Abstract
Determining the region of seizure onset is of critical importance for treating medically intractable epilepsy. Comparisons between an ictal and interictal Single Photon Emission Computed Tomography (SPECT) images have been shown to be successful in localizing focal epilepsy. The Ictal-Interictal Subtraction Analysis by Statistical Parametric Mapping (ISAS) algorithm remains one the more successful algorithms for comparing these images. However ISAS is limited by its statistical design. This design introduces a scan order bias in the estimation of the normal variance of sequential SPECT images. We have corrected this bias by estimating the normal variance with a half-normal distribution. In this paper we present an updated algorithm (ISAS HN) based on the original ISAS algorithm with a corrected estimate of the normal variance and an open-source utility for ISAS HN.
Index Terms: Epilepsy, SPECT, Change detection
1. INTRODUCTION
Epilepsy is a common disease. In many patients, medication does not fully or even partially suppress their seizures [1]. Surgical procedures involving the area of seizure foci offer the next hope for treatment. Furthermore, epilepsy is a complex disease which may arise from many regions of the brain and, before any surgery can take place, the regions of seizure foci must be determined. The current gold standard for localizing seizure foci, intracranial electroencephalography (EEG), is an invasive surgical procedure where EEG electrodes may either be placed on the cortical surface or inserted into sub-cortical structures. While this technique provides direct measurements of the neuronal activity, EEG is spatially limited by the number of electrodes that can be safely implanted.
Several non-invasive neuroimaging techniques are available to localize seizure foci. These techniques may fully localize a seizure or guide the placement of EEG electrodes to a more targeted area. Magnetic Resonance Imaging, Positron Emission Tomography, and Magnetic Resonance Spectroscopy have all shown some success in localizing seizure foci [2]. However, these imaging modalities can only acquire images between seizures as they are limited by patient movement during a seizure.
Single Photon Computed Tomography (SPECT) offers a unique advantage over other neuroimaging techniques with the ability to image cerebral blood flow (CBF) during a seizure. Several analysis method are available to compare ictal (during a seizure) and interictal (between seizures) SPECT images [3, 4, 5]. These methods generally revolve around subtracting the ictal SPECT from the interictal SPECT to locate areas of hyperperfusion (increased CBF during a seizure) which correspond to the areas of seizure foci.
One of the more successful analysis algorithms is Ictal-Interictal Subtraction Analysis by Statistical Parametric Mapping (ISAS). ISAS detects statistically significant differences between an ictal SPECT image and an interictal SPECT image for a patient by comparing this to the normal variation. The normal variation is estimated using a set of image-pairs acquired from 14 normal controls imaged twice under base-line conditions [6]. However, the current ISAS algorithm has several disadvantages including a challenging implementation with different SPM and MATLAB versions, a reliance on an inefficient general linear model, and a biased estimate of the normal variation. This paper presents an improved, unbiased, statistical model for the ISAS algorithm based on a half-normal distribution, preliminary results on patient with surgically confirmed epilepsy, and an open source application for the ISAS algorithm which is currently being used at our epilepsy research program.
2. METHODS
2.1. ISAS Review
The ISAS algorithm consists of several preprocessing steps in which the patient images and the control images are non-linearly registered to common space, smoothed, masked, and intensity normalized (described in section 3). All images are then used as inputs to a linear model to compute voxelwise significance based on a Student’s T-distribution model. The resulting t-map is clustered and corrected for multiple corrections with the largest cluster of positive t-values indicating the predicted area of seizure foci. All the preceding steps were implemented in an open source image processing package with modification (presented below) to the linear model used to compute voxel by voxel significance.
2.2. Our Improved Algorithm
Reformulation and Simplification of the Linear Model
The ISAS algorithm calculates significance based on the full general linear model (GLM),
| (1) |
with with inputs (Y̲ ) and design matrix (X̲) as shown in Figure 1. Here, β̲ is 19×1 column vector of images estimated by the GLM. Voxel by voxel T statistics are calculated from β̲ using
| (2) |
where σ̂2 is the residual variance and c̲ is the contrast term. Equation 2 can be expressed as a simpler t-test reducing the input images and eliminating the need to estimate the parameters β̲. The product of the contrast term, and the parameters β̲ in the numerator expands to −β0 + β1 + β2 − β3. From solving the GLM, β0 and β1 can be combined to equal the difference of the two patient images while β2 and β3 can be combined to equal the mean of the differences of each pair of control images. The control images are the same for every analysis; thus the numerator can be considered the difference of the two patient images plus a constant control term. The denominator can be simplified to a constant term. The residual variance can be expressed as a constant term as it is only a function of the control images and other constants. Furthermore, since the contrast and design matrix are the same for every analysis, the c̲T (X̲TX̲)−1c̲ term simplifies to a constant. This simplification leaves the denominator as a single constant term equaling the standard error of the differences of each pair of control images. Eqn. 2 can then be rewritten as
| (3) |
where x is the difference of the two patient images, x̂ is the mean of the differences of each pair of control images, and SE is the standard error of the differences of each pair of control images. Using this equation, no β̲ parameters need to be estimated. x̂ and SE can be determined before runtime reducing the number of input images from thirty to four.
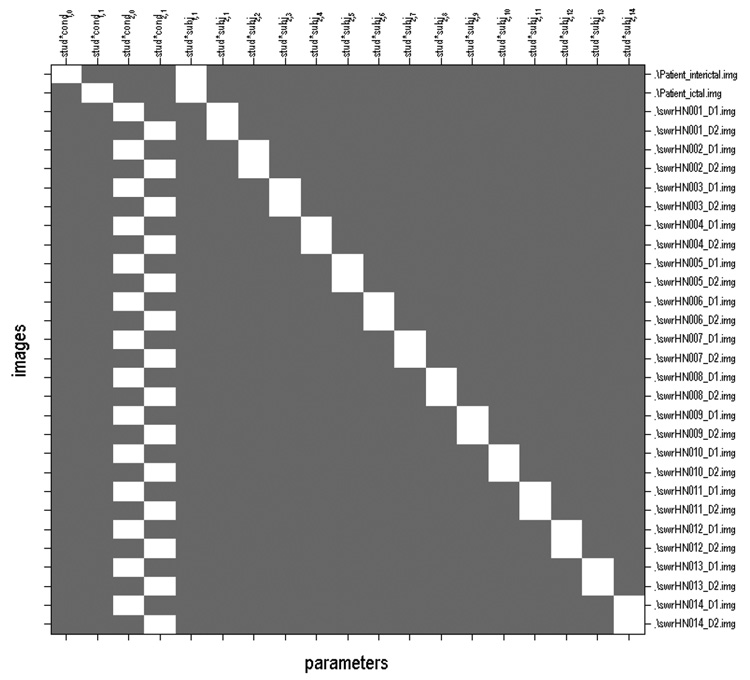
Fig. 1.
Figure 1 shows the inputs (Y̲ ) and the design matrix (X̲) used in the ISAS algorithm. Y̲ is a column vector of the images listed on the right of the design matrix.
Bias Elimination using a Half-Normal Distribution
In Equation 3, x̂ and SE are calculated from the second image subtracted from the first image for each control. However, the conditions for the control are not different between the two acquisitions (unlike the patient who is imaged during a seizure and at baseline) and could just as easily be swapped. However reversing the order of the control images will impact the estimates of both the mean and the standard deviation of the distribution of normal variation. This reveals that the existing ISAS method is biased. Hence the meaningful information that can be extracted from these image pairs is the absolute value of their difference (which is invariant to the scan order) as opposed to the actual difference.
Using this insight, we proceed to reformulate ISAS as follows. Given a pair of images under the same conditions for a control, each image (y1, y2) can be expressed as the sum of the true underlying value (x) and the normal variation, (n). The difference of the noise between the pair of images (d) would then be d = y1−y2 = x+n1−x−n2 = n1−n2. If we make the common assumption that n is normally distributed with zero mean and unknown standard deviation, then the distribution of d also has zero mean with an unknown standard deviation. From this assumption, the distribution of the absolute value of d is a half-normal distribution with mean of , where σd is the standard deviation of d. Thus, given a collections of d’s from N different patients, the standard deviation of d can be estimated, without any order bias, using the mean of |d| as follows:
| (4) |
The statistical t-test presented in Equation 3 can be reworked to incorporate the estimate of the noise based on a half-normal distribution. The x̂ term reduces to zero as the differences of the control images is just the system noise which is assumed to have a zero mean. The SE term now becomes Equation 4 divided by the appropriate sample size with y1i = H N1i, y2i = H N2i, and N = 14. Using the half-normal distribution to estimate the normal variation removes the bias associated with the specific order of the control images and further reduces the number of input images to three. The name ISAS HN is used in the remainder of the paper to refer to this modification to the ISAS algorithm. For completeness, the full equation used for significance in the ISAS HN algorithm is presented below.
| (5) |
2.3. Tool Implementation
The algorithms were implemented within BioImage Suite [7] – an open-source image processing software. Both Equation 3 and Equation 5 were implemented for significance calculation giving the user access to both the ISAS and ISAS HN algorithms. A custom GUI was designed to streamline many of the ISAS algorithms pre- and post-processing steps and to reduce user error.
3. RESULTS
Thirteen patients with either surgically confirmed epilepsy were use for preliminary testing of the methods outlined in Section 2. The injections for the ictal SPECT images were performed by a technologist at the start of a seizure. Injections for the interictal SPECT images were performed following a 24 hour seizure free period. [99m Tc]-labeled HMPAO (Medi-Physics; Amersham Healthcare, Arlington Heights, IL, U.S.) was used as the radiotracer allowing the perfusion at the time of injection to be imaged later. SPECT images were acquired within 90 min after injection. Projection data were acquired for 40 min on a Picker IRIX (Philips Medical Systems, Best, Netherlands), and reconstructed into transverse slices with Chang attenuation correction. Images were converted into the Analyze format for image analysis.
All images were transformed into common space (MNI space) using an intensity based B-spline tensor model with a normalized mutual information match metric [8, 9, 10]. The registered images were then smoothed with 16×16×16 mm Gaussian kernel, masked to remove the skull, and proportionally scaled to a image mean of 50. The control images were calculated from the healthy normal controls with the above preprocessing. T-map were calculated as described in section 2.2. Cluster level probabilities as well as multiple comparison correction were calculated using Random Field Theory.
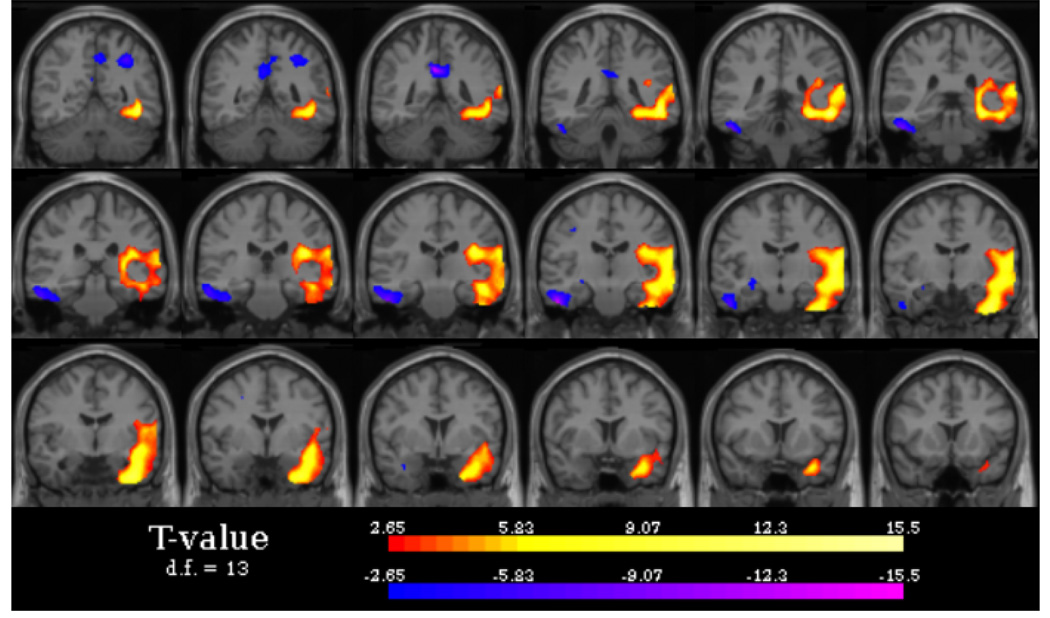
Preliminary results from these patients show no loss of accuracy in the localizing abilities of the ISAS algorithm from the elimination of the full GLM and the use of Equation 5. These results also show good concordance between the new ISAS HN algorithm and the areas of confirmed seizure foci. Figure 2 shows an example of a result from the ISAS algorithm. The largest area of hyperperfusion (red) is located in the left temporal lobe corresponding to the area of surgically confirmed epilepsy.
Fig. 2.
Typical hyper- and hypoperfusion (increase and decrease in CBF during a seizure) results for a patient with left temporal lobe epilepsy from the ISAS HN algorithm. Notes: images are displayed in radiological convention
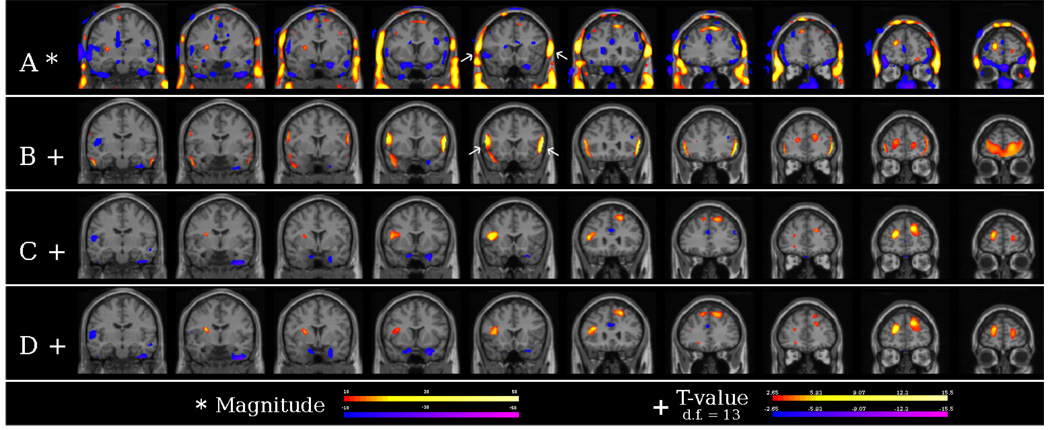
Registration has been improved by using the registration algorithms [9, 10] in BioImage Suite. Figure 3 shows the results of four different SPECT analysis methods: (A) the differences between an ictal and interictal SPECT with minimal preprocessing (rigid alignment and intensity normalization), (B) the results from the original ISAS implementation, and (C and D) the results from both implementations in our tool. From Figure 3A, a large bilateral frontal difference is shown be outside the brain. However, the original ISAS implementation shows this artifact to be inside the brain and mistakenly labels it as the area of seizure foci. Neither results from our tool show this artifact giving the correct localization. Run time of the statistical analysis was also reduced. Using a Windows XP PC, Intel Atom processor at 1.6 GHz, with 1 GB of RAM, the run time was reduced from 5 minutes for the original the ISAS algorithm to under 1 minute in our tool.
Fig. 3.
A) Difference imaging showing large artifact in the skull. B) Results from the original ISAS implementation showing the skull artifacts inside the brain. C) and D) Results from our tool for both ISAS and ISAS HN showing no skull artifact.
4. DISCUSSION
We have developed a new way to analysis the differences between ictal and interictal SPECT images base off of the ISAS algorithm. This new method follows the same step of the ISAS algorithm but estimates the noise variance based off a half-normal distribution. The original ISAS algorithm estimates the standard deviation based on a design matrix suited for a group versus group analysis. This design matrix leads to a scan order bias in the standard deviation. Rather than performing a group vs group analysis, we only need to establish the normal variation of sequential SPECT images in a normal control group. By using a half-normal distribution, we can estimate this normal variation without any scan order bias. More generally, this technique is applicable to other problems where the interest is to detect significant changes in some measure over time for a single patient using a set of repeated measurements from a normal group to establish the normal variation.
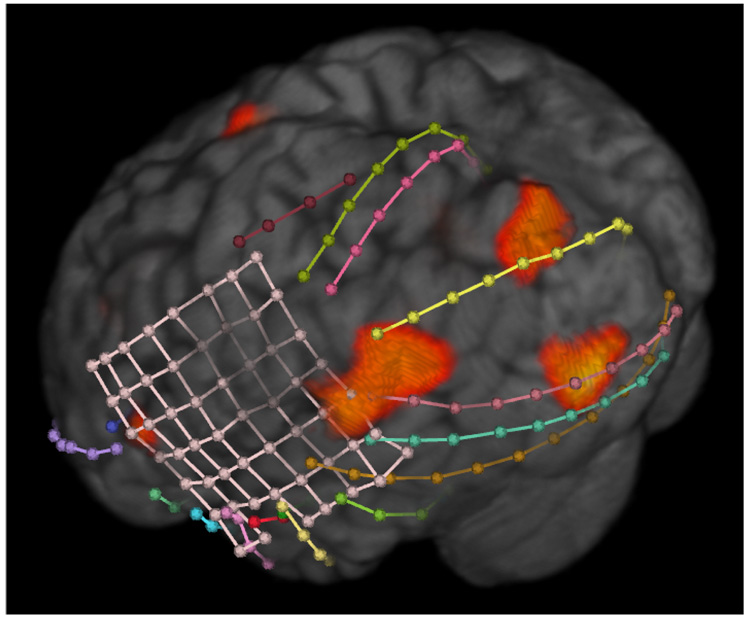
We have provide this algorithm as part of BioImage Suite, a comprehensive, multi-platform, open-source image analysis suite. With this software, users can combine the ISAS HN algorithm with other epilepsy related utilities as shown in Figure 4. Here, the results from the ISAS HN algorithm were use to help target intracranial electrodes to a possible seizure foci. While further work is needed to clinically verify the ISAS HN algorithm, we hope this tool makes ictal SPECT analysis more readily available and more inviting to use for both large and small epilepsy centers.
Fig. 4.
The results of the ISAS HN algorithm are overlayed on a volume rendering of a MRI image with intra-cranial electrodes localized from a CT image.
Acknowledgments
This work was supported in part by the NIH under grant R01-EB006494.
REFERENCES
- 1.Cascino GD. Surgical treatment for epilepsy. Epilepsy Research. 2004 July–August;vol. 60(no. 2–3):179–186. doi: 10.1016/j.eplepsyres.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Widjaja E, Raybaud C. Advances in neuroimaging in patients with epilepsy. Neurosurg Focus. 2008 September;vol. 25(no. 3):E3. doi: 10.3171/FOC/2008/25/9/E3. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien TJ, So EL, Mullan BP, Hauser MF, Brinkmann BH, Bohen NI, Hanson D, Cascino GD, Jack CR, Jr, Sharbrough FW. Subtraction ictal spect co-registered to mri improves clinical usefulness of spect in localizing the surgical seizure focus. Neurology. 1998 February;vol. 50(no. 2):445–454. doi: 10.1212/wnl.50.2.445. [DOI] [PubMed] [Google Scholar]
- 4.Knowlton RobertC, Lawn NicholasD, Mountz JamesM, Kuzniecky RubenI. Ictal spect analysis in epilepsy: Subtraction and statistical parametric mapping techniquess. Neurology. 2004 July;vol. 63(no. 1):10–15. doi: 10.1212/01.wnl.0000132885.83350.45. [DOI] [PubMed] [Google Scholar]
- 5.Lopes R, Viard R, Dewalle AS, Steinling M, Maouche S, Betrouni N. 3d mutifractal analysis: A new tool for epileptic fit sources detection in spect images; Conf Proc IEEE Eng Med Biol Soc; 2008. pp. 3912–3915. [DOI] [PubMed] [Google Scholar]
- 6.McNally KellyA, Lebron Paige A, Varghese George, Zhang Heping, Novotny EdwardJ, Spencer Spencer SusanS, George Zubal I, Blumenfeld Hal. Localizing value of ictal-interictal spect analyzed by spm (isas) Epilepsia. 2005 September;vol. 46(no. 9):1450–1464. doi: 10.1111/j.1528-1167.2005.06705.x. [DOI] [PubMed] [Google Scholar]
- 7.Papademetris X, Jackowski M, Rajeevan N, DiStasio M, Okuda H, Constable RT, Staib LH. BioImage Suite: An integrated medical image analysis suite: An update; ISC/NA-MIC Workshop on Open Science at MICCAI 2006; 2006. http://hdl.handle.net/1926/209. [PMC free article] [PubMed] [Google Scholar]
- 8.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Non-rigid registration using free-form deformations: Application to breast MR images. IEEE Trans. Med. Imag. 1999 doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 9.Papademetris X, Jackowski A, Schultz RT, Staib LH, Duncan JS. Integrated intensity and point-feature nonrigid registration. MICCAI. 2004:763–770. doi: 10.1901/jaba.2001.3216-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Studholme C, Hill D, Hawkes D. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimisation of voxel similarity measures. Medical Physics. 1997;vol. 24(no. 1):25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]