Abstract
Hospital-based infection control in veterinary medicine is emerging and the role of the environment in hospital-acquired infections (HAI) in veterinary hospitals is largely unknown. This study was initiated to determine the recovery of Escherichia coli and selected veterinary and zoonotic pathogens from the environments of 101 community veterinary hospitals. The proportion of hospitals with positive environmental swabs were: E. coli — 92%, Clostridium difficile — 58%, methicillin-resistant Staphylococcus aureus (MRSA) — 9%, CMY-2 producing E. coli — 9%, methicillin-resistant Staphylococcus pseudintermedius — 7%, and Salmonella — 2%. Vancomycin-resistant Enterococcus spp., canine parvovirus, and feline calicivirus were not isolated. Prevalence of antimicrobial resistance in E. coli isolates was low. Important potential veterinary and human pathogens were recovered including Canadian epidemic strains MRSA-2 and MRSA-5, and C. difficile ribotype 027. There is an environmental reservoir of pathogens in veterinary hospitals; therefore, additional studies are required to characterize risk factors associated with HAI in companion animals, including the role of the environment.
Résumé
Escherichia coli et certains pathogènes vétérinaires et zoonotiques isolés dans l’environnement des cliniques pour animaux de compagnie dans le Sud de l’Ontario. Le contrôle des infections à la clinique vétérinaire est un domaine émergent et le rôle de l’environnement pour la contraction d’infections nosocomiales (IN) dans les cliniques vétérinaires est largement inconnu. Cette étude a été entamée pour déterminer la récupération d’E. coli et de certains pathogènes vétérinaires et zoonotiques dans l’environnement de 101 cliniques vétérinaires communautaires. La proportion des cliniques avec des écouvillons environnementaux positifs était : E. coli — 92 %, Clostridium difficile — 58 %, Staphylococcus aureus résistant à la méthicilline (SARM) — 9 %, CMY-2 produisant E. coli — 9 %, Staphylococcus pseudintermedius résistant à la méthicilline — 7 % et Salmonella — 2 %. Les pathogènes Enterococcus spp. résistant à la vancomycine, parvovirus canin et calicivirus félin n’ont pas été isolés. La prévalence de la résistance antimicrobienne dans les isolats d’E. coli était faible. Des pathogènes vétérinaires et humains potentiels importants ont été récupérés, incluant des souches épidémiques canadiennes de SARM-2 et de SARM-5 et de C. difficile ribotype 027. Il y a un réservoir environnemental de pathogènes dans les cliniques vétérinaires; par conséquent, des études additionnelles sont requises pour caractériser les facteurs de risque associés aux infections nosocomiales chez les animaux de compagnie, y compris le rôle de l’environnement.
(Traduit par Isabelle Vallières)
Introduction
Hospital-acquired infections (HAI) are an important cause of morbidity and mortality in human and veterinary patients and are associated with multiple factors, including patient susceptibility and sources of exposure (1). In companion animals, HAI by antimicrobial resistant Escherichia coli (2), Clostridium difficile (3), and Acinetobacter baumanii (4) with an associated environmental reservoir have been reported. Environmental sources associated with hospital-acquired salmonellosis in horses have also been documented (5,6); however, the role and contribution of environmental pathogens in the epidemiology of HAI is not well understood.
In human medicine, environmental sites are not typically considered to be important sources of exposure to pathogens and routine surveillance of environmental sites is not recommended (7). This may not apply to companion animal veterinary medicine, however, because of the behaviors, housing, and management of companion animals. For example companion animals have close contact with floors during clinical examination, venipuncture, and recovery from anesthesia. Moreover, floors of veterinary hospitals are more likely to be contaminated with infectious material (feces, for example) and animal exploratory behavior may place animals’ noses and mouths in contact with these areas. Sites such as floors and perhaps other sites, therefore, may be of greater importance as environmental reservoirs of pathogens in companion animal medicine than in human medicine.
Veterinary hospitals are an intersection of human and animal interaction. Thus, when investigating agents associated with HAI in veterinary medicine, different types of agents need to be considered: animal pathogens, antimicrobial-resistant bacteria that may be potential pathogens to humans or animals, zoonotic pathogens, and microorganisms that are relatively resistant to environmental disinfection.
The objectives of this study were to determine: 1) the prevalence of environmental E. coli, Salmonella enterica, extended spectrum beta-lactamase (ESBL) producing E. coli, CMY-2 producing E. coli (blaCMY-2 E. coli), C. difficile, vancomycin-resistant Enterococcus spp. (VRE) methicillin-resistant Staphylococcus aureus (MRSA), methicillin-resistant Staphylococcus pseudinter-medius (MRSP), canine parvovirus (CPV), and feline calicivirus (FCV); 2) the antimicrobial susceptibility of environmental E. coli and Salmonella isolates; 3) the distribution of molecular types of C. difficile and MRSA; and 4) the associations between specific infection control practices and environmental recovery of E. coli, ampicillin-resistant E. coli (AMP-R E. coli), C. dif-ficile and MRSA.
Materials and methods
Sample size calculations
We sought to enrol 100 veterinary hospitals to enable detection of an individual organism at a prevalence of 10% among hospitals with a precision of 6%, 80% of the time (8).
Veterinary hospital selection
Southern Ontario companion animal hospitals or offices, including those with additional licensures for food animal or equine facilities (mixed animal hospitals), licensed by the College of Veterinarians of Ontario in 2005, were eligible for recruitment. Eligible veterinary hospitals (n = 766) were contacted by mail and invited to participate. Participants were then asked to respond by mail, fax, or telephone with a completed hospital-demographic survey.
Sampling
Environmental sampling in the hospitals was done in 2 phases. During phase 1, individual sites were sampled throughout the hospital without pooling. Sampling was performed in areas used for reception, treatment, examination, hospitalization, isolation, runs, boarding, and grooming. The sites sampled were table surfaces (examination rooms, treatment, surgery, grooming and boarding areas), floor surfaces (reception, treatment, kennel rooms, surgery, isolation, grooming and boarding areas), equipment (stethoscopes, thermometers, otoscopes/ophthalmoscopes, and otoscope tips), and areas of high human hand contact (telephone, computer keyboards, taps, and doorknobs). The median number of sites sampled in each hospital was 23 (range 17–27). Based on the results of phase 1, the sites sampled in phase 2 were pooled: 1) kennel areas and runs; 2) examination and treatment tables; 3) floors; 4) isolation areas; 5) kitchen and bathroom taps, keyboards and telephones; 6) examination and treatment room sink taps; 7) otoscope tips; 8) otoscopes, ophthalmoscopes, and stethoscopes; and 9) thermometers. Sites sampled for bacteriology were sampled using sterile, divided electrostatic cloths (Swiffer, Proctor and Gamble, Toronto, Ontario) and gloved hands. Gloves were changed between sampling of different sites. Samples for CPV and FCV were collected from kennel and isolation areas only and sampling was done using a sterile cotton swab (Becton, Dickinson, Franklin Lakes, New Jersey, USA).
Samples were collected Monday through Friday, between 9 a.m. and 4 p.m. during May 2005 and August 2005. Sample collection was not targeted to sites recently disinfected or sites that had been used, yet not cleaned or disinfected. Samples were taken irrespective of environmental disinfection within the practice.
Microbiology
Escherichia coli
Half of the electrostatic cloth was placed in 50 mL of buffered peptone water (BPW) (B-D) and incubated at 37°C for 18 to 24 h. Then, 25 mL of BPW was added to 25 mL of double strength E. coli (EC) broth (Becton, Dickinson) and incubated at 42°C for 18 to 24 h. Next, loops of broth were streaked onto Eosin Methylene Blue agar (Becton, Dickinson) and incubated at 37°C for 18 to 24 h, then 6 presumptive E. coli colonies were plated onto MacConkey agar (Becton Dickinson) and incubated at 37°C for 18 to 24 h. Then, 6 presumptive E. coli colonies were plated onto tryptic soy agar (Becton Dickinson) and incubated at 37°C for 18 to 24 h. Escherichia coli were confirmed indole-positive (Kovac’s reagent; PML Microbiologicals, Mississauga, Ontario) and citrate-negative (Simmons citrate agar; Becton Dickinson) reactions.
Extended spectrum β-lactamase Escherichia coli
The initial preparation of the electrostatic cloth was as described, except that the EC broth was streaked for isolated colonies onto MacConkey agar with 2 μg/mL of cefpodoxime (Oxoid Company, Nepean, Ontario) and incubated at 37°C for 48 h in accordance with the Clinical Laboratory Standards Institute (CLSI) guidelines (9). Escherichia coli were confirmed as described.
blaCMY-2 Escherichia coli
Escherichia coli isolates with the following antimicrobial resistance phenotype were selected for amplification, sequencing and hybridization using polymerase chain reaction (PCR) for the blaCMY-2 gene (10): ampicillin (minimum inhibitory concentration [MIC] ≥ 32 μg/mL) together with amoxicillin-clavulanic acid (MIC ≥ 32 μg/mL), and cefoxitin (MIC > 8 μg/mL) or ceftriaxone (MIC > 8 μg/mL) or ceftiofur (MIC > 2 μg/mL) (see Antimicrobial susceptibility testing).
Salmonella
Salmonella was isolated using 2 methods. In the first method, 1 mL of BPW pre-enrichment was added to 9 mL Rappaport Vassiliadis (RV) broth (Becton, Dickinson) and incubated at 42°C for 24 h. Then, 1 mL of the RV broth was added to 9 mL Tetrathionate broth (B-D) and incubated at 37°C for 18 to 24 h. Tetrathionate broth was streaked for isolated colonies onto a Xylose-lysine Tergitol 4 (XLT4) agar plate and incubated at 37°C for 18 to 24 h. Two colonies with morphologies typical of Salmonella were streaked on to MacConkey agar and were incubated at 37°C for 18 to 24 h. Presumptive Salmonella isolates were plated onto nutrient agar and incubated at 37°C for 18 to 24 h after which biochemical and serological testing was conducted on the isolates.
In the second method, 0.1 mL of the initial BPW pre-enrichment was inoculated onto Modified Semisolid Rappaport Vassiliadis (MSRV) agar (Becton, Dickinson) and incubated at 42°C for 24 to 72 h. Plates were examined for a Salmonella migratory pattern at 24, 48, and 72 h of incubation. Areas of growth with typical Salmonella migration were streaked onto MacConkey agar and incubated at 37°C for 18 to 24 h. Two presumptive Salmonella colonies were plated onto nutrient agar and incubated at 37°C for 18 to 24 h.
Salmonella were confirmed by reactions on triple sugar iron agar slants (Becton, Dickinson), Christensen’s urea agar (Becton Dickinson) slants and by slide agglutination in Salmonella O antiserum polyvalent A-I and Vi. Salmonella isolates were serotyped at the Laboratory for Foodborne Zoonoses Salmonella reference laboratory using standard methods (11).
Vancomycin-resistant Enterococcus spp
One quarter of the electrostatic cloth was placed in 50 mL BPW and incubated at 35°C for 24 h. Then, 1 mL of BPW was added to 9 mL VRE enrichment broth and incubated at 35°C for 24 h. This was plated onto selective agar (Oxoid) and incubated at 35°C for 48 h. Next, brown and black colonies were plated onto Columbia blood agar and incubated at 35°C for 24 h. Catalase-negative, gram-positive cocci were confirmed as enterococci using the API Strep 20 biochemical identification system (Oxoid).
Clostridium difficile
One quarter of the electrostatic cloth was placed in 50 mL of Clostridium difficile Moxalactam Norfloxacin broth (CDMN) and was incubated in an anaerobic chamber at 37°C for 7 d. Then, 2 mL of the CDMN broth were added to 2 mL of 95% ethanol, which was incubated at room temperature for 1 h, then centrifuged at 4400 g and the pellet was streaked onto blood agar and incubated in an anaerobic chamber at 37°C for 48 h. Clostridium difficile identification was confirmed by colony morphology, Gram stain appearance, characteristic odor and l-proline aminopeptidase activity (PRO Disc assay, Carr-Scarborough Microbiologicals, Decatur, Georgia, USA). Isolates were characterized for ability to produce toxins A, B, and CDT binary by PCR (12,13). Ribotyping was performed to further identify the strains (14) and patterns were compared with a collection of human and animal isolates archived by the investigators (15).
Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius
One quarter of the electrostatic cloth was placed in BPW and incubated at 35°C for 24 h. Then, 1 mL of the BPW rinse was added to 9 mL of MRSA enrichment broth (7.5% NaCl, 2.5 g/L yeast extract, 10 g/L tryptone, and 10 g/L mannitol) and was incubated at 35°C for 24 h. The broth was plated onto mannitol salt agar with 2 μg/mL oxacillin (Oxoid) and was incubated at 35°C for 48 h. Coagulase and catalase positive, gram-positive cocci were identified as Staphylococcus species. Methicillin resistance was confirmed using the PBP2 assay (Oxoid). Identification to the species level was done using the API STAPH system (BioMerieux Canada, St. Laurent, Quebec) and polymyxin B susceptibility. Isolates of MRSA were typed by SmaI pulsed-field gel electrophoresis (PFGE) and categorized as described by Mulvey et al (16), with 2 modifications: 1) The lysostaphin and lysozyme were added to the buffer, and 2) the switch times were 0.5 to 90 s.
Virus isolation
Swabs were placed in cooled viral transport medium and stored (2 to 6 h) in a portable cooler with an ice pack. Next, the swab and medium were vortexed and 1.25 mL of the mixed medium was placed in cryovials and frozen at −86°C. Before inoculation, the samples were centrifuged at 2100 to 2200 g for 10 min.
Canine parvovirus
Cell monolayers were prepared by seeding Lab-Tek 2-well Permanox slides (Nalge Nunc International, Rochester, New York, USA) with 1:4 to 1:5 dilutions of Crandall feline kidney cells (1.5 mL per chamber). Sample supernatant was added to the seeded chamber slides (50 μL per well) and incubated for 5 d at 37°C with 5% CO2. A positive control using canine parvovirus stock virus and a negative control using an uninoculated chamber was included in each run. After 5 d, media were aspirated and monolayers were air-dried and then heat-fixed at 60°C on a slide warmer for 30 min. The slides were fixed in 3:2 acetone and phosphate buffered saline mixture (BPS) for 15 min and then dipped in methanol prior to fan-drying.
Fixed monolayers were stained with canine parvovirus monoclonal antibody (TropBio Pty, Townsville, Queensland, Australia) and incubated in a moist chamber for 30 min at 37°C. Next, slides were rinsed 3 times for 4 min each time in PBS and then flooded with fluorescein isothiocyanate conjugated goat anti-mouse IgG (MP Biomedicals, Solon, Ohio, USA) for 30 min at 37°C. The slides were rinsed in PBS (3 × 4-min), followed by several rinses in reverse osmosis water. Slides were fan-dried and coverslipped with fluorescent antibody mounting medium before examining monolayers for viral-specific fluorescence.
Feline calicivirus
Cell monolayers were prepared as described except that 25 cm2 (5 mL) flasks were seeded in place of chamber slides. Samples were added (100 μL per flask) and incubated for 7 d with daily monitoring for cytopathic effects (CPE). A negative cell control was included with each run. Samples were considered negative if no CPE was evident after 7 d post-inoculation.
Antimicrobial susceptibility testing
For each sample, susceptibility testing was performed on 3 E. coli isolates, all presumptive ESBL-E. coli isolates, 2 Salmonella isolates from the first isolation method, and 1 from the second isolation method. Minimum inhibitory concentrations were determined by broth microdilution methodology (Sensititre; Trek Diagnostic Systems, Cleveland, Ohio, USA). The National Antimicrobial Resistance Monitoring System (NARMS) microtiter plate configuration (NARMS CMV1AGNF) was used with the following resistance breakpoints: amikacin (≥ 64 μg/mL), amoxicillin-clavulanic acid (≥ 32 μg/mL), ampicillin (≥ 32 μg/mL), cefoxitin (≥ 32 μg/mL), ceftiofur (≥ 8 μg/mL), ceftriaxone (≥ 64 μg/mL), chloramphenicol (≥ 32 μg/mL), ciprofloxacin (≥ 4 μg/mL), gentamicin (≥ 16 μg/mL), kanamycin (≥ 64 μg/mL), nalidixic acid (≥ 32 μg/mL), streptomycin (≥ 64 μg/mL), sulfamethoxazole (≥ 512 μg/mL), tetracycline (≥ 16 μg/mL), and trimethoprim-sulfamethoxazole (≥ 4 μg/mL). The CLSI breakpoints for resistance (9) were used for all antimicrobials except streptomycin for which the NARMS 2001 susceptibility breakpoint was used (17).
Generalized linear mixed models of factors potentially associated with recovery of environmental bacteria
The binary outcomes of interest were: recovery (or not) of E. coli, AMP-R E. coli, C. difficile, and MRSA. The categorical independent variables were: the type of disinfectant used for environmental disinfection, the type of hand cleansers used for hand washing, an isolation area, in-hospital antimicrobials, and documented “Standard Operating Procedures” (SOPs) for cleaning and disinfection of environmental areas, and cleaning, disinfection and sterilization of equipment. Potential confounding variables examined were type of hospital, number of staff in the veterinary hospital, number of hospitalized patients per day, number of appointments seen per day and recovery of other organisms. The data were captured through questionnaires administered to 1 veterinarian and 1 veterinary technician from each hospital.
Unconditional associations between infection control practices and the outcomes of interest were evaluated using Fisher’s exact test. Next, a single-level (hospital-level) multivariate logistic model was built for each outcome using a step-wise, forward selection process of predictors unconditionally associated with the outcome at P ≤ 0.2. The P-value for entry into the model was 0.2 and for removal was 0.05 for all predictors including potential confounding variables. Further modelling was performed manually. Potential confounding variables and other predictors that were removed during the step-wise forward selection were reintroduced to the initial model to examine their effect on coefficients of the other factors.
Following the primary logistic model building procedure, a mixed multilevel (level 1: site, level 2: hospital) modelling procedure was performed including site of bacterial recovery within the veterinary hospital as both random and fixed effects. The model was built by manual backwards selection using significant variables (P ≤ 0.05) in the primary logistic model and the potential confounders. Variables were retained in the final model if significantly associated with the outcome (P ≤ 0.05), significant by the likelihood ratio test (P ≤ 0.05), or their removal altered the coefficient of 1 or more of the other predictors by ≥ 10%. The distribution of the standardized residuals (hospital level) was assessed graphically with a normal (Q-Q) plot. Influential observations at the hospital level were assessed using Cook’s distance. The intraclass correlation (ICC) for bacterial recovery was calculated using the variance provided from the model (8).
Descriptive statistics were performed using Micosoft Excel 2003 (Microsoft Corporation, Redmond, Washington, USA) and Intercooled Stata 10.0 (StataCorp, Collage Station, Texas, USA). Univariate and multivariate analyses were performed using Intercooled Stata (StataCorp). Mixed multi-level modelling was performed using GLAAM (18,19) within Intercooled Stata (StataCorp).
The study procedures were approved by the Research Ethics Board at the University of Guelph.
Results
One hundred and twenty-one hospitals responded with interest to the recruitment letter (response rate 16%) and from these the study population of 101 hospitals was selected. Twelve could not be sampled because of time limitations and the other 8 were out of the geographic sampling region. Of the 101 hospitals, 90 were companion animal, 10 were mixed animal, and 1 hospital treated primarily exotic animals. The median number (and range) of full-time veterinarians per hospital was 2 (0 to 12), part-time veterinarians was 1 (0 to 5), and other staff was 10 (3 to 45). The median number (and range) of appointments per day was 10 (2 to 40), of dogs hospitalized each day was 3 (1 to 18) and cats was 3 (1 to 25).
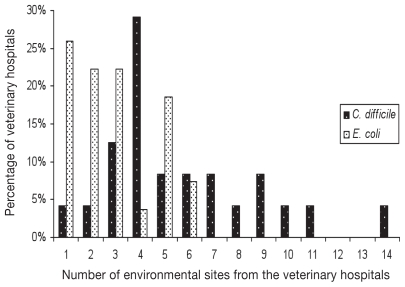
In phase 1, E. coli was recovered from 23 (96%) hospitals (n = 24) and from up to 13 different sites within individual hospitals (Figure 1). Clostridium difficile was recovered from 83% of hospitals (n = 27) from up to 5 sites (Figure 1). Combining data from phases 1 and 2, E. coli and/or C. difficile were recovered from at least 1 site in each veterinary hospital and each type of site sampled (Tables 1, 2). Overall, the hospital prevalence of recovery of E. coli and C. difficile (phase 1 and phase 2) was 92% (n = 93) and 58% (n = 58), respectively (Table 1). Forty-seven percent (n = 56) of the C. difficile ribotypes indentified were previously identified in humans, and 43% (n = 52) were previously identified in animals. The remaining 9% (n = 11) were newly identified ribotypes (Table 3). Among the 56 isolates of known human ribotypes, 36 (64%) produced toxins A and B, 19 (34%) produced toxins A, B, and CDT and 1 (2%) produced toxin B. Ninety-four percent (n = 49) of the isolates from ribotypes only associated with animals were non-toxigenic. Four percent (n = 2) of the isolates from known animal ribotypes had toxins A and B, and 2% (n = 1) had toxins A, B, and CDT. Five of the newly identified types (n = 11) were toxigenic; 4 produced toxins A and B and 1 produced toxins A, B, and CDT. Methicillin-resistant S. aureus and MRSP were identified in 9% (n = 9) and 7% (n = 7) of the hospitals, respectively (Table 1). Five of the MRSA isolates belonged to Canadian epidemic strain (cMRSA)-5 and 3 were cMRSA-2. The remaining 5 were non-typeable. Salmonella Typhimurium was isolated on the XLT4 medium and was recovered from an examination room floor. Salmonella Mbandaka was isolated on MSRV medium and was recovered from the run area of another hospital. The Salmonella isolates were susceptible to all antimicrobials tested. VRE, CPV, and FCV were not recovered from any sampled sites.
Figure 1.
Number of sites in the phase 1 component of the study where environmental E. coli (n = 24 hospitals) and C. difficile (n = 27 hospitals) were recovered from companion animal veterinary hospitals.
Table 1.
Prevalence (%) of bacterial recovery from environmental sites in companion animal veterinary hospitals
| Prevalence (%)(95% confidence interval) |
||||||
|---|---|---|---|---|---|---|
| Organism | Hospitals | Kennels | Isolationd | Runs | Floors | Tables |
| E. colia | 92 (85, 96) | 48 (38, 59) | 66 (48, 81) | 27 (18, 36) | 72 (62, 81) | 26 (18, 35) |
| C. difficileb | 58 (48, 69) | 17 (10, 26) | 29 (15, 46) | 11 (6, 19) | 16 (9, 25) | 7 (3, 14) |
| Salmonellaa | 2 (0.2, 7) | 0d (0, 4)d | 0 (0, 10)e | 1 (0.02, 5) | 1 (0.02, 5) | 0 (0, 4)e |
| MRSAb | 9 (4, 16) | 1 (0.02, 5) | 3 (0.07, 15) | 0 (0, 4)e | 3 (0.6, 8) | 4 (1, 10) |
| MRSPc | 7 (3, 14) | 1 (0.02, 5) | 3 (0.07, 15) | 0 (0, 4)e | 3 (0.6, 9) | 2 (0.2, 7) |
| blaCMY-2E. colia | 9 (4, 16) | 2 (0.2, 7) | 3 (0.07, 15) | 0 (0, 4)e | 5 | 0 (0, 4)e |
Hospital n = 101;
n = 100;
n = 99;
n = 35;
One-sided 97.5% confidence interval. Isolation was not present from every hospital. All other sites were present in each of the sampled hospitals.
Table 2.
Prevalence (%) of bacteria on equipment in companion animal veterinary hospitals
| Prevalence (95% confidence interval) |
||||
|---|---|---|---|---|
| Organism | Telephone, keyboards and taps | Otoscope tips | Stethoscopes, Oto/ Ophthalmoscopes | Thermometers |
| E. colia | 34 (25, 44) | 7d (2, 16) | 22 (14, 31) | 28c (20, 38) |
| C. difficileb | 15 (9, 24) | 8e (2, 17) | 10 (5, 18) | 11c (6, 19) |
| MRSAb | 2 (0.2, 7) | 2f (0.04, 9) | 1 (0.02, 5) | 0c (0, 4)h |
| MRSPc | 1 (0.03, 5) | 0g (0, 6) | 1 (0.03, 5) | 0 (0, 4) |
| blaCMY-2E. colia | 0 (0, 4) | 0 (0, 4) | 0 (0, 4) | 2 (0.2, 7) |
n = 101 (unless noted);
n = 100 (unless noted);
n = 99 (unless noted);
n = 55;
n = 66;
n = 61;
n = 56;
One-sided, 97.5% confidence interval.
Table 3.
Ribotypes and toxin profiles of environmental Clostridium difficile isolates (n = 119) from companion animal veterinary hospitals
| Ribotype | Speciesa | Toxin A | Toxin B | CDT | Number of isolates | Percentage of isolatesb |
|---|---|---|---|---|---|---|
| V | Human | + | + | − | 25 | 21 |
| Y | Human | + | + | + | 8 | 7 |
| W | Human | + | + | − | 7 | 6 |
| 027 | Human | + | + | + | 6 | 5 |
| C | Human | + | + | + | 2 | 2 |
| K | Human | + | + | + | 2 | 2 |
| N | Human | + | + | + | 1 | 0.8 |
| L | Human | + | + | − | 1 | 0.8 |
| MS E | Human | + | + | − | 1 | 0.8 |
| MS I | Human | + | + | − | 1 | 0.8 |
| AK | Human | + | + | − | 1 | 0.8 |
| AA | Human | − | + | − | 1 | 0.8 |
| OVC B | Canine | − | − | − | 27 | 23 |
| OVC C | Canine | − | − | − | 21 | 18 |
| OVC Cc | Canine | + | + | − | 1 | 0.8 |
| OVC H | Canine | − | − | − | 1 | 0.8 |
| SL 3 | Canine | + | + | + | 1 | 0.8 |
| SL 4 | Canine | + | + | − | 1 | 0.8 |
| CM 65 | NDd | − | − | − | 3 | 3 |
| CM 53 | NDd | − | − | − | 3 | 3 |
| CM 57 | NDd | + | + | − | 2 | 2 |
| CM 76 | NDd | + | + | + | 1 | 0.8 |
| CM 103 | NDd | + | + | − | 1 | 0.8 |
| CM 92 | NDd | + | + | − | 1 | 0.8 |
Known animal host where the ribotype has been identified;
Sum does not equal 100 due to rounding;
Ribotype had 2 toxin gene patterns;
ND — not described — newly identified ribotype that has not been described in humans or an animal species.
The prevalence of antimicrobial resistance in E. coli isolates was low (0% to 13%), yet at least 1 resistant isolate was observed to each class of antimicrobials tested (Table 4). Combined resistance to ampicillin and amoxicillin-clavulanic acid was only observed with resistance to a third-generation cephalosporin or to a cephamycin. Sixty-eight E. coli isolates were selected for testing for the blaCMY-2 gene including 47% (n = 32) of the E. coli isolates obtained using ESBL selection methodology. Sixty percent (n = 41) of the tested isolates were PCR positive for the blaCMY-2 gene. These isolates were recovered from 9% (n = 9) of the hospitals (Table 1). Ninety-five percent (n = 39) of the blaCMY-2 E. coli isolates were resistant to ampicillin, amoxicillin-clavulanic acid and cefoxitin. Although only 2 isolates were resistant to ampicillin, amoxicillin-clavulanic acid, cefoxitin and ceftriaxone, isolates positive for the blaCMY-2 gene had a significantly higher proportion (P < 0.01) with a MIC ≥ 8 μg/mL for ceftriaxone.
Table 4.
Distribution (%) of antimicrobial minimum inhibitory concentrations of environmental Escherichia coli isolates (n = 1554) from veterinary hospitals.
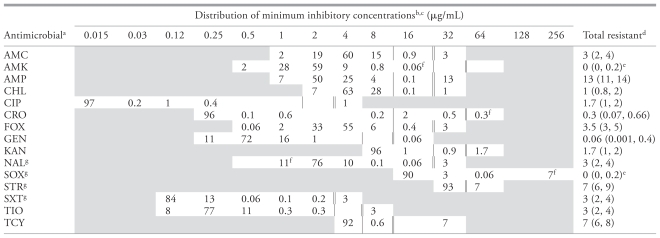 |
Antimicrobial abbreviations: AMC — amoxicillin-clavulanic acid, AMK — amikacin, AMP — ampicillin, CHL — chloramphenicol, CIP — ciprofloxacin, CRO — ceftriaxone, FOX — cefoxitin, GEN — gentamicin, KAN — kanamycin, NAL — nalidixic acid, SOX — sulfizoxazole, STR — streptomycin, SXT — trimethoprim-sulphamethoxazole, TIO — ceftiofur, TCY — tetracycline.
Minimum Inhibitory Concentration distribution: The unshaded fields indicate the MIC range tested for each antimicrobial in the plate configuration. The MICs at the upper or lower bound of the distribution are censored. The values at the upper bound are ≥ to the value presented and the values at the lower bound are ≤ the value presented.
Double bar represents the resistant breakpoint. Single bar represents the susceptible breakpoint.
Values in brackets are 95% confidence intervals. Total resistant may not equal values presented in table due to rounding error.
One sided 97.5% confidence interval.
Exact MIC.
Susceptible breakpoint-NAL: ≤ 16 μg/mL, Susceptible breakpoint-SOX: ≤ 256 μg/mL, Resistant breakpoint-SOX: ≤ 512 μg/mL, Susceptible breakpoint-STR: ≤ 32 μg/mL, Susceptible breakpoint-SXT: ≤ 2 μg/mL.
Generalized linear mixed models of potential factors associated with recovery of environmental bacteria
Models of potential factors associated with the recovery of MRSA and AMP-R E. coli could not be generated because of insufficient power. A model could not be generated for potential factors associated with the recovery of E. coli because of insufficient heterogeneity in the outcome. Using a multi-level logistic modeling procedure, the use of in-hospital parenteral trimethoprim-sulfonamide combinations was positively associated with the recovery of C. difficile [odds ratio (OR) = 2.73; P = 0.008; 95% confidence interval (CI): 1.30, 5.77] (Table 5). The use of in-hospital oral enrofloxacin was negatively associated with the recovery of C. difficile (OR = 0.51; P = 0.036; 95% CI: 0.27, 0.96). There were significantly higher odds of C. difficile recovery from kennel areas (OR = 2.17; P = 0.051; 95% CI: 1.01, 4.74) and borderline significantly lower odds of recovery from table surfaces (OR = 0.37; P = 0.057, 95% CI: 0.14, 1.03) when compared to floors. Hospital demographic data, disinfectant use, hand cleansers, availability of SOPs for environmental cleaning and disinfection use or presence of an isolation unit were not significantly associated with recovery of C. difficile. The ICChospital of C. difficile recovery was 0.25, indicating that there was low to moderate within-hospital clustering of environmental C. difficile. The distribution of residuals was bounded between −3.4 and 1.4. The upper tail of the normal plot was skewed since few hospitals had strongly positive residuals when compared to the tail of the distribution. However, Cook’s distance revealed no observations that were influential to the model.
Table 5.
Results from a generalized linear mixed modela of factors potentially associated with the recovery of Clostridium difficile from the environment in veterinary hospitals
| Variable | Odds ratio | Standard error | z-value | P-value | 95% confidence interval |
|---|---|---|---|---|---|
| Use of in-hospital enrofloxacin | 0.51 | 0.16 | −2.10 | 0.036 | 0.27, 0.96 |
| Use of in-hospital parenteral trimethoprim-sulfonamide combinations | 2.73 | 1.04 | 2.65 | 0.008 | 1.30, 5.77 |
| Sitesb | |||||
| Tables | 0.37 | 0.19 | −1.90 | 0.057 | 0.14, 1.03 |
| Kennels | 2.17 | 0.87 | 1.93 | 0.051 | 1.01, 4.74 |
| Isolation | 0.58 | 0.27 | −1.14 | 0.255 | 0.23, 1.47 |
| Telephones, computer keyboards, door knobs, and taps on kitchen and bathroom sinks | 1.06 | 0.46 | 0.14 | 0.891 | 0.45, 2.47 |
| Taps on examination and treatment room sinks | 1.11 | 0.54 | 0.21 | 0.832 | 0.43, 2.87 |
| Otoscopes, ophthalmoscopes, and stethoscopes | 0.58 | 0.27 | −1.15 | 0.250 | 0.23, 1.46 |
| Otoscope tips | 0.45 | 0.27 | −1.35 | 0.177 | 0.14, 1.43 |
| Thermometers | 0.59 | 0.28 | −1.10 | 0.269 | 0.24, 1.50 |
Number of observations: Level 1 (sites within veterinary hospitals) n = 807; Level 2 (veterinary hospitals) n = 92.
Number of iterations: 4. Log likelihood = −295.025. LR chi-squared value = 41.88 (P-value = 0.0000). Pseudo R2 = 0.0657. Level 2 variance = 1.045.
Referent: Floors.
Discussion
The bacteria recovered from environmental surfaces in this study are potential pathogens (E. coli, C. difficile) and also represent a pool of antimicrobial resistance genes (such as, MRSA, MRSP, blaCMY-2 E. coli) that could place human and animal health at risk. The recovered organisms may contribute to hospital acquired infections and possibly to the epidemiology of opportunist infections in the hospital and community including zoonoses. These bacteria can colonize or infect companion animals and people, and this could lead to dissemination of bacteria in the environment. In the absence of clinical signs, the identification of potentially infectious patients or staff, for practical purposes, is difficult. Therefore, implementation of appropriate measures to control the spread of these organisms in the veterinary hospital environment is important.
Methicillin-resistant S. aureus is an important antimicrobial resistant zoonotic organism. The cMRSA-2 clone is a common North American human clone associated with human hospital and community MRSA infections and colonization (20), companion animal clinical infections (20–22), colonization of human contacts of animal patients (21) and has been recovered from companion animal veterinarians (23). The cMRSA-5 clone is the predominant one associated with equine colonization and infections in North America (24) and can also colonize veterinary personnel (23,25). With both human and animal reservoirs, there is an opportunity for MRSA transmission in veterinary hospitals through contact between individuals. However, as demonstrated in this study an environmental reservoir of MRSA could be an additional source for colonization or infection of both humans and animals.
Methicillin-resistant S. pseudintermedius is an emerging antimicrobial resistant opportunist pathogen of companion animals. The relatively high prevalence of MRSP from environmental sites within hospitals (hospital prevalence 7%) is concerning especially given the low frequency of colonization and infection in dogs (26–28). Although it is a recognized zoonosis, no large human reservoir is known, yet it has been identified in veterinary personnel (29,30). The observed high frequency of recovery may result from poor environmental disinfection within hospitals. We were unable to test this hypothesis because of limited statistical power. However, given the recent emergence of highly multi-drug resistant MRSP (28,31,32) and the high frequency of recovery from veterinary hospitals, there is a need for further work to describe the epidemiology of MRSP.
In addition, blaCMY-2 E. coli was recovered from environ-mental sites. This organism has been described in humans (33) and many animal species (10,34–36) including dogs (2,37,38); therefore, the recovery of blaCMY-2 E. coli was not surprising. There are previous reports of HAI associated with this organism in dogs where an environmental reservoir was documented (2). Additionally, addressing many aspects of infection control, including environmental disinfection, aided in control of outbreaks of HAI associated with this pathogen.
Salmonella was recovered infrequently from hospitals. This may be due to the low level prevalence of carriage in healthy companion animals (37,39–41). Comparable studies in healthy humans have not been performed; however, S. Typhimurium was the second most common serovar reported from clinical human isolates in Canada (42). Salmonella Mbandaka infections have been described in humans (43) and food animals (44,45), but not in companion animals. Outbreaks of salmonellosis associated with S. Typhimurium in people and companion animals linked with veterinary hospitals or animal shelters have been reported (41,46). In 2 outbreaks, environmental samples yielded the strain/phage type of S. Typhimurium identified in the outbreak (41).
The high prevalence of C. difficile, including human and animal associated ribotypes, is noteworthy; especially the identification of ribotype 027, an epidemiologically important ribo-type, which has been associated with outbreaks of C. difficile-associated disease in North America and Europe (47) and has been recovered from a healthy hospital visitation dog in southern Ontario (48). The recovery of C. difficile is not unexpected since humans and animals can be asymptomatically colonized, and as a spore-forming bacterium, it is resistant to disinfectants. We were not able to demonstrate significant statistical associations between the recovery of C. difficile and the use of specific disinfectants, hand-cleansers, and the presence of an isolation area; however, the statistical power to determine any association was < 10%. Furthermore, the format of capturing data on the hypothesized risk factors for bacterial recovery using a questionnaire may have been inadequate. This method relied on veterinarians and veterinary technicians to accurately and precisely describe environmental disinfection (such as, contact time for disinfectants) and other infection control practices (duration of hand washing for example). There are no publications evaluating the accuracy of data on the evaluated infection control practices gathered in this manner. Inaccuracies or imprecision of the responses may result in the misclassification of hypothesized risk factors leading to an inability to determine associations between the recovered organisms and the hypothesized infection control practices.
The recovery of C. difficile was significantly higher from kennels and tables than from floors. Kennels are frequently contaminated with fecal material, including diarrhea. Tables in veterinary practices are high contact areas by animals and humans and can easily be contaminated with fecal material. Quaternary ammonium compounds (QAC) were the most frequently reported disinfectants used on table and kennels by veterinarians and veterinary technicians from the investigated practices (49). Clostridium difficile is resistant to disinfection with QACs. The data from these studies suggest that veterinary practices may need to consider high-level disinfectants for routine disinfection of kennels and tables.
The observed associations between C. difficile recovery and the use of in-hospital parenteral trimethoprim-sulfonamide combinations (positive association) and oral enrofloxacin (negative association) were difficult to interpret. In human medicine, use of many antimicrobials have been identified as risk factors for C. difficile infection, including fluoroquinolones, clindamycin and cephalosporins (50); however, their role as risk factors for human hospital environmental contamination with C. difficile has not been published (47,51,52). Alternatively, the observed associations may be driven by other unmeasured factors that were highly correlated with antimicrobial use, such as other infection control practices, patient population characteristics or patient care and management factors. Further studies are required to clarify the role, if any, of antimicrobial use on environmental contamination with C. difficile.
Given the high prevalence of C. difficile, it is perhaps surprising that CPV and FCV were not recovered from any hospitals, since they all require high level disinfection for removal from the environment. However, CPV and FCV are limited to dogs and cats, respectively, and shedding may have been uncommon in the study population. Companion animals and people can be asymptomatically colonized with C. difficile which may lead to a greater opportunity for dissemination of C. difficile in the veterinary hospital environment. The differences in apparent prevalence could also be due to infection control practices used in the management of known or suspected cases of CPV, or cases and carriers of FCV. Veterinarians from the investigated hospitals reported that animals with known CPV or FCV associated disease, animals with clinical signs of disease associated with the gastrointestinal or respiratory tract and animals without vaccinations would require specific infection control measures (49). Adequate infection control practices and low frequency of shedding, therefore, could account for the lack of recovery of CPV and FCV. However it is possible that sensitivity of the sampling methodology was too low to detect CPV and FCV.
This study demonstrated that recovery of environmental bacteria was possible using sterile electrostatic cloths, which are inexpensive, easily accessible, and simple to use and sterilize. Other authors have had similar experiences (53). Standardized sampling and microbiological methods for environmental organisms from the hospital environment have not been established. In addition, the sensitivity of recovery using different tools (such as, cotton swabs, electrostatic cloths, contact plates) is unknown. Determining the sensitivity of recovery and development of sampling and microbiological standards will improve interpretation of results and reproducibility of study design. Another challenge in determining the role of environmental pathogens in the epidemiology of HAI, is that environmental contamination may be a consequence of infection, rather than a source of infection. Selecting the appropriate study group, such as secondary cases rather than primary cases in an epidemic, may assist in determining real associations between an environmental reservoir of pathogens and HAI.
The issue of infection control in companion animal veterinary medicine is in the early stages. This study demonstrated that E. coli, blaCMY-2 E. coli, C. difficile, MRSA, and MRSP were present in environmental sites within community veterinary hospitals. Although this study did not attempt to correlate HAI with an environmental reservoir of organisms, the high frequency of recovery of potential pathogens combined with inadequate infection control policies provides an opportunity for an excess of HAI in community companion animal veterinary hospitals. This preliminary study points to the need for research in infection control in community companion animal veterinary medicine. This includes studies that quantify the frequency of HAI, enhance understanding of endemic and epidemic HAI, describe the scope of conditions and associated organisms contributing to HAI, describe factors associated with HAI, including the contribution of an environmental reservoir, and factors associated with reducing the environmental burden.
Acknowledgments
We acknowledge the following for their contributions to this study: Alyssa Calder, Karlee Thomas, and Adriana Sage for clinic sampling; staff of the Canadian Research Institute for Food Safety, especially Gabriel Jantzi and Virginia Young for E. coli and Salmonella isolation; Meredith Craig for data entry, Joyce Rousseau for C. difficile, MRSA and MRSP isolation and typing; Rebeccah Travis and Gerry Lazarek for PCR of the blaCMY-2 gene; staff of the Laboratory for Foodborne Zoonoses for susceptibility testing and typing of Salmonella isolates; and staff at the Prairie Diagnostics Services for virus isolation. CVJ
Footnotes
This study was funded by the OVC Pet Trust Fund and the Public Health Agency of Canada.
Reprints will not be available from the authors.
Use of this article is limited to a single copy for personal study. Anyone interested in obtaining reprints should contact the CVMA office (hbroughton@cvma-acmv.org) for additional copies or permission to use this material elsewhere.
References
- 1.Griffin FA. 5 Million Lives Campaign. Reducing methicillin-resistant Staphylococcus aureus (MRSA) infections. Jt Comm J Qual Patient Saf. 2007;33:726–731. doi: 10.1016/s1553-7250(07)33087-0. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez S, McCrackin Stevenson MA, Hudson CR, et al. Characterization of multidrug-resistant Escherichia coli isolates associated with nosocomial infections in dogs. J Clin Microbiol. 2002;40:3586–3595. doi: 10.1128/JCM.40.10.3586-3595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weese JS, Armstrong J. Outbreak of Clostridium difficile-associated disease in a small animal veterinary teaching hospital. J Vet Intern Med. 2003;17:813–816. doi: 10.1111/j.1939-1676.2003.tb02519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerlin P, Eugster S, Gaschen F, Straub R, Schawalder P. Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet Microbiol. 2001;82:347–359. doi: 10.1016/s0378-1135(01)00396-0. [DOI] [PubMed] [Google Scholar]
- 5.Ward MP, Brady TH, Couetil LL, Liljebjelke K, Maurer JJ, Wu CC. Investigation and control of an outbreak of salmonellosis caused by multidrug-resistant Salmonella Typhimurium in a population of hospitalized horses. Vet Microbiol. 2005;107:233–240. doi: 10.1016/j.vetmic.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Schott HC, Ewart SL, Walker RD, et al. An outbreak of salmonellosis among horses at a veterinary teaching hospital. J Am Vet Med Assoc. 2001;218:1152–1159. doi: 10.2460/javma.2001.218.1152. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC) 2003. [Last accessed July 13, 2010]. Available from http://www.cdc.gov/ncidod/dhqp/gl_environinfection.html.
- 8.Dohoo I, Martin W, Stryhn H. Veterinary Epidemiologic Research. Charlottetown, Prince Edward Island, Canada: AVC Inc; 2003. [Google Scholar]
- 9.NCCLS. Performance standards for antimicrobial susceptibility testing. National Committee for Clinical Laboratory Standards; Wayne, Pennsylvania: 2002. Approved standard M100-S15 M7-A-MIC Testing Section. [Google Scholar]
- 10.Allen KJ, Poppe C. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by beta-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can J Vet Res. 2002;66:137–144. [PMC free article] [PubMed] [Google Scholar]
- 11.Government of Canada. Canadian integrated program for antimicrobial resistance surveillance (CIPARS) 2004. Guelph, (Ontario): Public Health Agency of Canada; 2006. [Google Scholar]
- 12.Kato H, Kato N, Watanabe K, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupnik M, Grabnar M, Geric B. Binary toxin producing Clostridium difficile strains. Anaerobe. 2003;9:289–294. doi: 10.1016/j.anaerobe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 15.Martin H, Willey B, Low DE, et al. Characterization of Clostridium difficile strains isolated from patients in Ontario, Canada, from 2004 to 2006. J Clin Microbiol. 2008;46:2999–3004. doi: 10.1128/JCM.02437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulvey MR, Chui L, Ismail J, et al. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J Clin Microbiol. 2001;39:3481–3485. doi: 10.1128/JCM.39.10.3481-3485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CDC. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): 2001 Annual Report. U.S. Department of Health and Human Services, CDC; 2003. [Google Scholar]
- 18.Rabe-Heskethe S, Skrondal A, Pickles A. Reliable estimation of generalized linear mixed models using adaptive quadrature. The Stata Journal. 2002;2:1–21. [Google Scholar]
- 19.Rabe-Hesketh S, Skrondal A. Multilevel and longitudinal modeling using Stata. 2nd ed. College Station, Texas: Stata Press; 2008. [Google Scholar]
- 20.Christianson S, Golding GR, Campbell J, Mulvey MR. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45:1904–1911. doi: 10.1128/JCM.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weese JS, Dick H, Willey BM, et al. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet Microbiol. 2006;115:148–155. doi: 10.1016/j.vetmic.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Weese JS, Faires M, Rousseau J, Bersenas AM, Mathews KA. Cluster of methicillin-resistant Staphylococcus aureus colonization in a small animal intensive care unit. J Am Vet Med Assoc. 2007;231:1361–1364. doi: 10.2460/javma.231.9.1361. [DOI] [PubMed] [Google Scholar]
- 23.Hanselman BA, Kruth SA, Rousseau J, et al. Methicillin-resistant Staphylococcus aureus colonization in veterinary personnel. Emerg Infect Dis. 2006;12:1933–1938. doi: 10.3201/eid1212.060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weese JS, Rousseau J, Willey BM, Archambault M, McGeer A, Low DE. Methicillin-resistant Staphylococcus aureus in horses at a veterinary teaching hospital: Frequency, characterization, and association with clinical disease. J Vet Intern Med. 2006;20:182–186. doi: 10.1892/0891-6640(2006)20[182:msaiha]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Anderson ME, Lefebvre SL, Weese JS. Evaluation of prevalence and risk factors for methicillin-resistant Staphylococcus aureus colonization in veterinary personnel attending an international equine veterinary conference. Vet Microbiol. 2008;129:410–417. doi: 10.1016/j.vetmic.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 26.Hanselman BA, Kruth S, Weese JS. Methicillin-resistant staphylococcal colonization in dogs entering a veterinary teaching hospital. Vet Microbiol. 2008;126:277–281. doi: 10.1016/j.vetmic.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Griffeth GC, Morris DO, Abraham JL, Shofer FS, Rankin SC. Screening for skin carriage of methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi in dogs with healthy and inflamed skin. Vet Dermatol. 2008;19:142–149. doi: 10.1111/j.1365-3164.2008.00663.x. [DOI] [PubMed] [Google Scholar]
- 28.Ruscher C, Lubke-Becker A, Wleklinski CG, Soba A, Wieler LH, Walther B. Prevalence of methicillin-resistant Staphylococcus pseudin-termedius isolated from clinical samples of companion animals and equidaes. Vet Microbiol. 2009;136:197–201. doi: 10.1016/j.vetmic.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J Clin Microbiol. 2007;45:1118–1125. doi: 10.1128/JCM.02193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Duijkeren E, Houwers DJ, Schoormans A, et al. Transmission of methicillin-resistant Staphylococcus intermedius between humans and animals. Vet Microbiol. 2008;128:213–215. doi: 10.1016/j.vetmic.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Wettstein K, Descloux S, Rossano A, Perreten V. Emergence of methicillin-resistant Staphylococcus pseudintermedius in Switzerland: Three cases of urinary tract infections in cats. Schweiz Arch Tierheilkd. 2008;150:339–343. doi: 10.1024/0036-7281.150.7.339. [DOI] [PubMed] [Google Scholar]
- 32.Loeffler A, Linek M, Moodley A, et al. First report of multiresistant, mecA-positive Staphylococcus intermedius in Europe: 12 cases from a veterinary dermatology referral clinic in Germany. Vet Dermatol. 2007;18:412–421. doi: 10.1111/j.1365-3164.2007.00635.x. [DOI] [PubMed] [Google Scholar]
- 33.Pitout JD, Gregson DB, Church DL, Laupland KB. Population-based laboratory surveillance for AmpC beta-lactamase-producing Escherichia coli, Calgary. Emerg Infect Dis. 2007;13:443–448. doi: 10.3201/eid1303.060447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer RS, Patterson SK, Wallace RL. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl Environ Microbiol. 2008;74:6956–6962. doi: 10.1128/AEM.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smet A, Martel A, Persoons D, et al. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob Agents Chemother. 2008;52:1238–1243. doi: 10.1128/AAC.01285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanc V, Mesa R, Saco M, et al. ESBL- and plasmidic class C beta-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet Microbiol. 2006;118:299–304. doi: 10.1016/j.vetmic.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Murphy CP. MSc thesis. Guelph, Ontario: University of Guelph; 2004. Occurrence of antimicrobial resistance in selected bacteria in healthy dogs and cats presented to private veterinary clinics in Southern Ontario. [Google Scholar]
- 38.Carattoli A, Lovari S, Franco A, Cordaro G, Di Matteo P, Battisti A. Extended-spectrum beta-lactamases in Escherichia coli isolated from dogs and cats in Rome, Italy, from 2001 to 2003. Antimicrob Agents Chemother. 2005;49:833–835. doi: 10.1128/AAC.49.2.833-835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefebvre SL, Waltner-Toews D, Peregrine AS, et al. Prevalence of zoo-notic agents in dogs visiting hospitalized people in Ontario: Implications for infection control. J Hosp Infect. 2006;62:458–466. doi: 10.1016/j.jhin.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Van Immerseel F, Pasmans F, De Buck J, et al. Cats as a risk for transmission of antimicrobial drug-resistant Salmonella. Emerg Infect Dis. 2004;10:2169–2174. doi: 10.3201/eid1012.040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright JG, Tengelsen LA, Smith KE, et al. Multidrug-resistant Salmonella Typhimurium in four animal facilities. Emerg Infect Dis. 2005;11:1235–1241. doi: 10.3201/eid1108.050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Government of Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2007-Preliminary Results. Guelph, (Ontario): Public Health Agency of Canada; 2008. [Google Scholar]
- 43.Whichard JM, Gay K, Stevenson JE, et al. Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg Infect Dis. 2007;13:1681–1688. doi: 10.3201/eid1311.061438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerin MT, Martin SW, Darlington GA, Rajic A. A temporal study of Salmonella serovars in animals in Alberta between 1990 and 2001. Can J Vet Res. 2005;69:88–99. [PMC free article] [PubMed] [Google Scholar]
- 45.Liebana E, Guns D, Garcia-Migura L, Woodward MJ, Clifton-Hadley FA, Davies RH. Molecular typing of Salmonella serotypes prevalent in animals in England: Assessment of methodology. J Clin Microbiol. 2001;39:3609–3616. doi: 10.1128/JCM.39.10.3609-3616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherry B, Burns A, Johnson GS, et al. Salmonella Typhimurium outbreak associated with veterinary clinic. Emerg Infect Dis. 2004;10:2249–2251. doi: 10.3201/eid1012.040714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuijper EJ, Coignard B, Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12 (Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 48.Lefebvre SL, Arroyo LG, Weese JS. Epidemic Clostridium difficile strain in hospital visitation dog. Emerg Infect Dis. 2006;12:1036–1037. doi: 10.3201/eid1206.060115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy CP, Reid-Smith RJ, Weese JS, McEwen SA. Evaluation of specific infection control practices used by companion animal veterinarians in community veterinary practices in southern. Ontario Zoonoses and Public Health. doi: 10.1111/j.1863-2378.2009.01244.x. In press. [DOI] [PubMed] [Google Scholar]
- 50.Deshpande A, Pant C, Jain A, Fraser TG, Rolston DD. Do fluoroquinolones predispose patients to Clostridium difficile associated disease? A review of the evidence. Curr Med Res Opin. 2008;24:329–333. doi: 10.1185/030079908x253735. [DOI] [PubMed] [Google Scholar]
- 51.Elliott B, Chang BJ, Golledge CL, Riley TV. Clostridium difficile-associated diarrhoea. Intern Med J. 2007;37:561–568. doi: 10.1111/j.1445-5994.2007.01403.x. [DOI] [PubMed] [Google Scholar]
- 52.Blossom DB, McDonald LC. The challenges posed by reemerging Clostridium difficile infection. Clin Infect Dis. 2007;45:222–227. doi: 10.1086/518874. [DOI] [PubMed] [Google Scholar]
- 53.Burgess BA, Morley PS, Hyatt DR. Environmental surveillance for Salmonella enterica in a veterinary teaching hospital. J Am Vet Med Assoc. 2004;225:1344–1348. doi: 10.2460/javma.2004.225.1344. [DOI] [PubMed] [Google Scholar]