting that the GW182-PABC interaction contributes to microRNA-mediated gene silencing.
MicroRNAs (miRNAs) are short endogenous RNAs that regulate a variety of eukaryotic cell processes, including cell growth, proliferation and differentiation. In Metazoa, miRNAs associate with Argonaute (Ago) proteins in miRNA-induced silencing complexes (miRISCs) to silence target mRNAs by a mechanism involving translational repression and concurrent stimulation of mRNA deadenylation and subsequent degradation1, 2.
In addition to Ago proteins, miRNA-mediated silencing requires proteins of the GW182 family3. Tethering of GW182 proteins to mRNA reporters induces translational repression and mRNA decay in the absence of miRNAs and Ago, suggesting that GW182 proteins have silencing functions downstream of Ago4. A direct interaction between Ago and GW182, mediated by glycine-tryptophan (GW) motifs found in the N-terminal region of GW182, is essential for miRNA-driven repression4–6. Apart from the N-terminal GW region, GW182 proteins also contain a ubiquitin-associated (UBA) domain followed by a glutamine-rich (Q-rich) region and an RNA recognition motif (RRM) domain. The sequence connecting the Q-rich region and the RRM domain contains a conserved motif known as DUF (domain of unknown function) in mammalian GW182 proteins7 or motif III in Drosophila melanogaster dGW1828. The silencing activity of GW182 proteins can be attributed to a C-terminal region containing the DUF and RRM domains7–10.
Polyadenylate-binding protein 1 (PABPC1) is present in Ago- and GW182-containing complexes immunoprecipitated from mammalian cells11–13. A bona fide miRNA coactivator in mammalian cells, PABPC1 directly interacts with the C-terminal silencing region of the human GW182 paralog TNRC6C13. Consistently, PABPC1 co-immunoprecipitates with the silencing domain of Drosophila melanogaster dGW18214. Both full-length human TNRC6C and a fragment containing the DUF and RRM domains could be co-precipitated with glutathione-S-transferase (GST)-tagged PABPC1 from transiently transfected HEK293T cells (Supplementary Fig. 1). A TNRC6C fragment lacking the DUF domain did not interact with GST-PABPC1, indicating that the DUF domain in TNRC6C is required to mediate the TNRC6C-PABPC1 interaction.
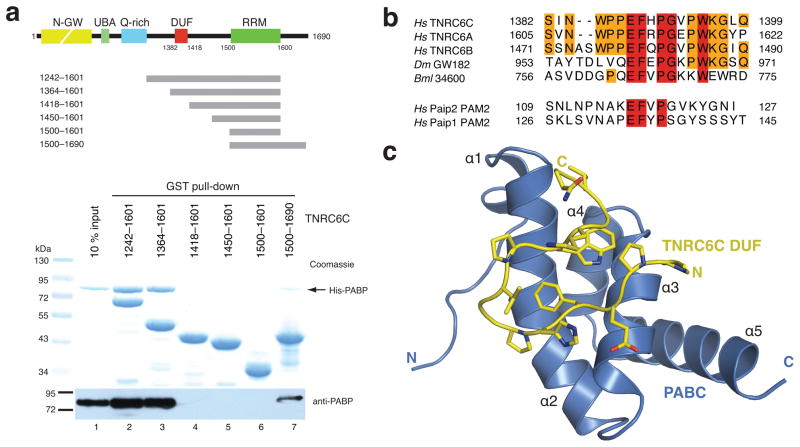
To confirm a direct interaction, we performed in vitro co-precipitations using recombinant PABPC1 and a series of GST-fused TNRC6C fragments. Fragments encompassing residues 1242-1601 and 1364-1601 efficiently precipitated PABPC1, while shorter fragments spanning residues 1418-1601, 1450-1601 and 1500-1601 did not, indicating that the DUF domain harbors a PABPC1 interaction site (Fig. 1a). Additionally, a TNRC6C fragment covering residues 1500-1690 also precipitated PABPC1, albeit less efficiently (lane 7, Fig. 1a), but did not precipitate His-tagged T7 RNA polymerase (Supplementary Fig. 2). This suggests that the region in TNRC6C downstream of the RRM may constitute a second interaction site that binds PABPC1 independently of the DUF domain. The relative weakness of this interaction might explain why it was not detectable in co-precipitations from mammalian cell lysates.
Figure 1.
The DUF domain of TNRC6C interacts with the C-terminal domain of PABPC1. (a) Top: Domain structure of the human GW182 paralog TNRC6C and schematic representation of TNRC6C fragments. Bottom: Recombinant glutathione-S-transferase (GST)-fused TNRC6C fragments were immobilized on glutathione sepharose beads and incubated with recombinant full-length His-tagged human PABPC1. Precipitated proteins were resolved by SDS-PAGE and analyzed by staining with Coomassie blue and immunoblotting using an anti-PABPC1 antibody. (c) Alignment of GW182 DUF domain sequences from human (Hs), Drosophila melanogaster (Dm) and Brugia malayi (Bml) GW182 proteins with the PAM2 motifs from human Paip2 and Paip1. Strictly conserved residues are highlighted in red; substantially conserved residues are orange. (d) Structure of the human TNRC6C (yellow)-PABC (blue) complex.
A highly conserved motif in the DUF domain of TNRC6C resembles the PAM2 motif found in the PABPC1-interacting proteins 1 and 2 (Paip1 and Paip2)13, proteins that positively and negatively regulate translation, respectively15, 16 (Fig. 1b). The PAM2 motif is required for Paip1 and Paip2 to interact with the C-terminal domain of PABPC1 (hereafter referred to as PABC)17–19, implying a similar role for the DUF domain of GW182-family proteins in binding to the PABC domain13.
To obtain structural insights into the PABC-GW182 interaction, we determined the 1.5 Å crystal structure of a reconstituted complex consisting of the PABC domain (PABPC1 residues 545-619) and an 18-amino acid peptide (residues 1382-1399) derived from the DUF domain of TNRC6C (Supplementary Methods and Supplementary Table 1). The DUF peptide forms a beta-turn hairpin that contacts helices α2 and α3 of the PABC domain (Fig. 1c). The TNRC6C peptide binds to the same face of the PABC domain as recognized by the PAM2 motifs of both Paip1 and Paip218 (Supplementary Fig. 3a). However, in contrast to the Paip1 and Paip2 complexes, the PABC domain does not appear to undergo a conformational change upon TNRC6C peptide binding, as helix α2 of the PABC domain remains uninterrupted in the TNRC6C-PABC complex (Supplementary Fig. 3b,c).
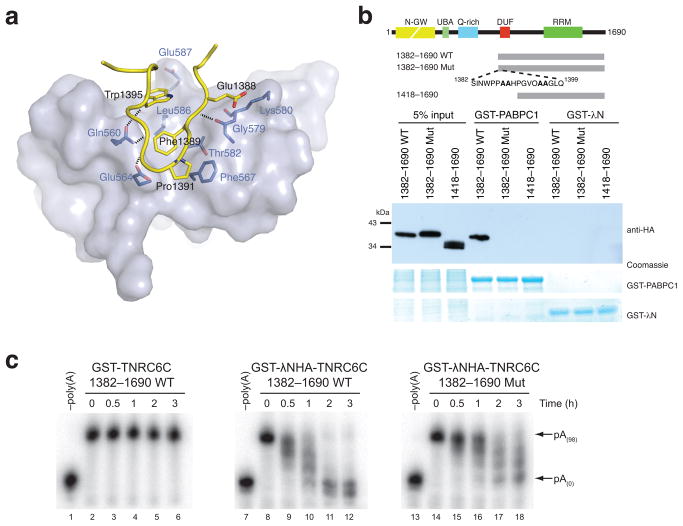
The 567 Å2 complex interface features an extensive network of hydrophobic and hydrogen-bonding interactions. The side chains of the invariant TNRC6C residues Phe1389 and Trp1395 pack against each other and insert into a hydrophobic groove between helices α2 and α3 of the PABC domain lined with the side chains of PABPC1 residues Phe567, Gly579, Thr582 and Leu586 (Fig. 2a). The beta-turn conformation adopted by the invariant TNRC6C residues Pro1391 and Gly1392 is specifically recognized via a hydrogen bond between the backbone amide of Gly1392 and the side chain of PABPC1 Glu564 and through a van der Waals contact between TNRC6C Pro1391 and PABPC1 Phe567. Additionally, the invariant TNRC6C Glu1388 forms a salt bridge with PABPC1 Lys580.
Figure 2.
Disruption of the TNRC6C-PABC interaction interferes with mRNA deadenylation in vitro. (a) A view of the interface between the TNRC6C DUF peptide (yellow, interacting side chains in stick format) and PABC domain (light blue surface, interacting residues highlighted in dark blue). Dashed lines indicate hydrogen-bonding interactions. (b) Hemagglutinin epitope (HA)-tagged wild type (WT) and mutant (Mut) TNRC6C fragments were transiently expressed in HEK293T cells. Cell lysates were incubated with recombinant GST-PABPC1 or GST-λN (negative control) immobilized on glutathione sepharose. Resulting complexes were analyzed by SDS-PAGE and anti-HA immunoblotting. (c) 5-BoxB-pA RNA deadenylation in Krebs extract in the presence of recombinant wild-type (WT) or mutant (Mut) GST-λNHA-tagged TNRC6C(1382-1690), or a control construct lacking the λNHA tag. GST-λNHA-TNRC6C(1382-1690)Mut carries the same point mutations as 1382–1690Mut in Fig. 2b. –Poly(A) RNA was prepared by incubating 5-BoxB-pA with oligo-d(T) and RNaseH.
Alanine substitutions at positions Glu1388-Phe1389 or Trp1395-Lys1396 in the TNRC6C DUF domain abolished the interaction with PABPC1 in vitro (Supplementary Fig. 4). Corroborating this, a mutant TNRC6C fragment harboring alanine substitutions in place of the above dipeptides (1382-1690 Mut) could not be precipitated with GST-PABPC1 from HEK293T cell lysates, in contrast to a wild-type TNRC6C 1382-1690 fragment (Fig. 2b). This indicates that the TNRC6C-PABC interaction relies upon the presence of the invariant aromatic residues in the DUF domain.
Addition of Paip2-derived PAM2 peptide, which competes with TNRC6C for binding to the PABC domain, inhibits miRNA-mediated deadenylation of a let-7-targeted RNA in a mammalian in vitro extract that faithfully recapitulates miRNA-mediated repression13. To determine the significance of the TNRC6C-PABC interaction in miRISC-mediated deadenylation, we utilized the λN-BoxB system20 to tether recombinant wild-type and mutant TNRC6C fragments to an RNA reporter containing five BoxB stem-loops and a 98-nucleotide poly(A) tail (5-BoxB-pA) in Krebs extract (Supplementary Fig. 5a,b). Wild-type GST-λNHA-TNRC6C(1382-1690) facilitated complete deadenylation of 5-BoxB-pA RNA within three hours (Fig. 2c, lane 12). Deadenylation was specific, as recombinant GST-TNRC6C(1382-1690) lacking the λNHA tag was unable to mediate deadenylation of 5-BoxB-pA RNA (Fig. 2c, lanes 3–6). In contrast, when a mutant construct carrying alanine substitutions of the invariant residues Glu1388-Phe1389 and Trp1395-Lys1396, GST-λNHA-TNRC6C(1382-1690)Mut, was tethered to 5-BoxB-pA RNA (compare lane 18 to lane 12 in Fig. 2c), the rate and processivity of deadenylation was impaired. Taken together, these data suggest that GW182 must interact with PABPC1 via the DUF-PABC interface in order to mediate complete and efficient deadenylation of target mRNAs in vitro.
The structure of the TNRC6C-PABC complex reveals that key features of the intermolecular interface between TNRC6C and PABC are maintained throughout evolution. The PABC-interacting motif (PIM) in the DUF domain is conserved in all three mammalian GW182 paralogs, in Drosophila melanogaster and in the nematode Brugia malayi7 (Fig. 1c), implying functional conservation in miRISC activity. Our tethering experiments, and also previous studies in mammalian cell extracts13, indicate that the TNRC6C-PABC interaction is required for efficient miRISC-mediated mRNA deadenylation in vitro. Together, these data suggest that the TNRC6C-PABC interaction contributes to the assembly of a repressive complex with target mRNAs that ultimately directs their deadenylation (Supplementary Fig. 6).
In tethering assays in mammalian cells7 and in tethering as well as complementation assays in Drosophila S2 cells14, the DUF domain appears to be dispensable for GW182-mediated repression. It is possible that the role of the DUF domain in GW182-mediated silencing was overlooked in cell-based studies, as these experiments rely on protein overexpression, and the outcome of repression is assayed hours or days after target mRNA recognition. Intriguingly, recent co-immunoprecipitation experiments also suggest that Drosophila dGW182 interacts with the N-terminal RRM motifs of PABPC1 but not the C-terminal domain14. Conceivably, secondary interactions between GW182 proteins and PABPC1, potentially mediated by the polypeptide sequence downstream of the RRM in GW182, may be sufficient for repressive activity in mammalian cells and might be the dominant mode of PABPC1-GW182 interaction in Drosophila. Further studies will be required to resolve these apparent differences.
The results presented here extend our understanding of the central role of GW182-family proteins in miRNA-mediated gene silencing by elucidating the structural basis for GW182 protein binding to PABPC1. The observation that GW182 proteins exploit a similar binding site in PABPC1 as Paip1 and Paip2 hints that miRNA-mediated silencing functions, at least in part, by mimicking protein interactions utilized by canonical protein regulators of translation.
Supplementary Material
Acknowledgments
We thank D. King (HHMI Mass Spectrometry Laboratory, UC Berkeley) for peptide synthesis and mass spectrometry, and C. Ralston and J. Holton (Beamlines 8.2.1 and 8.3.1, Advanced Light Source, LBNL) for assistance with X-ray data collection. We are indebted to W. Filipowicz for discussions and to members of the Doudna laboratory for critical reading of the manuscript. M.J. is supported by a Human Frontiers Science Program fellowship. M.R.F. is supported by a Terry Fox Foundation fellowship from the Canadian Cancer Society. This work was funded in part by a Canadian Institutes of Health Research grant to N.S. N.S. is an International Scholar of the Howard Hughes Medical Institute. J.A.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
Accession codes. Protein Data Bank: Structure factors and coordinates for the TNRC6C-PABC complex have been deposited with accession code 2X04.
AUTHOR CONTRIBUTIONS
M.J., M.R.F., N.S. and J.A.D. designed experiments. M.J. performed binding assays, crystallized the TNRC6C-PABC complex and determined its structure. M.R.F. performed in vitro deadenylation assays. S.M.C. assisted with X-ray data collection and structure determination. M.J. and J.A.D. wrote the manuscript.
References
- 1.Filipowicz W, Bhattacharyya SN, Sonenberg N. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Eulalio A, Huntzinger E, Izaurralde E. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Eulalio A, Tritschler F, Izaurralde E. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behm-Ansmant I, et al. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Till S, et al. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 6.Eulalio A, Huntzinger E, Izaurralde E. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 7.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chekulaeva M, Filipowicz W, Parker R. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazzaretti D, Tournier I, Izaurralde E. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Höck J, et al. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landthaler M, et al. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabian MR, et al. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. Mol Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig AW, Haghighat A, Yu AT, Sonenberg N. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 16.Khaleghpour K, et al. Mol Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 17.Khaleghpour K, et al. Mol Cell Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozlov G, et al. EMBO J. 2004;23:272–281. doi: 10.1038/sj.emboj.7600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy G, et al. Mol Cell Biol. 2002;22:3769–3782. doi: 10.1128/MCB.22.11.3769-3782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron-Benhamou J, Gehring NH, Kulozik AE, Hentze MW. Methods Mol Biol. 2004;257:135–154. doi: 10.1385/1-59259-750-5:135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.