Abstract
Objectives
Efavirenz is extensively metabolized by CYP2B6, and associations between CYP2B6 polymorphisms and plasma efavirenz exposure have been reported. The objective of this study was to investigate CYP2B6 haplotype structure and functional consequences in a Latin American population.
Patients and methods
Two hundred and nineteen patients were recruited at Fundación Arriarán, Chile, between September and December 2008. Plasma efavirenz concentrations were determined using liquid chromatography with mass spectrometry. Genotyping for 30 single nucleotide polymorphisms (SNPs) with a minor allele frequency of >0.05 in the HapMap CEU population at intervals of ∼1 kb across the CYP2B6 locus was conducted using Sequenom iPLEX MALDI-TOF.
Results
Thirteen SNPs passed quality control and, of these, statistically significant associations (P < 0.001) with plasma efavirenz concentrations were observed for 11. Pairwise tagging SNP analysis (R2 > 0.8) identified 3 SNPs (rs10403955, rs2279345 and rs8192719) representative of the 11 associated SNPs. A composite genetic model of these three alleles was constructed, and an association between carriers of four to six of these alleles and the risk of efavirenz plasma concentrations >4 µg/mL was identified with an odds ratio of 48.1 (95% confidence interval: 13.5–207.7). This represents a positive predictive value of 80.9% and a negative predictive value of 91.8%, with sensitivity of 57.9% and specificity of 97.2%.
Conclusions
A composite genetic model of CYP2B6 SNPs in a Chilean HIV-positive cohort may have value in predicting concentrations of efavirenz associated with a higher likelihood of CNS toxicity. Further investigation of the functional basis of these associations is now required.
Keywords: antiretrovirals, pharmacokinetics, genetics, single nucleotide polymorphisms
Introduction
Efavirenz is a first-line therapy for the treatment of HIV globally. Despite the potency and tolerability of efavirenz-containing regimens, some patients can fail to achieve durable viral suppression or develop treatment-limiting toxicity.1,2 Efavirenz plasma concentrations >4 µg/mL have been associated with an increased risk of CNS adverse effects, while concentrations <1 µg/mL have been associated with virological failure.3 Although other enzymes are involved, efavirenz is predominantly metabolized by CYP2B6.4
The most commonly studied CYP2B6 single nucleotide polymorphism (SNP) is c.516G→T (rs3745274), which encodes a Gln172His amino acid substitution. This polymorphism is associated with lower hepatic expression and enzymatic activity for CYP2B6.5,6 Many studies have shown an association between the c.516G→T allele and higher efavirenz exposure in different ethnic groups.7–10 In addition to c.516G→T, CYP2B6 polymorphisms, such as the c.983T→C SNP, have also been shown to impact on plasma efavirenz concentrations in black populations.11–13
To date, there is little information on the frequency and functional consequences of CYP2B6 polymorphisms in Latin American populations. Therefore, the aim of this study was to investigate the CYP2B6 haplotype structure in a Chilean HIV cohort and to identify associations with efavirenz plasma concentrations. The ability of CYP2B6 genotypes to predict individuals at risk of high efavirenz plasma concentrations was also assessed.
Methods
Patients
Two hundred and nineteen HIV-positive patients were recruited at Fundación Arriarán, Santiago, Chile, between September and December 2008. Of these individuals, 214 were self-described white or mestizos (mixed European and Amerindian ancestry), with 5 of native ancestry. All individuals were >18 years of age and taking 600 mg of efavirenz once daily for ≥2 weeks. The median [interquartile range (IQR)] duration of therapy was 3.6 years (1.6–5.8 years). The median (IQR) time since last dose was 11.7 h (10.8–12.7 h) and, as such, all efavirenz plasma concentration measurements can be considered as Ctrough. Patients were excluded if informed consent could not be obtained or if rifampicin was used as a co-medication. Ethical approval for the study was obtained from the Comité de Ética de Investigación en Seres Humanos de la Facultad de Medicina Universidad de Chile.
Plasma efavirenz analysis
Sample preparation consisted of protein precipitation with 0.1% acetic acid in methanol followed by centrifugation at 2000 g for 5 min. An aliquot of 250 µL of supernatant was used and 100 µL of an internal standard consisting of 2 mg/L 6,7-dimethyl-2,3-di(2-pyridyl)-quinoxaline (Aldrich, Milwaukee, WI, USA) was added. All solvents and chemicals were obtained from Sigma (Oakville, ON, Canada) and were of HPLC grade. The liquid chromatography with mass spectrometry system consisted of a HP1100 LC system (Agilent Technologies, Wilmington, DE, USA) with a Supelcosil™ ABZ+ (15 cm × 4.6 mm, 3 µm) C18 column (Supelco, Bellefonte, PA, USA) coupled to an API-2000 mass spectrometer (AB/MDS/Sciex, Concord, ON, Canada) with a turbo ion-spray source. The efavirenz calibration curve ranged from 0.05 to 5 mg/L. The intraday accuracy and precision were −2.6%–2.0% and 5.4%–11.2%, respectively. The interday accuracy and precision were −5.3% to −0.9% and 8.4%–13.0%, respectively. More details on this assay can be found elsewhere.14
SNP selection
SNPs with a minor allele frequency (MAF) of >0.05 were identified within dbSNP (http://www.ncbi.nlm.nih.gov/SNP) at intervals of ∼1 kb across the CYP2B6 locus at chr19q13.2 and an additional 5 kb up- and downstream of the gene. Where no SNPs with validated MAFs were available, the nearest positional SNP was included.
DNA extraction and genotyping
DNA was extracted using a QIAamp whole blood mini kit (Qiagen, UK), according to the manufacturer's protocol. Thirteen DNA samples were of insufficient quality or quantity to acquire reliable genotype data and so were omitted. Therefore, a total of 206 individuals were genotyped for 32 SNPs using the MALDI-TOF-based Sequenom iPLEX system (Sequenom Inc., San Diego, CA, USA), according to the manufacturer's protocol. Sequence-specific PCR and extend reaction oligonucleotides were obtained from Metabion GmbH (Martinsried, Germany). Multiplex assays were designed using the software available at https://mysequenom.com/default.aspx. SNPs were divided into a 21- and an 11-plex assay.
Statistical analysis
SNPs with MAF <0.05 or call rate <90% were omitted. Statistical analysis was undertaken using SPSS 17.0. Continuous variable analysis by genotype was by one-way analysis of variance (ANOVA). Binary outcomes by genotype were analysed by Fisher's exact test. P < 0.05 was regarded as significant. Tagging SNP analysis was carried out using the Tagger function within Haploview 4.1 software (www.broad.mit.edu/mpg/haploview/).
Results
Association of individual CYP2B6 SNPs with plasma efavirenz concentrations
Thirteen SNPs were carried forward for further analysis (Table 1) and all were in Hardy–Weinburg equilibrium. Ten SNPs were rejected for MAF <0.05 and seven for call rate <90%. One-way ANOVA of efavirenz plasma concentration with genotype for individual SNPs indicated statistically significant associations (P < 0.001) for 11/13 SNPs within the CYP2B6 gene. The exceptions were rs4802100 (5′ upstream) (P = 0.08) and rs34083050 (intron 4) (P = 0.91). The statistical significance of the genotype associations with efavirenz plasma concentration ranged from P = 2.6 × 10−8 (rs2279345) to P = 3.6 × 10−22 (rs8192719). Only one associated SNP was exonic [c.516G→T (rs3745274)] (P = 5.6 × 10−20).
Table 1.
Genotype and allelic frequencies of CYP2B6 single nucleotide polymorphisms and median plasma efavirenz concentrations
| Allele frequency |
Genotype frequency |
Median efavirenz plasma concentration (µg/mL) (range) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession number | CYP allele | Reference sequence position NT_011109.15 | Gene position | Alleles (A1/A2) | A1 | A2 | A1/A1 | A1/A2 | A2/A2 | A1/A1 | A1/A2 | A2/A2 | ANOVA P value |
| rs4802100 | 13764243 | 5′ upstream | G/C | 0.22 | 0.78 | 0.04 | 0.37 | 0.59 | 3.00 (0.68–3.52) | 2.51 (0.31–7.24) | 2.91 (0.13–10.2) | 0.082 | |
| rs7250601 | 13771622 | intron 1 | A/C | 0.65 | 0.35 | 0.44 | 0.43 | 0.13 | 2.29 (0.13–4.67) | 2.92 (0.78–9.58) | 5.07 (1.91–10.2) | 9.1 × 10−20 | |
| rs16974799 | 13772295 | intron 1 | C/T | 0.63 | 0.37 | 0.42 | 0.42 | 0.16 | 2.29 (0.13–3.64) | 2.93 (0.72–9.58) | 4.95 (1.91–10.2) | 1.5 × 10−17 | |
| rs10500282 | 13776660 | intron 1 | T/C | 0.63 | 0.37 | 0.39 | 0.48 | 0.13 | 2.26 (0.13–2.26) | 2.67 (0.69–9.58) | 5.05 (1.91–10.2) | 2.0 × 10−18 | |
| rs10403955 | 13777656 | intron 1 | T/G | 0.64 | 0.36 | 0.41 | 0.47 | 0.12 | 2.27 (0.13–4.84) | 2.87 (0.72–9.58) | 5.54 (1.91–10.2) | 5.4 × 10−20 | |
| rs3745274 | 2B6*6(PO)/*9 | 13781059 | exon 4 | G/T | 0.65 | 0.35 | 0.43 | 0.42 | 0.15 | 2.24 (0.13–4.44) | 2.92 (0.78–9.58) | 4.95 (1.37–10.2) | 5.6 × 10−20 |
| rs34083050 | 13782845 | intron 4 | A/G | 0.14 | 0.84 | 0.01 | 0.32 | 0.67 | 2.49 (2.49–2.49) | 2.66 (0.69–10.2) | 2.79 (0.31–9.79) | 0.906 | |
| rs2279344 | 13783701 | intron 5 | C/A | 0.23 | 0.77 | 0.05 | 0.35 | 0.60 | 1.80 (1.05–2.96) | 2.14 (0.13–7.21) | 3.14 (0.31–10.2) | 4.3 × 10−8 | |
| rs2279345 | 13783920 | intron 5 | T/C | 0.23 | 0.77 | 0.04 | 0.34 | 0.62 | 1.80 (1.05–2.96) | 2.14 (0.13–4.84) | 3.12 (0.31–10.2) | 2.6 × 10−8 | |
| rs8192719 | 13786991 | intron 8 | C/T | 0.65 | 0.35 | 0.42 | 0.45 | 0.13 | 2.26 (0.13–4.43) | 2.87 (0.72–9.58) | 5.32 (1.91–10.2) | 3.6 × 10−22 | |
| rs11671243 | 13787933 | intron 8 | A/C | 0.22 | 0.78 | 0.05 | 0.35 | 0.60 | 1.79 (1.05–2.96) | 2.14 (0.13–4.84) | 3.18 (0.31–10.2) | 6.8 × 10−9 | |
| rs11673270 | 13789062 | intron 8 | A/C | 0.66 | 0.34 | 0.43 | 0.46 | 0.11 | 2.27 (0.13–4.84) | 2.90 (0.78–10.2) | 4.95 (1.91–9.79) | 2.9 × 10−13 | |
| rs10853744 | 13790157 | intron 8 | G/T | 0.63 | 0.37 | 0.41 | 0.45 | 0.14 | 2.28 (0.13–4.43) | 2.87 (0.72–9.58) | 5.05 (1.91–10.2) | 7.2 × 10−20 | |
Statistical significance as determined by one-way ANOVA is indicated.
CYP2B6 haplotype structure in Chilean HIV-positive population
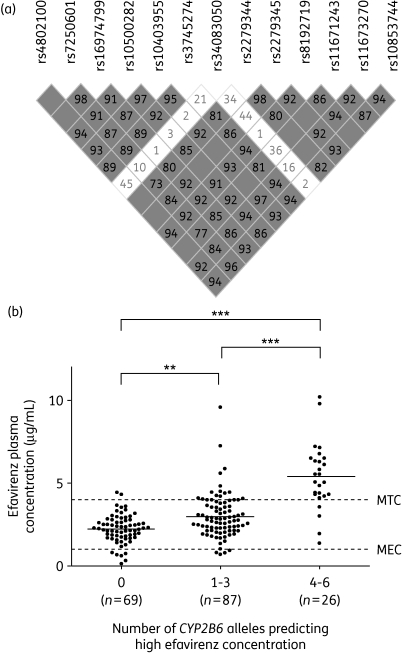
Linkage disequilibrium (LD) analysis of the 13 CYP2B6 polymorphisms showed a significant degree of LD (Figure 1a). Pairwise tagging SNP analysis (R2 > 0.8) identified 3 SNPs (rs10403955, rs2279345 and rs8192719) that were representative of the 11 SNPs associated with plasma efavirenz concentration. These three SNPs were therefore used to construct a composite genetic model. Inferred haplotype analysis of the three SNPs showed that the GCT haplotype was more frequent in Chilean patients (0.34) than in the HapMap (http://hapmap.ncbi.nlm.nih.gov/) Caucasian population (0.27), but less frequent than in the HapMap Yoruba population (0.40). No data were available for rs10403955 in the HapMap Mexican ancestry in Los Angeles cohort and, as such, a direct comparison of inferred haplotype frequency was not possible. However, no significant differences were observed between Chilean patients and the HapMap Mexican population for the frequency of other genotyped SNPs.
Figure 1.
LD and association of composite CYP2B6 genotype with efavirenz plasma concentrations. (a) Data are shown as represented in Haploview software. White squares, D′ < 1 and logarithm of odds (LOD) < 2; grey squares, D′ < 1 and LOD ≥ 2; and grey squares with no D′ value displayed, D′ = 1 and LOD ≥ 2. (b) Data represent efavirenz plasma concentrations of patients with 0, 1–3 and 4–6 alleles associated with high concentration. Median plasma efavirenz concentrations are given for each group and are represented by continuous horizontal lines. Broken horizontal lines indicate the minimum effective concentration (MEC) and minimum toxic concentration (MTC). Statistical significance was determined by a Mann–Whitney test (**P < 0.01, ***P < 0.001).
Association of CYP2B6 composite genetic model with plasma efavirenz concentrations
The number of alleles associated with high efavirenz concentration (rs10403955 G, rs2297345 C and rs8192719 T) possessed by each individual was tallied and analysed in a composite genetic model (Figure 1b). Individuals possessing zero of six associated alleles (n = 69) had a median (range) efavirenz plasma concentration of 2.26 µg/mL (0.13–4.43 µg/mL). Median (range) plasma efavirenz concentrations were significantly higher in those with one to three alleles (n = 87) at 2.76 µg/mL (0.72–9.18 µg/mL) (P < 0.01). At 5.32 µg/mL (1.37–10.1 µg/mL), individuals with four to six associated alleles had a significantly higher median efavirenz plasma concentration than either the zero or one to three allele groups (P < 0.001 in both instances).
An association between carriers of four to six risk alleles with the risk of an efavirenz plasma concentration greater than the proposed minimal toxic concentration (MTC) (4 µg/mL) was also identified. Of the 38 individuals with an efavirenz plasma concentration >MTC, 22 possessed four to six associated alleles compared with 4/140 of those with an efavirenz plasma concentration <MTC. This represents an odds ratio of 48.1 (95% confidence interval: 13.5–207.7) (P < 0.0001). If the prevalence of >MTC is taken to be 17% (38/219) then the positive predictive value for this association was 80.9% and the negative predictive value was 91.8%, with a sensitivity of 57.9% and a specificity of 97.2%. The positive and negative predictive values of homozygous (TT) carriers of the top associated SNP (rs8192719) were 80.8% and 91.1%, though the sensitivity was marginally lower than the composite genetic model at 53.6%.
Discussion
Many studies have identified associations between polymorphisms in CYP2B6 and efavirenz pharmacokinetics.7–10 The data reported show that strong LD exists across the CYP2B6 locus. Our report is the first to show the haplotype structure of CYP2B6 in Chile. The frequency of indigenous Chilean ethnicity in our cohort (2.3%) is comparable to the Chilean 2002 census (www.ine.cl/cd2002/sintesiscensal.pdf; 4.6%), suggesting that our cohort is ethnically representative. Given that >97% of the study population are of white European or mestizo ancestry (mixed European and Amerindian), comparisons with Los Angeles residents with Mexican ancestry are valid. Allelic frequencies for functional CYP2B6 polymorphisms appear comparable to those of Mexican ancestry (MEX) in the HapMap. For example, the 516T allele frequency in our cohort was 0.35 versus 0.29 in the MEX cohort (P > 0.05). However, the 516T frequency was statistically significantly higher (P < 0.05) than that of the Caucasian (CEU) HapMap cohort (0.27). It should, however, be noted that the HapMap MEX cohort is smaller (n = 100) than the Chilean cohort in the present study (and the HapMap CEU cohort).
This study identified 11 CYP2B6 SNPs associated with increased efavirenz exposure in Chilean patients. Based on LD across these 11 SNPs, 3 tagging SNPs were identified, which, when combined in a composite model, strengthened the observed association and had higher positive and negative predictive values for efavirenz concentrations above the proposed MTC (80.9% and 91.9%, respectively) than the c.516G→T TT genotype alone (71.1% and 90.2%, respectively) in this cohort. Given that individuals with a plasma concentration >4 µg/mL are three times more likely to develop CNS toxicity,3 a test with a high positive predictive value may be of clinical value. One of the three SNPs within the composite genetic model (rs10403955) tagged the c.516G→T SNP, which goes some way to explaining its contribution. However, it is less clear what the contribution of the other two SNPs within the composite model is. Both rs8192719 and rs2279345 are located in intronic enhancer sites, but have a low likelihood of directly exerting functional effects. Therefore, it is likely that their contribution is indirect, by tagging other functional variants within the CYP2B6 gene.
The frequency of the GCT inferred haplotype was higher in the Chilean patients than in the HapMap Caucasian population, but of lower frequency than in the HapMap Yoruba (Ibadan, Nigeria) population. It is therefore interesting to note that the plasma efavirenz concentrations in the present study (median = 2.6; range = 0.7–9.8 µg/mL) were ∼23% higher than those reported previously in a Caucasian population (P = 0.008; median = 2.1; range = 0.7–17.4 µg/mL; n = 113) recruited in Germany.12 Curiously, however, concentrations were also higher than in a Black population (P = 0.04; median = 2.1; range = 0.5–26.0 µg/mL; n = 54) recruited as part of the same study,12 indicating that genetic factors other than CYP2B6 also contribute to the observed differences. Aside from CYP2B6, recent studies have identified the influence on efavirenz pharmacokinetics of polymorphisms within the CYP2A6 and UGT2B7 genes, which also warrant further investigation.7,15,16
One limitation of this study worthy of mention is the potential for the introduction of selection bias towards those patients more likely to need antiretroviral treatment and, crucially, those more likely to tolerate efavirenz. This may also have implications for the predictive values for MTC >4 µg/mL, since patients maintained on standard-dose efavirenz may not be representative of an antiretroviral-naive population. Unpublished data from C. P. Cortes from the national Chilean AIDS Cohort (ChiAC) from October 2001 to March 2008 identified 3107 patients starting an antiretroviral regimen with efavirenz. Of these, 639 (20.6%) had an adverse event requiring a change of drug, 3% of which were CNS related. The mean number of days to efavirenz discontinuation due to CNS issues for Fundación Arriarán, the major contributing centre of ChiAC, was 99 days (13–200 days) (C. P. Cortes, unpublished data).
In conclusion, these data indicate that a composite genetic model that includes multiple CYP2B6 SNPs is more strongly associated with efavirenz plasma concentrations than the c.516G→T polymorphism alone. These observations now warrant confirmation in other studies.
Funding
This work was supported by the NHS R&D Health Technology Assessment (HTA) Programme (project number 04/35/08) for the position of D. F. C. The National Institute of Health Research (NIHR–Department of Health) and the Northwest Development Agency (NWDA) provided infrastructural and project support.
Transparency declarations
None to declare.
References
- 1.Arribas JR, Pozniak AL, Gallant JE, et al. Tenofovir disoproxil fumarate, emtricitabine, and efavirenz compared with zidovudine/lamivudine and efavirenz in treatment-naive patients: 144-week analysis. J Acquir Immune Defic Syndr. 2008;47:74–8. doi: 10.1097/QAI.0b013e31815acab8. [DOI] [PubMed] [Google Scholar]
- 2.Gallant JE, DeJesus E, Arribas JR, et al. Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine, and efavirenz for HIV. N Engl J Med. 2006;354:251–60. doi: 10.1056/NEJMoa051871. doi:10.1056/NEJMoa051871. [DOI] [PubMed] [Google Scholar]
- 3.Marzolini C, Telenti A, Decosterd LA, et al. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1-infected patients. AIDS. 2001;15:71–5. doi: 10.1097/00002030-200101050-00011. doi:10.1097/00002030-200101050-00011. [DOI] [PubMed] [Google Scholar]
- 4.Ward BA, Gorski JC, Jones DR, et al. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther. 2003;306:287–300. doi: 10.1124/jpet.103.049601. doi:10.1124/jpet.103.049601. [DOI] [PubMed] [Google Scholar]
- 5.Desta Z, Saussele T, Ward B, et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics. 2007;8:547–58. doi: 10.2217/14622416.8.6.547. doi:10.2217/14622416.8.6.547. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann MH, Blievernicht JK, Klein K, et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325:284–92. doi: 10.1124/jpet.107.133306. doi:10.1124/jpet.107.133306. [DOI] [PubMed] [Google Scholar]
- 7.Kwara A, Lartey M, Sagoe KW, et al. CYP2B6 (c.516G→T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67:427–36. doi: 10.1111/j.1365-2125.2009.03368.x. doi:10.1111/j.1365-2125.2009.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran G, Hemanth Kumar AK, Rajasekaran S, et al. CYP2B6 G516T polymorphism but not rifampin coadministration influences steady-state pharmacokinetics of efavirenz in human immunodeficiency virus-infected patients in South India. Antimicrob Agents Chemother. 2009;53:863–8. doi: 10.1128/AAC.00899-08. doi:10.1128/AAC.00899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rotger M, Tegude H, Colombo S, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81:557–66. doi: 10.1038/sj.clpt.6100072. doi:10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 10.Tsuchiya K, Gatanaga H, Tachikawa N, et al. Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319:1322–6. doi: 10.1016/j.bbrc.2004.05.116. doi:10.1016/j.bbrc.2004.05.116. [DOI] [PubMed] [Google Scholar]
- 11.Gatanaga H, Hayashida T, Tsuchiya K, et al. Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6 *6 and *26. Clin Infect Dis. 2007;45:1230–7. doi: 10.1086/522175. doi:10.1086/522175. [DOI] [PubMed] [Google Scholar]
- 12.Wyen C, Hendra H, Vogel M, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61:914–8. doi: 10.1093/jac/dkn029. doi:10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas DW, Gebretsadik T, Mayo G, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis. 2009;199:872–80. doi: 10.1086/597125. doi:10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorello G, la Porte C, Pilon R, et al. Discordance in HIV-1 viral loads and antiretroviral drug concentrations comparing semen and blood plasma. HIV Med. 2009;10:548–54. doi: 10.1111/j.1468-1293.2009.00725.x. doi:10.1111/j.1468-1293.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 15.Arab-Alameddine M, Di Iulio J, Buclin T, et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Ther. 2009;85:485–94. doi: 10.1038/clpt.2008.271. doi:10.1038/clpt.2008.271. [DOI] [PubMed] [Google Scholar]
- 16.di Iulio J, Fayet A, Arab-Alameddine M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics. 2009;19:300–9. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]