Abstract
Objectives
To determine the genetic composition of the first VanA-type plasmid (pIP816) reported, which was isolated from a clinical Enterococcus faecium (BM4147) strain in France in 1986, and to reveal the genetic units responsible for the dissemination of the vanA gene cluster by comparisons with current, published and additionally generated vanA-spanning plasmid sequences obtained from a heterogeneous E. faecium strain collection (n = 28).
Methods
Plasmid sequences were produced by shotgun sequencing using ABI dye chemistry and primer walking, and were subsequently annotated. Comparative sequence analysis of the vanA region was done with published plasmids, with a partial vanA plasmid (pVEF4) reported here and to >140 kb of sequence obtained from a collection of vanA-harbouring plasmid fragments.
Results
Bioinformatic analyses revealed that pIP816 from 1986 and contemporary vanA plasmids shared a conserved genetic fragment of 25 kb, spanning the 10.85 kb vanA cluster encoded by Tn1546, and that the larger unit is present in both clinical and animal complexes of E. faecium. A new group II intron in pVEF4 was characterized.
Conclusions
Comparative DNA analyses suggest that Tn1546 disseminates in and between clonal complexes of E. faecium as part of a larger genetic unit, possibly as a composite transposon flanked by IS1216 elements.
Keywords: glycopeptide drug resistance, mobile elements, horizontal gene transfer, vanA plasmids, poultry
Introduction
The mammalian gut-dwelling genus Enterococcus, particularly Enterococcus faecium and Enterococcus faecalis, have rapidly emerged as troublesome nosocomial pathogens. This has primarily occurred due to a combination of favourable antibiotic selective pressures with respect to intrinsic resistances,1 as well as an inherent propensity for inter- and intraspecies gene transfer2 (for an excellent review, see Willems and Bonten3). Plasmids and transposons have been shown to play key roles in the acquisition and dissemination of drug resistance in the genus Enterococcus.4–12 The emergence and rapid spread of glycopeptide-resistant E. faecium (GREF) has been a particular challenge, as there are few remaining options for antimicrobial treatment.1 The first clinical GREF was discovered in France in 1986 and harboured the VanA-type plasmid pIP816.13 Subsequently, nosocomial GREF has been reported worldwide, with an increasing prevalence in the USA in particular,14 but also in European countries in recent years (http://www.rivm.nl/earss/). In addition to the limited options for GREF treatment, there is also a concern for further horizontal transfer of glycopeptide resistance determinants into more pathogenic Gram-positive species, such as Staphylococcus aureus and Clostridium difficile.15 This is underscored by nine cases of VanA-type vancomycin-resistant S. aureus isolated in the USA since 2002, and the evidence for an enterococcal origin as well as plasmid-mediated transfer is compelling.16 More sequence information on VanA-type plasmids from different reservoirs is necessary to clarify their role and function in the maintenance and dissemination of glycopeptide resistance determinants in Enterococcus spp.
The use of the glycopeptide avoparcin as an animal growth promoter in European countries provided the opportunity for a build-up of a community reservoir of GREF.17,18 Avoparcin resistance mediates cross-resistance to vancomycin, a clinically important antibiotic,19 and avoparcin was thus prohibited for further use in animal husbandry. However, diverse GREF strains have persisted on farms several years after the ban.20–22 The persistent GREF populations carry vanA plasmids harbouring the non-conjugative transposon Tn1546.21,23 Tn1546 is also frequently located on plasmids in GREF strains isolated from hospitalized patients and healthy volunteers in the community.24–26 The presence of conserved Tn1546 elements in genomically heterogeneous E. faecium isolates from various environments suggests the spread of resistance by horizontal gene transfer.27 We have previously reported the presence of a common 372 bp DNA stretch immediately flanking Tn1546 in a polyclonal E. faecium population.21,22 It was hypothesized that the vanA gene cluster (Tn1546) disseminates as a larger genetic unit than the transposon, but smaller than a plasmid, due to the heterogeneous plasmid sizes and restriction patterns observed in the strain collection examined.
The aim of this study was to increase the knowledge on enterococcal plasmid population genetics and dynamics, with an emphasis on E. faecium vanA plasmids. We present: (i) the complete sequence of the Tn1546-containing pIP816,13 first isolated in France 1986; (ii) a partial sequence of the Tn1546-containing E. faecium pVEF4, isolated from a Norwegian poultry farm previously exposed to avoparcin; and (iii) 28 Tn1546-containing genetic fragments amplified and sequenced from diverse E. faecium strains. Comparative analyses suggest that a genetic unit larger than the 10.85 kbp Tn1546 has facilitated the horizontal spread of plasmid-encoded glycopeptide resistance between different E. faecium clonal lineages. Sequence data indicate horizontal dissemination as a composite transposon. Lastly, a novel enterococcal group II intron was identified within the composite transposon of several of the vanA plasmids and is functionally analysed here. Group II introns are ribozymes that catalyse their own excision and ligation of flanking exon sequences.28
Materials and methods
Bacterial strains and plasmids
The bacterial strains and plasmids, and their relevant characteristics are given in Table 1. All strains were grown at 37°C using brain heart infusion (BHI) agar or broth (Fluka BioChemika). The VanA-positive strains of E. faecium were grown in media supplemented with 10 mg/L vancomycin (Sigma).
Table 1.
The vanA-containing E. faecium strains used in this study and their characteristics
| Strain | Geographical origin | Sample source | Epidemiologya | MLST/CCb | PFGE | Reference/sourcec |
|---|---|---|---|---|---|---|
| 399/F99/H8 | Norway | human | cs | 195/CC9 | 7 | 21 |
| 399/F99/A9 | Norway | animal | cs | 241/CC9 | 10A | 21 |
| 399/S99/A7 | Norway | animal | cs | ND | 11 | 21 |
| BM4147 | France | human | ci | 95/CC22 | ND | 13 |
| 399/F98/A4 | Norway | animal | cs | ND | 5 | 21 |
| BM4147-1 | France | — | — | 95/CC22 | — | 13 |
| 399/F98/A1 | Norway | animal | cs | ND | 9 | 21 |
| 399/F99/A8 | Norway | animal | cs | 9/CC9 | 11 | 21 |
| 64/F98/H2 | Norway | human | cs | 242/ND | 14 | 21 |
| 356/98/H | Norway | human | cs | ND | ND | 22 |
| 31/F01/H | Norway | human | cs | ND | ND | 22 |
| 399/F99/A10 | Norway | animal | cs | 310/CC9 | 5A | 21 |
| 64/F99/A6 | Norway | animal | cs | 146/ND | 15 | 21 |
| 64/F99/H6 | Norway | human | cs | 246/CC9 | 17 | 21 |
| 58/F01/H | Norway | human | cs | ND | ND | 22 |
| 399/F98/H2 | Norway | human | cs | 8/CC9 | 2 | 21 |
| 399/S99/H6 | Norway | human | cs | 2/CC1 | 6 | 21 |
| 64/F98/H1 | Norway | human | cs | 48/CC9 | 13 | 21 |
| 64/F98/A3 | Norway | animal | cs | 8/CC9 | 20 | 21 |
| K8-61 | Norway | human | ci | ND | ND | K-resc |
| 64/F98/A2 | Norway | animal | cs | 195/CC9 | 19 | 21 |
| TUH32-76 | Portugal | human | ci | 132/CC17 | ND | 57 |
| 32/F02/H | Norway | human | cs | ND | ND | 22 |
| K9-72 | Norway | human | ci | ND | ND | K-resc |
| 64/S99/A5 | Norway | animal | cs | ND | 21 | 21 |
| TUH2-8 | Sweden | human | ci | ND | ND | K-resc |
| TUH32-64 | UK | human | ho | 18/CC17 | ND | 57 |
| TUH32-71 | Netherlands | human | ho | 16/CC17 | ND | 57 |
| TUH32-72 | USA | human | ho | 17/CC17 | ND | 57 |
| TUH32-74 | USA | human | ho | 20/ND | ND | 57 |
| TUH32-77 | Portugal | human | ci | 125/ND | ND | 57 |
| TUH32-78 | Netherlands | human | ci | 21/CC9 | ND | 57 |
| TUH32-79 | Netherlands | human | hs | 5/CC5 | ND | 57 |
| TUH43-7 | Norway | human | cs | ND | ND | 22 |
MLST, multilocus sequence type; PFGE, pulsed-field gel electrophoresis; CC, clonal complex; ND, not determined.
aci, clinical isolate; cs, community survey; ho, hospital outbreak; hs, hospital survey.
cK-res, Reference Centre for Detection of Antimicrobial Resistance, Department of Microbiology and Infection Control, University Hospital of North Norway.
DNA sequencing and analyses of pIP816 and pVEF4
Plasmid DNA was isolated by alkaline lysis, as previously described.21,29 The fragmentation of plasmid pIP816 and subsequent cloning in Escherichia coli was done with the TOPO shotgun subcloning kit, as described by the manufacturer (Invitrogen). Plasmid DNA was purified prior to sequencing with the Perfectprep Plasmid 96 Vac system (Eppendorf). Custom primers (Sigma–Genosys) were used in PCRs for gap closure. The sequencing was done using ABI BigDye terminator chemistry (Perkin–Elmer Applied Biosystems Inc.) with ABI3130XL automated sequencers. Assembly of the sequence data was done using the Staden package30 and Phrap (http://www.phrap.org/). The initial plasmid sequence of pIP816 was presented as a poster at the International Symposium on Plasmid Biology, 2006.31 The draft contig sequences of pVEF4 were provided by Macrogen, Korea, using BigDye chemistry and with a sequence depth of >14× coverage. Further extensive primer walk and gap closure experiments were done; however, we did not succeed in plasmid closure. Artemis was used to annotate the nucleotide sequence of pIP816 and pVEF4.32 GLIMMER was used to predict coding sequences (CDSs)33 that were checked manually by correlation scores of the open reading frames with ≥50 amino acids. Sequence similarity searches by FASTA and BLASTP refined the predictions.34,35
Five complete E. faecium vanA plasmids were published in the nucleotide databases EMBL, GenBank and DDBJ as of May 2010 (Table 2). These plasmids as well as the partial pVEF4 sequence were analysed according to genetic composition and synteny using the Artemis Comparison Tool.32
Table 2.
The completely sequenced vanA plasmids of enterococci as of May 2010
| Plasmid | Host strain | Size (kbp) | Reference | Accession number |
|---|---|---|---|---|
| pIP816 | E. faecium BM4147 | 34.6 | This study | AM296544 |
| pVEF1 | E. faecium 399/F99/H8 | 39.6 | 11 | AM410096 |
| pVEF2 | E. faecium 399/F99/A9 | 39.6 | 11 | AM410096 |
| pVEF3 | E. faecium 399/S99/A7 | 63.1 | 58 | AM931300 |
| pHTβa | E. faecium FH | 63.7 | 59 | AB183714 |
aThe vanA plasmid (pHTβ) isolated from an E. faecium strain in Japan was included in the presented analysis. However, no DNA sequence identity was found, except from the presence of Tn1546.
PCR amplification and DNA sequencing of fragments of vanA plasmids
The DNA regions flanking the vanA gene cluster were also determined for 28 different vanA plasmids isolated from genomically diverse E. faecium strains of human or animal origin (Table 1). Isolated plasmid DNA was treated with PlasmidSafe DNase (Epicentre), and subsequently digested separately with PstI, EcoRI and BamHI according to the manufacturer's protocol (New England BioLabs), and separated by agarose gel electrophoresis. Undigested plasmid DNA was used in PCR assays, where rTth DNA polymerase (GeneAmp XL PCR Kit, Applied Biosystems) and JumpStart Taq DNA Polymerase (JumpStart ReadyMix Taq, Sigma) were used according to the manufacturers' protocols to amplify DNA fragments up- and downstream of Tn1546 using the primers specified in Table 3. Four PCR assays with the following primer combinations were used: PCF1 and PCF2 (7.1 kb amplicon); PCF1 and PCF3 (7.6 kb amplicon); PCF1 and PCF4 (6.0 kb amplicon); and PCF5 and PCF6 (1.2 kb amplicon). The PCR cycling parameters used in the amplifications of long fragments were initial denaturation at 94°C for 3 min followed by 25 cycles of 94°C for 1 min and 64°C for 11 min, and a final extension at 72°C for 10 min. For the JumpStart Taq PCRs, the cycling parameters were denaturation at 94°C for 2 min followed by 30 cycles of denaturation (94°C, 30 s), annealing (58°C, 30 s) and extension (72°C, 1 min 30 s), and a final extension at 72°C for 5 min. PCR products were analysed by agarose gel electrophoresis. Positive PCR products were confirmed by sequencing, as described above. Custom sequencing primers were used for primer walking when necessary (primers not given).
Table 3.
PCR and RT–PCR primers used in the study
| Primer | Sequence (5′–3′) | Binding site or reference |
|---|---|---|
| PCF1 | AGGGATTCGTCAGGAAAATAGG | pVEF1 nt 19439–19460 |
| PCF2 | AGCGTGTATGGTTTCAATTCC | pVEF1 nt 20705–20685 |
| PCF3 | TCTCTTACGATTTTCTCATCCACA | pVEF1 nt 26932–26909 |
| PCF4 | TCGTGACAATCGGAACTAAAACT | pIP816 nt 27553–27575 |
| PCF5 | AGTAACAAAGAAAGCCCAATTATCA | pVEF1 nt 8748–8724 |
| PCF6 | ACTTTTAGTTGGCTTGGACTGAAC | pVEF1 nt 2742–2765 |
| giiF | TGGAATGATAGGGTAACG | Intron (5′ end, forward) |
| giiF5 | TGGTTGCGAGACTTAGGAAAAC | Intron (3′ end, forward) |
| giiR3 | AYACGGCGTTCCATCAA | Intron (3′ end, reverse) |
| giiR7 | TAAGGTATAAGGTGGGCGTTTG | topo (3′ end, reverse) |
| giiR8 | TGTTCTACCCGACACATTTCTG | Intron (5′ end, reverse) |
| ip3F | AGACCCACTATTTACAGATG | topo (5′ end, forward) |
| Ent1 | TACTGACAAACCATTCATGATG | 60 |
| Ent2 | AACTTCGTCACCAACGCGAAC | 60 |
Group II intron characterization
Intron RNA secondary structure predictions were performed by the mfold server v. 3.236,37 and classification was based on group II intron consensus structures.38 The broader distribution of the group II intron, and its presence in the topoisomerase I gene, was investigated among the 28 vanA plasmids isolated from the E. faecium strain collection (Table 1) by PCR with the primers giiF/giiR3 and ip3F/giiR7 (Table 3) and JumpStart Taq DNA Polymerase, as described above. RNA extraction and subsequent RT–PCR were used to confirm intron splicing. From overnight cultures of E. faecium 399/S99/A7 and E. faecium 399/F98/A4, 10 µL of each culture was transferred to 10 mL fresh BHI broth and further cultured to a density of ∼1 × 108 cells. The cell cultures were treated with RNAlater (Ambion), according to the supplier's protocol, followed by total RNA extraction by QIAGEN RNeasy Mini kit and DNaseI treatment with QIAGEN RNase-Free DNase (QIAGEN). RNA extracts were verified free of DNA contamination by using 1 µg of total RNA as template in a PCR with the DyNAzyme II DNA polymerase (Finnzymes). cDNA syntheses were done using the SuperScript III enzyme (Invitrogen) on ∼1 µg of total RNA as template and, in other respects, according to protocol. RT–PCRs were carried out with the primers given in Table 3. The RT–PCR cycling parameters used were denaturation at 94°C for 2 min followed by 30 PCR cycles of 94°C for 45 s, 58°C for 30 s and 72°C for 30 s to 1.5 min (depending on the length of the product), and a final extension at 72°C for 5 min. The products were analysed by agarose gel electrophoresis and stained with ethidium bromide. To confirm the splicing boundaries, the PCR fragments of ligated exons were sequenced.
Results
General features of the VanA-type plasmids pIP816 and pVEF4
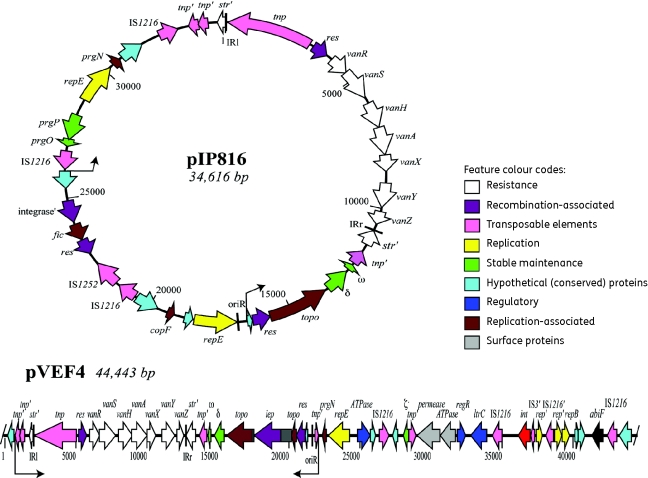
The complete sequence of the plasmid pIP816 was determined and a partial sequence of pVEF4 is presented. pIP816 was extracted from the first glycopeptide-resistant E. faecium reported (strain BM4147) and was sampled from a patient with leukaemia in France in 1986.13 pVEF4 was isolated from an E. faecium strain (399/F98/A4) sampled from poultry in 1998 on a Norwegian poultry farm where avoparcin was previously used as a growth promoter.21 pIP816 (EMBL accession no. AM932524) is 34 616 bp in size, and has 36 CDSs and an average G + C content of 36.9% (Figure 1 and Table 4). The partial sequence of pVEF4 (EMBL accession no. FN424376) is 44 443 bp in length, and has a G + C content of 36.2% and 46 CDSs (Figure 1 and Table 5). Repeated attempts were made to achieve gap closure, but we did not succeed. We do, however, include the 44.4 kb partial, single sequence in this study, because it provides valuable additional information for the analysis of Tn1546 junctions (see below) as well as on the composition of the overall E. faecium plasmid pool. Similarity searches identified 31 and 39 CDSs with known or predicted functions in pIP816 and in pVEF4, respectively. pIP816 and pVEF4 both encode a pseudo-streptomycin resistance gene (str) that the Tn1546 transposon has transposed into, as well as genes involved in plasmid partitioning, a resolvase, truncated transposases and a hypothetical protein (Figure 1). A topoisomerase is also present on both plasmids, but in pVEF4 a group II intron with an intron-encoding protein (CDS19) was identified inserted into the topo gene (see below). The entire Tn1546 transposon, with the 38 bp inverted repeats together with the 5 bp direct repeats (5′-GTCCT-3′) of the Tn1546 target site in str, is conserved in both plasmids.
Figure 1.
Genetic map of pIP816 and pVEF4. Coding regions are represented by arrows indicating the direction of transcription and are coloured according to their predicted functions. The inverted repeats (IR) of the Tn1546 transposon and the predicted origin of replication (oriR) of the plasmids are given as black boxes. The group II intron En.fm.I2 of pVEF4 is shown as dark grey boxes flanking the intron-encoding protein. Thin arrows indicate the 25 kb larger genetic unit. Truncated CDSs are indicated with a prime symbol (e.g. tnp′).
Table 4.
Coding sequences (CDSs) of the circular vanA plasmid pIP816
| CDSa |
Protein length (amino acids) | Database match | Amino acid identity (%) | |
|---|---|---|---|---|
| 1 | tnp Tn1546 | 988 | pIP816 Tn1546 transposase (Q06238) | 100 |
| 2 | res Tn1546 | 191 | pIP816 resolvase (Q06237) | 100 |
| 3 | vanR | 231 | pIP816 VanR protein (Q06239) | 100 |
| 4 | vanS | 384 | pIP816 VanS protein (Q06240) | 100 |
| 5 | vanH | 322 | pIP816 VanH protein (Q05709) | 100 |
| 6 | vanA | 343 | pIP816 VanA protein (P25051) | 100 |
| 7 | vanX | 202 | pIP816 VanX protein (Q06241) | 100 |
| 8 | vanY | 303 | pIP816 VanY protein (P37711) | 100 |
| 9 | vanZ | 161 | pIP816 VanZ protein (Q06242) | 100 |
| 10 | str′ | 188 | N-terminal part of pS194 streptomycin resistance protein, Staphylococcus aureus (P12055) | 95 |
| 11 | tnp′ | 176 | pLI100 transposase, Listeria innocua (CAC42047) | 98 |
| 12 | ω | 71 | pSM19035 transcriptional repressor ω, Streptococcus pyogenes (YP_232757) | 100 |
| 13 | δ | 298 | pIlo8 δ protein, Oenococcus oeni (CAD70616) / pSM19035 active partitioning δ protein, S. pyogenes (YP_232765) | 100/98 |
| 14 | topo | 714 | pAMβ1 type 1 topoisomerase, Enterococcus faecalis (AAC38606) | 97 |
| 15 | resIP | 205 | pGB354 resolvase, Streptococcus agalactiae (AAB48454) | 100 |
| 16 | hcp | 56 | pRE25 orf7 hypothetical conserved protein, E. faecalis (CAC29163) | 100 |
| 17 | repE | 497 | pAMβ1 RepE replication protein, E. faecalis (Q52249) | 100 |
| 18 | hcp | 99 | pAMβ1 orfD, E. faecalis (Q52248) | 100 |
| 19 | copF | 83 | pAMβ1 copy number repressor, CopF, E. faecalis (Q52247) | 100 |
| 20 | hcp | 274 | pAMβ1 orfC, E. faecalis (Q52246) | 100 |
| 21 | IS1216 | 228 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 100 |
| 22 | IS1252 | 319 | pHKK701 IS1252 transposase (AAB42161) | 99 |
| 23 | res | 190 | pRE25 orf53 resolvase, E. faecalis (Q9AKZ9) | 100 |
| 24 | fic | 201 | pRE25 orf52 cell filament protein, Fic, E. faecalis (Q9AL00) | 100 |
| 25 | Integrase′ | 215 | Integrase, Enterococcus faecium DO (Q3Y1H6) | 81 |
| 26 | hcp′ | 214 | Plasmid2 hypothetical protein, Nitrosomonas eutropha C71 (Q3N6Z6) | 39 |
| 27 | IS1216 | 228 | IS1216, E. faecium (Q9KI43) / IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786, pUW1965 and pE418 (e.g. CAC29206) | 100/99 |
| 28 | prgO | 91 | pRE25 PrgO protein, E. faecalis (Q9AKZ5) | 100 |
| 29 | prgP | 317 | pRE25 PrgP protein, E. faecalis (Q9AKZ4) | 100 |
| 30 | repE | 498 | pRE25 ORF1, putative replication protein, E. faecalis (Q9AL28) | 100 |
| 31 | prgN | 98 | pRE25 PrgN protein, E. faecalis (Q9AL27) | 100 |
| 32 | hcp | 283 | Hypothetical protein of E. faecium DO (EAN10371) / pRE25 hypothetical protein ORF4, E. faecalis (Q9AL25) | 100/99 |
| 33 | IS1216 | 228 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 100 |
| 34 | tnp′ | 118 | pLI100 pLI0071 protein, putative transposase of L. innocua (Q925W6) | 85 |
| 35 | tnp′ | 114 | pLI100 pLI0020 protein, putative transposase of L. innocua (Q926N5) | 87 |
| 36 | str′ | 94 | C-terminal part of pS194 streptomycin resistance protein, S. aureus (P12055) | 95 |
aPrime indicates truncated CDS; hcp, hypothetical conserved protein.
Table 5.
Coding sequences (CDSs) of the vanA plasmid pVEF4 (partial)
| CDSa |
Protein length (amino acids) | Database match (accession number) | Amino acid identity (%) | |
|---|---|---|---|---|
| 1 | hp | 135 | — | |
| 2 | tnp′ | 118 | pVEF2 truncated transposase, C-terminal part (CAL90948) | 100 |
| 3 | tnp′ | 114 | pVEF1/pVEF2 truncated transposase, N-terminal part (CAL36541/CAL90947) | 100 |
| 4 | str′ | 94 | pS194 streptomycin resistance protein, Staphylococcus aureus (P12055) | 95 |
| 5 | tnp Tn1546 | 988 | pIP816 Tn1546 transposase (Q06238) | 100 |
| 6 | res Tn1546 | 191 | pIP816 resolvase (Q06237) | 100 |
| 7 | vanR | 231 | pIP816 VanR protein (Q06239) | 100 |
| 8 | vans | 384 | pIP816 VanS protein (Q06240) | 100 |
| 9 | vanH | 322 | pIP816 VanH protein (Q05709) | 100 |
| 10 | vanA | 343 | pIP816 VanA protein (P25051) | 100 |
| 11 | vanX | 202 | pIP816 VanX protein (Q06241) | 100 |
| 12 | vanY | 303 | pIP816 VanY protein (P37711) | 100 |
| 13 | vanZ | 161 | pIP816 VanZ protein (Q06242) | 100 |
| 14 | str′ | 188 | pS194 streptomycin resistance protein, S. aureus (P12055) | 95 |
| 15 | tnp′ | 176 | pVEF1/pVEF2 truncated transposase, C-terminal part (CAL36529/CAL90935) | 100 |
| 16 | ω | 71 | pVEF1/pVEF2 putative transcriptional repressor ω (CAL36528/CAL90934) | 100 |
| 17 | δ | 298 | pVEF1/pVEF2 putative δ protein (CAL36527/CAL90933) | 100 |
| 18 | topo | 715 | pVEF1/pVEF2 putative topoisomerase I (CAL36526/CAL90932) | 100 |
| 19 | iep | 628 | pBc10987 group II reverse transcriptase, Bacillus cereus ATCC 10987 | 59 |
| 20 | res | 205 | pVEF1/pVEF2 putative resolvase (CAL36525/CAL90931) | 100 |
| 21 | hcp | 56 | pRE25 and pVEF1/pVEF2 hypothetical conserved protein (Q9AL24, CAL36524/CAL90930) | 100 |
| 22 | IS1216′ | 82 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 97 |
| 23 | prgN | 95 | pVEF1/pVEF2 putative PrgN protein (CAL36522/CAL90928) | 100 |
| 24 | repE | 499 | pVEF1/pVEF2 putative replication protein (CAL36521/CAL90927) | 100 |
| 25 | ATPase | 303 | pVEF1/pVEF2 putative ATPase (CAL36563/CAL90969) | 100 |
| 26 | hcp | 123 | pVEF1/pVEF2 hypothetical protein (CAL36562/CAL09068) | 100 |
| 27 | IS1216 | 228 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 100 |
| 28 | hcp | 121 | pIP501 orf7, Streptococcus agalactiae (Q7AYQ0) | 100 |
| 29 | ζ′ | 84 | pVEF1/pVEF2 ζ toxin (CAL36553/CAL90959) | 95 |
| 30 | hcp | 196 | pVEF1/pVEF2 hypothetical protein (CAL36558/CAL909064) | 99 |
| 31 | permease | 537 | Putative tetronasin resistance transmembrane protein, Streptococcus pyogenes MGAS10750 (YP_603196) | 69 |
| 32 | ATPase | 293 | Putative tetronasin resistance ATP-binding protein, S. pyogenes MGAS10750 (YP_603195) | 85 |
| 33 | regR | 198 | pVEF1/pVEF2 putative regulatory protein, TetR family (CAL36555/CAL90961) | 97 |
| 34 | ltrC | 374 | pMRC01 low temperature requirement C protein LtrC, Lactococcus lactis (AAC56005) | 58 |
| 35 | IS1216 | 228 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 100 |
| 36 | int | 278 | Putative integrase, catalytic region, Enterococcus faecium DO (EAN09812) | 99 |
| 37 | IS3/IS911 | 96 | Putative IS3/IS911, E. faecium DO (EAN09811) | 100 |
| 38 | repA′ | 137 | pEF418 putative replication protein, Enterococcus faecalis (AAL05545) | 98 |
| 39 | IS1216′ | 179 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 100 |
| 40 | rep′ | 172 | pEFR putative replication protein, E. faecium (Q8KSS2) | 66 |
| 41 | repB | 174 | pB82 replication protein RepB, E. faecium (A0JBS1) | 49 |
| 42 | hcp | 93 | Hypothetical conserved protein, E. faecium DO (EAN10162) | 60 |
| 43 | hcp | 138 | pEF1 orf33 hypothetical conserved protein, E. faecium 6T1a (A3QN12) | 55 |
| 44 | abiF | 258 | pNP40 abortive infection bacteriophage resistance protein, L. lactis DRC3 (AAB52386) | 53 |
| 45 | IS1216 | 228 | IS1216 on pRE25, pRUM, pTEF1, pTEF3, pUW786 and pE418 (e.g. CAC29206) | 100 |
| 46 | hp | 293 | Hypothetical protein | — |
aPrime indicates truncated CDS; hcp, hypothetical conserved protein; hp, hypothetical protein.
The plasmids pIP816 and pVEF4 contain putative CDSs involved in their own replication and maintenance. The replication of pIP816 is probably ensured through the replication protein RepE (CDS17) described in the Inc18 plasmid pAMβ1 from E. faecalis.39 The origin of replication (oriR), copy number repressor (CopF), and hypothetical proteins ORFC and ORFD of pAMβ1 are also 100% conserved in pIP816 (Table 4). Thus, pIP816 is predicted to replicate by a DNA polymerase I-dependent θ mechanism, such as pAMβ1.40,41 pVEF4 encodes a replication protein (CDS24) previously reported in pVEF1 and pVEF2,11 and an oriR identical to oriR of pAMβ1 is found downstream of the Rep protein. Recently, Jensen et al.42 presented a classification system for plasmids from enterococci and other Gram-positive bacteria based on the conserved areas of the replication initiation genes (rep). According to this classification system, pIP816 groups into rep families 1 and 2, whereas pVEF4 is a member of group 1 (but also harbours an additional rep gene with limited sequence similarity to group 11). The presence of more than one rep gene may indicate increased host range, and groups 1 and 2 contain strains from Enterococcus, Staphylococcus and Streptococcus.
Genes involved in site-specific recombination (resolvases) and active partition processes of plasmids are found on pIP816 and pVEF4, putatively contributing to stable inheritance. Thus, the segregational stability of pIP816 is putatively under the control of the active partitioning systems delta-omega (δ-ω), as described for plasmid pSM19035 from Streptococcus pyogenes,43 and/or the prgP-prgO gene products as described for pCF10 from E. faecalis.44 Complete and fragmented insertion sequences (ISs) and transposases constitute 21% (n = 8) and 18% (n = 11) of the DNA sequence of pIP816 and the partial pVEF4, respectively. Inverted repeats of the IS6 (5′-ggttctgttgcaaagttttaaatctactatcaaa-3′) or IS30 families (5′-cgccgattgtaaaattaagctagacaaata-3′) are found along with most of the IS elements.
Comparative analysis of vanA plasmids
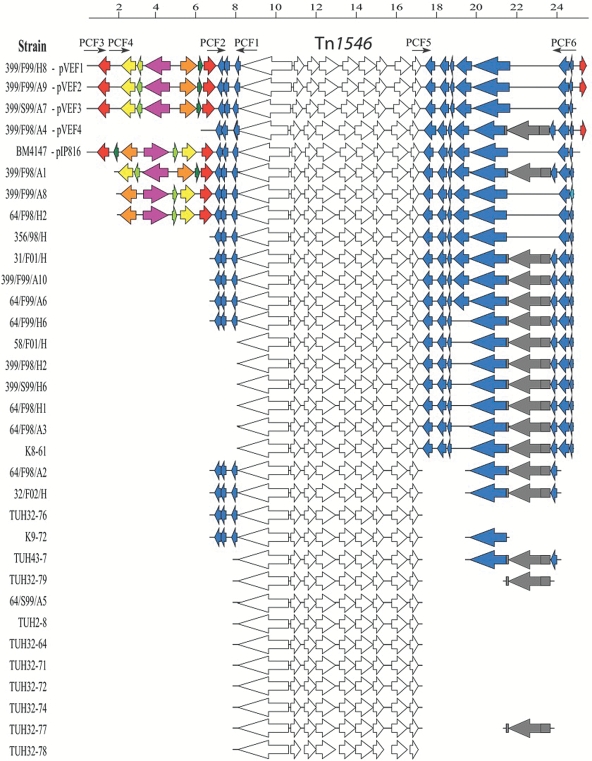
A genetic element (∼18 kb) including the non-conjugative Tn1546 was found conserved between the vanA plasmids pIP816, pVEF1, pVEF2, pVEF3 and partial pVEF4. An additional 7 kb element, flanked by two IS1216, was present immediately upstream in pIP816 (inverted), pVEF1, pVEF2 and pVEF3 (Figure 2). Thus, a common conserved genetic fragment of ∼25 kb was present in three vanA plasmids with a separate evolutionary history over >13 years.
Figure 2.
Gene organization of Tn1546 and flanking areas in vanA plasmids from genomically different E. faecium strains of human or animal origin. Identical coding regions are colour-coded to highlight similarities in the Tn1546 flanking regions. Similar colour indicates identity. White, Tn1546; red, IS1216; dark grey, intron En.fm.I2 and iep. Note that the ∼7 kbp region flanked by IS1216 (red arrows) in pVEF1–pVEF3 is inverted in pIP816, and that similar organization was found in plasmids from two GREF from two Norwegian poultry farms (strains 399/F99/A8 and 64/F98/A1). The top line indicates the size of the aligned Tn1546 flanking regions, with positional marks in kbp.
The presence of the conserved 25 kb vanA-containing fragment was further examined among a collection of 28 heterogeneous vanA plasmids, as determined by restriction fragment length polymorphism analyses (data not shown), from genomically different E. faecium strains of diverse epidemiology and geographical origin (Table 1). The complete 25 kb fragment was only present in the completely sequenced pVEF1, pVEF2 and pVEF3. These three plasmids are present in genomically diverse strains, as previously shown by PFGE.21 In pVEF1 and pVEF2, the 25 kb fragment is flanked by IS1216. Parts of the elements flanking the vanA gene cluster were also found on 15 additional plasmids. Of these, 12 were extracted from E. faecium strains sampled from Norwegian poultry farms (four poultry strains and eight poultry farmer strains at six different timepoints from 1998–2002) and three extracted from clinical strains (two from Norway and one from Portugal). Available multilocus sequence typing (MLST) data revealed that the different strains with intact or parts of the common Tn1546-containing fragment group into clonal complexes 1, 9, 17 and 22 (Table 1).
DNA sequence alignment of the amplified plasmid fragments demonstrates high sequence identity, but a deletion of the δ gene was found for eight plasmids (Figure 2). The 5′ end of Tn1546 is inverted in pIP816 compared with pVEF1, pVEF2 and pVEF3, and both gene order patterns are found on other plasmids (Figure 2).
pVEF4 encodes the group II intron En.fm.I2
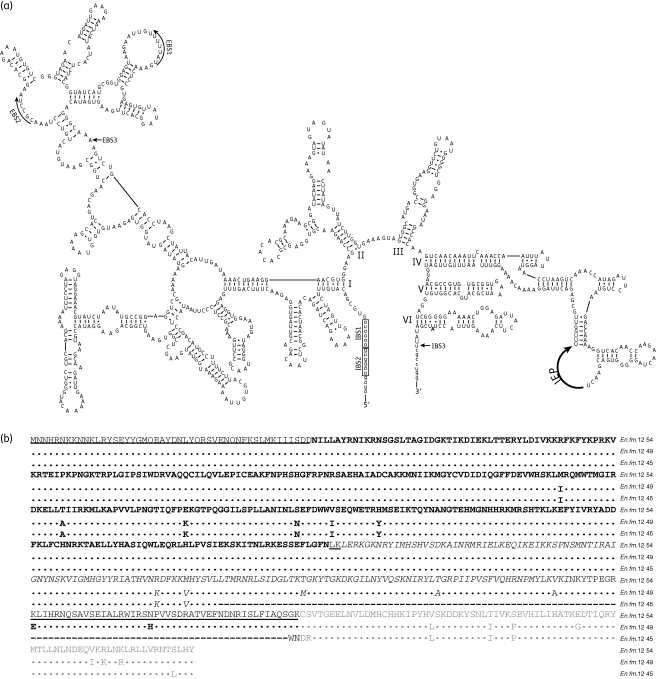
The annotation of the partial pVEF4 from E. faecium 399/F98/A4 identified a 2770 bp group II intron inserted into the topoisomerase I (topo) gene and it was named En.fm.I2 according to the mobile group II intron database nomenclature.45 The predicted secondary RNA structure (Figure 3a) shows that En.fm.I2 displays structural features similar to introns belonging to the bacterial class B of group II introns. En.fm.I2 carries a putative intron-encoded protein (IEP) (CDS19, Table 5) located in domain IV, with domains responsible for the maturase function, reverse transcriptase function and endonuclease function (Figure 3b).
Figure 3.
Structural features of En.fm.I2 and its intron-encoded protein (IEP). (a) Predicted secondary RNA structure of En.fm.I2. Intron nucleotides are written in capital letters and exon sequences are written in lowercase letters. Roman numerals denote the domains I–VI. The IEP is found in domain IV. Intron-binding sites (IBSs) 1 and 2 along with the exon-binding sites (EBSs) 1 and 2 are marked by arrows and boxes, respectively. IBS/EBS3 is a single nucleotide interaction and denoted by pointing arrows. The bulged A (branch site) is located in domain VI and shown in bold. (b) The putative IEP displays a reverse transcriptase (RT) domain (bold letters), a maturase (X) domain (italics) and an endonuclease (En) domain (grey). All introns analysed, except two, had identical amino acid composition to En.fm.I2 of pVEF4 (no. 54, top). The non-synonymous substitutions in En.fm.I2 from E. faecium strains 31/F01/H (no. 49) and TUH32-79 (no. 45) are shown in the alignment (identical amino acids are represented by a dot; a dash indicates gaps or substitutions).
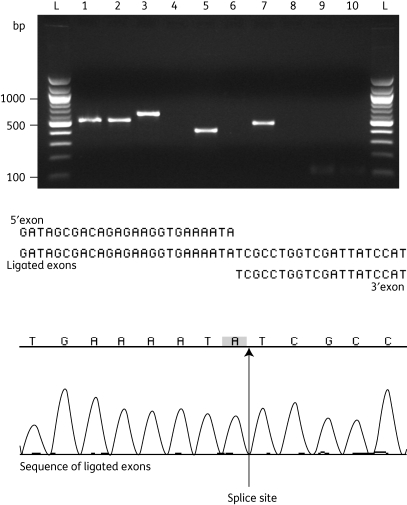
To test if the En.fm.I2 is a functional ribozyme and splices in vivo, total RNA was extracted, and RT–PCR was run with exon–exon-specific primers on DNA of pVEF4 and on pVEF3 as a negative control. En.fm.I2 intron splicing was established (Figure 4, lane 1) and the ligated exons were confirmed by sequencing (data not shown). Specific primers targeting the intron–exon junctions (both 5′ and 3′) were used in RT–PCR reactions and confirmed the presence of unspliced intron in pVEF4, but not in the negative control (pVEF3) (Figure 4). Primers amplifying the 2′–5′ junction allowed the identification of the splicing pathway of En.fm.I2 (Figure 4, lanes 7 and 8). DNA sequencing of the generated PCR product showed that the En.fm.I2 forms a lariat structure in vivo with the 5′ end docking into the bulged A in domain VI.
Figure 4.
RT–PCR analyses of the topoisomerase, intron, intron splicing products and enterococcal elongation factor. RT–PCR products from pVEF4 and pVEF3 are given on alternate lanes 1–10. PCR products are shown as follows: topo mRNA without intron (lanes 1 and 2, primer pair ip3F/giiR7); 5′ intron–exon junction (lanes 3 and 4, primer pair ip3F/giiR8); 3′ intron–exon junction (lanes 5 and 6, primer pair giiF5/giiR7); intron lariat structure (lanes 7 and 8, primer pair giiF5/giiR8); and positive RT–PCR control (lanes 9 and 10, primer pair Ent1/Ent2). Ladder (lanes L), 100 bp DNA molecular size marker from New England Biolabs. The sequence data of the ligated exon with the indicated splice site is shown in the lower half of the figure.
Distribution of the intron En.fm.I2 in vanA plasmids
Because the topo gene and En.fm.I2 of pVEF4 were found adjacent to Tn1546, the wider presence of En.fm.I2 was determined among the 28 heterogeneous vanA plasmids (Figure 2). In total, 15 En.fm.I2-encoding plasmids were identified, for which 14 of the En.fm.I2 introns were found localized in the topo gene. The intron-positive PCR products were confirmed by sequencing (data not shown). A highly conserved DNA sequence identity over the entire intron, including the IEP, was found. Only two of the En.fm.I2 introns (from E. faecium strains 31/F01/H and TUH32-79) displayed non-synonymous substitutions in the IEP protein (Figure 3b).
Discussion
We present the complete sequence of pIP816, the first vanA plasmid isolated in France 1986, as well as a partial, annotated sequence of the fourth plasmid isolated from a single Norwegian poultry farm, pVEF4. The comparative analyses with other completely sequenced vanA plasmids reveal that pVEF1–pVEF4 share a common genetic element of ∼18 kb that spans the entire Tn1546 transposon, a topoisomerase I (topo) gene and the genes encoding the active partitioning system δ-ω of plasmid pSM1903543 upstream of the vanA gene cluster. Three shared CDSs are located downstream of Tn1546.
Surprisingly, an identical (>99% identity at the nucleotide level) 18 kb fragment is present in pIP816. When the inverted fragment located upstream is included, the size of the common fragment present in pIP816, pVEF1, pVEF2 and pVEF3 is ∼25 kb. The observation that the same plasmid-borne DNA fragment (harbouring Tn1546) was found on a Norwegian poultry farm 13 years after the original isolation of pIP816 in a clinical isolate in France13 suggests either successful clonal spread or horizontal transfer between E. faecium strains of different origins. As determined by Werner and colleagues,46 the pIP816 host BM4147 belongs to the MLST clonal complex (CC) 22, one of three CCs considered to be host-specific for humans (CC17, CC22 and CC94).47–49 In contrast, most of the strains isolated from the Norwegian poultry farms (of both poultry and farmer origin) belong to CC9, considered to be host-specific for poultry (including the hosts of pVEF1 and pVEF2).47,48 This observation is consistent with the suggested horizontal transfer of a mobile genetic unit larger than Tn1546 between different clonal complexes of E. faecium, although the direction and frequency of transfer is uncertain.
Tn1546 was found inserted into a streptomycin resistance gene and conserved direct repeats were found in all vanA plasmids studied. The majority of these host strains were from Norwegian poultry farms previously exposed to avoparcin. Available MLST typing data22,46,47,50 showed that the different MLST types clustered into CC9. These data further extend previous reports of the successful spread of a common plasmid-mediated genetic element surrounding Tn1546 on Norwegian poultry farms.21,22 Our observation is also consistent with the original pIP816 characterization with respect to insertion junctions,23 and a recent Tn1546 junction fragment analysis of E. faecium strains from the UK, Denmark, the Netherlands and Norway;51 presenting evidence of the geographical distribution of two different Tn1546–plasmid insertion junctions. Garcia-Migura et al.51 hypothesize that a common genetic element has spread across different clonal lines within the European broiler sector. The data presented here shed light on the composition and size of this element. Moreover, our data also suggest genetic interactions between E. faecium in animal reservoirs and CC17, the only E. faecium clinical high-risk CC.47,49 Strain TUH32-76, a clinical CC17 GREF isolate from Portugal, displays Tn1546 insertion in the streptomycin resistance gene and two immediately adjacent CDSs are linked to the 5′ end of the vanA gene cluster.
Tn1546 belongs to the non-conjugative class II transposable elements of the Tn3 family52 and transfers intracellularly by replicative transposition to diverse sites.23 Thus, the expected transfer by replicative transposition is inconsistent with the genetic patterns observed flanking the Tn1546 transposon reported here and elsewhere.51 Our data, as well as previous experimental results,23 do not support the hypothesis that the streptomycin resistance gene represents a hotspot for Tn1546 integration.
A detailed look at the plasmid sequences analysed here reveals that IS1216 is present in one or both flanking regions of the larger glycopeptide resistance-encoding region (of 18–25 kb), suggesting movement of Tn1546 as part of a composite transposon. Composite transposons are well known to mobilize antibiotic resistance genes in enterococci, e.g. Tn5281 flanked by IS256, Tn1547 flanked by IS16- and IS256-like elements, and Tn5385 flanked by IS1216.8,9,53 Composite transposition enabled by the IS1216 elements flanking the vanA operon has also previously been described.54 The authors showed that IS1216 elements facilitated the transfer of Tn1546 from a non-conjugative plasmid to a pheromone-responsive conjugative plasmid present in the same enterococcal cell, with subsequent intercellular transfer of the vanA gene cluster.
The broader comparative analysis of the genetic composition and gene order (synteny) of pIP816 and the pVEF-type vanA plasmids reveals multiple copies of plasmid replication and maintenance genes interspersed with several copies of IS elements. The mosaic plasmid structures indicate frequent formations of plasmid hybrids, recombination and transposition events. Interestingly, no known conjugal transfer genes have been found in any of the completely sequenced vanA plasmids from Norwegian poultry farms. This, however, does not mean that the plasmids or parts of them are constrained to a single strain. Plasmid co-transfer by the conjugative transposon Tn916 has been reported in enterococci.55 Conjugative mobilization of enterococcal plasmids has also been reported.5,10
The sequence comparison of the E. faecium vanA plasmids enabled the discovery of the novel group II intron En.fm.I2 inserted into the topo gene of pVEF4. Mobile genetic group II intron elements are commonly found in bacteria, although they are not well characterized in enterococci.45 The intron En.fm.I2 belongs to the group II bacterial class B introns, has an IEP that displays features of a multifunctional protein that might enable its mobility and follows the standard group II intron splicing pathway.28 Bacterial mobile group II introns are generally site-specific on insertion and rapidly spread horizontally.56 However, vanA plasmids encoding intron-free topo genes were detected in E. faecium strains (64/F98/H2 and 64/F98/H1) of human origin (one farmer) sampled at one poultry farm on the same occasion, where both plasmids are topo-encoding but only one is invaded by En.fm.I2.
In conclusion, comparative analysis of the original vanA plasmid pIP816 with the recently sequenced pVEF-type plasmids revealed a conserved genetic fragment (including Tn1546) of 18–25 kb. Taken together, the data show that VanA-type glycopeptide resistance is present in different clonal complexes of E. faecium and suggests that glycopeptide resistance can be disseminated through IS1216-facilitated composite transposition.
Funding
This work was in part supported by the Research Council of Norway, the European Commission (QLK2-CT-2002-00843 ‘ARTRADI’ and LSHE-CT-2007-037410 ‘ACE’) and the Medical Research Foundation, North-Norway.
Transparency declarations
None to declare.
References
- 1.Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–21. doi: 10.1056/NEJM200003093421007. doi:10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 2.Paulsen IT, Banerjei L, Myers GS, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–4. doi: 10.1126/science.1080613. doi:10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 3.Willems RJ, Bonten MJ. Glycopeptide-resistant enterococci: deciphering virulence, resistance and epidemicity. Curr Opin Infect Dis. 2007;20:384–90. doi: 10.1097/QCO.0b013e32818be63d. doi:10.1097/QCO.0b013e32818be63d. [DOI] [PubMed] [Google Scholar]
- 4.De Boever EH, Clewell DB, Fraser CM. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol Microbiol. 2000;37:1327–41. doi: 10.1046/j.1365-2958.2000.02072.x. doi:10.1046/j.1365-2958.2000.02072.x. [DOI] [PubMed] [Google Scholar]
- 5.Francia MV, Clewell DB. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol Microbiol. 2002;45:375–95. doi: 10.1046/j.1365-2958.2002.03007.x. doi:10.1046/j.1365-2958.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- 6.Francia MV, Haas W, Wirth R, et al. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid. 2001;46:117–27. doi: 10.1006/plas.2001.1533. doi:10.1006/plas.2001.1533. [DOI] [PubMed] [Google Scholar]
- 7.Hirt H, Manias DA, Bryan EM, et al. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol. 2005;187:1044–54. doi: 10.1128/JB.187.3.1044-1054.2005. doi:10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quintiliani R, Jr, Courvalin P. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. doi:10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 9.Rice LB, Carias LL. Transfer of Tn5385, a composite, multiresistance chromosomal element from Enterococcus faecalis. J Bacteriol. 1998;180:714–21. doi: 10.1128/jb.180.3.714-721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz FV, Perreten V, Teuber M. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid. 2001;46:170–87. doi: 10.1006/plas.2001.1544. doi:10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- 11.Sletvold H, Johnsen PJ, Simonsen GS, et al. Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob Agents Chemother. 2007;51:736–9. doi: 10.1128/AAC.00557-06. doi:10.1128/AAC.00557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomita H, Tanimoto K, Hayakawa S, et al. Highly conjugative pMG1-like plasmids carrying Tn1546-like transposons that encode vancomycin resistance in Enterococcus faecium. J Bacteriol. 2003;185:7024–8. doi: 10.1128/JB.185.23.7024-7028.2003. doi:10.1128/JB.185.23.7024-7028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclercq R, Derlot E, Duval J, et al. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–61. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 14.Hidron AI, Edwards JR, Patel J, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. doi:10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 15.Weigel LM, Donlan RM, Shin DH, et al. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother. 2007;51:231–8. doi: 10.1128/AAC.00576-06. doi:10.1128/AAC.00576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perichon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:4580–7. doi: 10.1128/AAC.00346-09. doi:10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aarestrup FM. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb Drug Resist. 1995;1:255–7. doi: 10.1089/mdr.1995.1.255. doi:10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 18.Klare I, Heier H, Claus H, et al. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol Lett. 1995;125:165–71. doi: 10.1111/j.1574-6968.1995.tb07353.x. doi:10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 19.Aarestrup FM, Ahrens P, Madsen M, et al. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;40:1938–40. doi: 10.1128/aac.40.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borgen K, Simonsen GS, Sundsfjord A, et al. Continuing high prevalence of VanA-type vancomycin-resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J Appl Microbiol. 2000;89:478–85. doi: 10.1046/j.1365-2672.2000.01137.x. doi:10.1046/j.1365-2672.2000.01137.x. [DOI] [PubMed] [Google Scholar]
- 21.Johnsen PJ, Østerhus JI, Sletvold H, et al. Persistence of animal and human glycopeptide-resistant enterococci on two Norwegian poultry farms formerly exposed to avoparcin is associated with a widespread plasmid-mediated vanA element within a polyclonal Enterococcus faecium population. Appl Environ Microbiol. 2005;71:159–68. doi: 10.1128/AEM.71.1.159-168.2005. doi:10.1128/AEM.71.1.159-168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sørum M, Johnsen PJ, Aasnes B, et al. Prevalence, persistence, and molecular characterization of glycopeptide-resistant enterococci in Norwegian poultry and poultry farmers 3 to 8 years after the ban on avoparcin. Appl Environ Microbiol. 2006;72:516–21. doi: 10.1128/AEM.72.1.516-521.2006. doi:10.1128/AEM.72.1.516-521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arthur M, Molinas C, Depardieu F, et al. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–27. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biavasco F, Foglia G, Paoletti C, et al. VanA-type enterococci from humans, animals, and food: species distribution, population structure, Tn1546 typing and location, and virulence determinants. Appl Environ Microbiol. 2007;73:3307–19. doi: 10.1128/AEM.02239-06. doi:10.1128/AEM.02239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawalec M, Gniadkowski M, Hryniewicz W. Outbreak of vancomycin-resistant enterococci in a hospital in Gdask, Poland, due to horizontal transfer of different Tn1546-like transposon variants and clonal spread of several strains. J Clin Microbiol. 2000;38:3317–22. doi: 10.1128/jcm.38.9.3317-3322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werner G, Klare I, Fleige C, et al. Increasing rates of vancomycin resistance among Enterococcus faecium isolated from German hospitals between 2004 and 2006 are due to wide clonal dissemination of vancomycin-resistant enterococci and horizontal spread of vanA clusters. Int J Med Microbiol. 2008;298:515–27. doi: 10.1016/j.ijmm.2007.05.008. doi:10.1016/j.ijmm.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Willems RJ, Top J, van den Braak N, et al. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999;43:483–91. doi: 10.1128/aac.43.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robart AR, Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet Genome Res. 2005;110:589–97. doi: 10.1159/000084992. doi:10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Russel DW. Preparation of Plasmid DNA by Alkaline Lysis with SDS. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 30.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–30. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 31.Sletvold H, Johnsen PJ, Simonsen GS, et al. Complete DNA sequence of the Enterococcus faecium vanA plasmid pIP816, and comparative analysis to the vanA plasmids pVEF1/pVEF2. Plasmid. 2007;57:239. doi:10.1016/j.ijmm.2007.05.008. [Google Scholar]
- 32.Carver TJ, Rutherford KM, Berriman M, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;16:3422–3. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 33.Delcher AL, Harmon D, Kasif S, et al. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–41. doi: 10.1093/nar/27.23.4636. doi:10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–8. doi: 10.1073/pnas.85.8.2444. doi:10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter AE, Turner DH, Kim J, et al. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–22. doi: 10.1073/pnas.91.20.9218. doi:10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. doi:10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–52. doi: 10.1017/s1355838201010251. doi:10.1017/S1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swinfield TJ, Oultram JD, Thompson DE, et al. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAM beta 1. Gene. 1990;87:79–90. [PubMed] [Google Scholar]
- 40.Bidnenko V, Ehrlich SD, Janniere L. In vivo relations between pAMβ-encoded type I topoisomerase and plasmid replication. Mol Microbiol. 1998;28:1005–16. doi: 10.1046/j.1365-2958.1998.00862.x. doi:10.1046/j.1365-2958.1998.00862.x. [DOI] [PubMed] [Google Scholar]
- 41.Bruand C, Ehrlich SD. Transcription-driven DNA replication of plasmid pAMβ1 in Bacillus subtilis. Mol Microbiol. 1998;30:135–45. doi: 10.1046/j.1365-2958.1998.01044.x. doi:10.1046/j.1365-2958.1998.01044.x. [DOI] [PubMed] [Google Scholar]
- 42.Jensen LB, Garcia-Migura L, Valenzuela AJ, et al. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods. 2010;80:25–43. doi: 10.1016/j.mimet.2009.10.012. doi:10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Dmowski M, Sitkiewicz I, Ceglowski P. Characterization of a novel partition system encoded by the δ and ω genes from the streptococcal plasmid pSM19035. J Bacteriol. 2006;188:4362–72. doi: 10.1128/JB.01922-05. doi:10.1128/JB.01922-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedberg PJ, Leonard BA, Ruhfel RE, et al. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid. 1996;35:46–57. doi: 10.1006/plas.1996.0005. doi:10.1006/plas.1996.0005. [DOI] [PubMed] [Google Scholar]
- 45.Dai L, Toor N, Olson R, et al. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–6. doi: 10.1093/nar/gkg049. doi:10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner G, Klare I, Witte W. The current MLVA typing scheme for Enterococcus faecium is less discriminatory than MLST and PFGE for epidemic-virulent, hospital-adapted clonal types. BMC Microbiol. 2007;7:28. doi: 10.1186/1471-2180-7-28. doi:10.1186/1471-2180-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freitas AR, Novais C, Ruiz-Garbajosa P, et al. Dispersion of multidrug-resistant Enterococcus faecium isolates belonging to major clonal complexes in different Portuguese settings. Appl Environ Microbiol. 2009;75:4904–8. doi: 10.1128/AEM.02945-08. doi:10.1128/AEM.02945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Homan WL, Tribe D, Poznanski S, et al. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002;40:1963–71. doi: 10.1128/JCM.40.6.1963-1971.2002. doi:10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006;5:454–60. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Rosvoll TC, Pedersen T, Sletvold H, et al. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin–antitoxin systems. FEMS Immunol Med Microbiol. 2010;58:254–68. doi: 10.1111/j.1574-695X.2009.00633.x. doi:10.1111/j.1574-695X.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Migura L, Hasman H, Svendsen C, et al. Relevance of hot spots in the evolution and transmission of Tn1546 in glycopeptide-resistant Enterococcus faecium (GREF) from broiler origin. J Antimicrob Chemother. 2008;62:681–7. doi: 10.1093/jac/dkn265. doi:10.1093/jac/dkn265. [DOI] [PubMed] [Google Scholar]
- 52.Merlin C, Mahillon J, Nesvera J, et al. Gene recruiters and transporters: the modular structure of bacterial mobile elements. In: Thomas M,, editor. The Horizontal Gene Pool. Amsterdam: Harwood Academic Publishers,; 2000. pp. 363–409. [Google Scholar]
- 53.Hodel-Christian SL, Murray BE. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob Agents Chemother. 1991;35:1147–52. doi: 10.1128/aac.35.6.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heaton MP, Discotto LF, Pucci MJ, et al. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-responsive plasmid. Gene. 1996;171:9–17. doi: 10.1016/0378-1119(96)00022-4. doi:10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 55.Flannagan SE, Clewell DB. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J Bacteriol. 1991;173:7136–41. doi: 10.1128/jb.173.22.7136-7141.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. doi:10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 57.Willems RJ, Top J, van Santen M, et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–8. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sletvold H, Johnsen PJ, Hamre I, et al. Complete sequence of Enterococcus faecium pVEF3 and the detection of an omega-epsilon-zeta toxin–antitoxin module and an ABC transporter. Plasmid. 2008;60:75–85. doi: 10.1016/j.plasmid.2008.04.002. doi:10.1016/j.plasmid.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Tomita H, Ike Y. Genetic analysis of transfer-related regions of the vancomycin resistance Enterococcus conjugative plasmid pHTβ: identification of oriT and a putative relaxase gene. J Bacteriol. 2005;187:7727–37. doi: 10.1128/JB.187.22.7727-7737.2005. doi:10.1128/JB.187.22.7727-7737.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ke D, Picard FJ, Martineau F, et al. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]