Abstract
Microdeletions of 3q29 have previously been reported, but the postulated reciprocal microduplication has only recently been observed. Here, cases from four families, two ascertained in Toronto (Canada) and one each from Edinburgh (UK) and Leiden (Netherlands), carrying microduplications of 3q29 are presented. These families have been characterized by cytogenetic and molecular techniques, and all individuals have been further characterized with genome-wide, high density single nucleotide polymorphism (SNP) arrays run at a single centre (The Centre for Applied Genomics, Toronto). In addition to polymorphic copy-number variants (CNV), all carry duplications of 3q29 ranging in size from 1.9 to 2.4 Mb, encompassing multiple genes and defining a minimum region of overlap of about 1.6 Mb bounded by clusters of segmental duplications that is remarkably similar in location to previously reported 3q29 microdeletions. Consistent with other reports, the phenotype is variable, although developmental delay and significant ophthalmological findings were recurrent, suggesting that dosage sensitivity of genes located within 3q29 is important for eye and CNS development. We also consider CNVs found elsewhere in the genome for their contribution to the phenotype. We conclude by providing preliminary guidelines for management and anticipatory care of families with this microduplication, thereby establishing a standard for CNV reporting.
Contiguous gene syndromes involving small chromosomal duplications are typically less frequently reported in comparison to their microdeletion counterparts. Although these rearrangements can both arise from a common mechanism involving nonallelic homologous recombination with region-specific low copy repeats (Lupski, 2004), microduplication syndromes are usually less commonly recognized, possibly due to ascertainment bias, milder and more variable phenotype, and technical limitations of cytogenetics and fluorescentin situhybridization (FISH). Well characterized chromosomal regions shown to involve these reciprocal duplication and deletion events include duplication of 17p11.2 causing a phenotype associated with moderate mental retardation and behavioural disturbances (Potocki et al., 2000), with the reciprocal microdeletion resulting in Smith-Magenis syndrome; microduplication of 22q11.2 (Ensenauer et al., 2003) having a somewhat variable phenotype with cardiac malformation and features similar to the classical microdeletion 22q11.2 syndrome; microduplication of 15q11→q13 characterized by developmental delay and autism, reciprocal to deletions causing Prader-Willi/Angelman syndromes (Dimitropoulos and Schultz, 2007), and microduplication of 7q11.23, which has been related to severe expressive language delay (Somerville et al., 2005; Merritt and Lindor, 2008; Orellana et al., 2008; Torniero et al., 2008), while the corresponding deletion causes Williams-Beuren syndrome (Osborne et al., 1996). Most recently, copy number variations (CNVs) in the form of microdeletions and microduplications of chromosome 16p11.2 have also been observed in autism spectrum disorder (Kumar et al., 2008; Marshall et al., 2008; Weiss et al., 2008).
With the use of microarray-based techniques, increasing numbers of novel copy number variants are being discovered both in apparently healthy control individuals (Redon et al., 2006; Pinto et al., 2007), and in patients with genetic disorders such as autism (Autism Genome Project Consortium 2007; Sebatet al., 2007; Marshall et al., 2008) and schizophrenia (Walsh et al., 2008; Xu et al., 2008). Improved resolution of these microarray platforms is resulting in greater power to detect ever smaller events, well below the level of resolution of conventional cytogenetic examination (Feuk et al., 2006; Carter, 2007).
A microdeletion syndrome on chromosome 3q29 was originally described in six patients (Willatt et al., 2005). The common phenotypic features included a long narrow face, short philtrum, high nasal bridge, developmental and significant speech delay. The microdeletion was approximately 1.5 Mb in length and was between identical low copy repeat sequences on either side of the deletion breakpoints. This suggests that this region is susceptible to nonallelic homologous recombination, which could result in reciprocal exchange events at chromosome 3q29. Two recent reports describe the apparent reciprocal microduplication event: the first, in the heterozygous state in five individuals of a three-generation pedigree (Lisi et al., 2008), and the second including 19 cases, five of which appear to be the reciprocal event with the remainder overlapping this region (Ballif et al., 2008). Here, we describe index cases from four pedigrees (Case 1 apparently de novo, Case 2 a mother-child inheritance, Case 3 a nuclear family with multiple members carrying the duplication, and Case 4 an adopted child from whom information about the biological parents is unavailable). These cases all have microduplication of chromosome 3q29, validated by fluorescent in situ hybridization (FISH), array-CGH, MLPA and/or high-resolution DNA SNP microarrays. Regardless of the initial discovery and validation techniques, we have also analyzed these individuals with genome-wide Affymetrix 500K SNP arrays in order to provide fine-map duplication breakpoints and ascertain other CNV events in their genomes. The clinical phenotypes of these patients are described in detail. Of interest, two have significant ophthalmological findings and developmental delay was frequent, suggesting that dosage sensitivity of genes located within 3q29 might be important for eye and cognitive development.
Clinical report
Case 1 (Toronto)
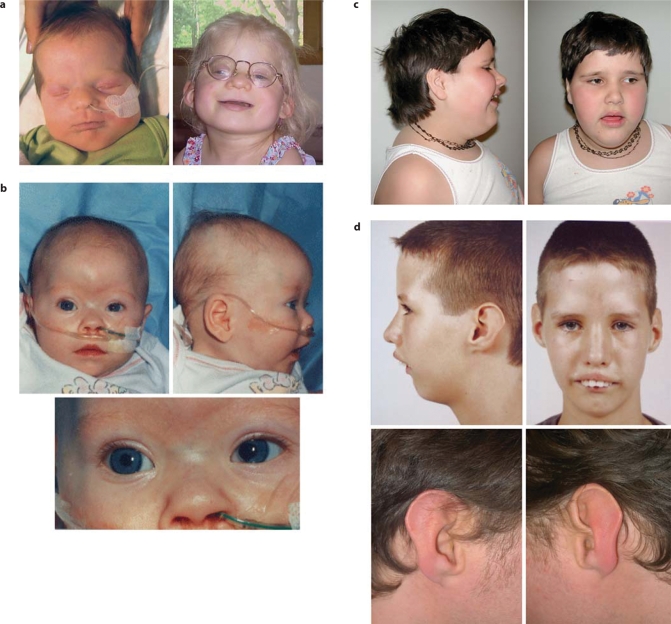
This patient is a 23 month old girl (Fig. 1a), who was born to healthy, non-consanguineous parents. The family history was negative for congenital anomalies (see pedigree, Fig. 2a). The pregnancy was complicated by hyperemesis for the first five months and hypertension for the last two weeks. There were no known teratogenic exposures. Fetal ultrasounds at 9 and 20 weeks of gestation were reportedly normal. The patient was born at 36 weeks gestation via spontaneous vaginal delivery. Labour and delivery were uncomplicated with no neonatal resuscitation required. Apgars were nine at one and five minutes. The birth weight was 2,580g (50th–75th centile), length was 50 cm (90th centile), and head circumference was 31.5 cm (25th centile). Multiple congenital anomalies noted at birth included a large anterior fontanel, a high forehead with bitemporal narrowing, a downslanting right palpebral fissure, simple low-set ears, a broad nasal root and slit-like nares, a deeply grooved philtrum, thin upper lip and short neck with redundant nuchal skin (Fig. 1a). She had a U-shaped cleft of the secondary palate. Extensive ophthalmologic abnormalities included bilateral microphthalmia, a right iris coloboma, right corneal clouding consistent with a Peter's anomaly, and a cataract of the left eye. There was a 2 cm umbilical hernia. The anus was simple and anteriorly displaced. An abdominal ultrasound revealed a cyst of unknown etiology located at the right crest of the diaphragm. Examination of the extremities revealed partial 2–3 toe syndactyly bilaterally, sandle-gap bilaterally, and camptodactyly of the toes. A skeletal survey in the newborn period revealed bilateral proximal radial-ulnar synostosis. MRI of the brain at birth revealed absence of the inferior cerebellar vermis with an enlarged cisterna magna, consistent with a Dandy-Walker variant. There were also multiple small cystic changes of the periventricular white matter within the frontal horns of the lateral ventricles. An echocardiogram at birth was reported as normal; however re-evaluation at approximately one month of age for a persistent murmur revealed an 8 mm secundum atrial septal defect with left to right shunting, which has remained asymptomatic since birth.
Fig. 1.
Clinical presentation photographs. Consent for publication was obtained in all cases. (a) Case 1, ascertained in Toronto, Canada (newborn, left panel; at age 23 months, right panel). (b) Case 2, ascertained in Edinburgh, UK. Detail of left eye abnormalities shown (lower panel). (c) Case 3, ascertained in Leiden, The Netherlands. (d) Case 4, ascertained in Toronto, Canada, at age 11 (top panels). Details of ears are shown (bottom panels).
Fig. 2.
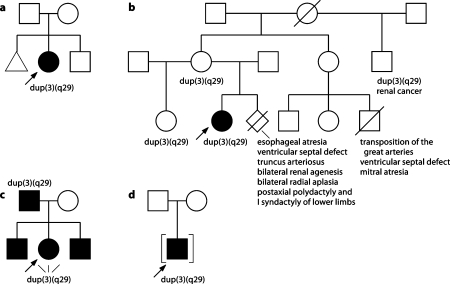
Pedigrees of cases. (a) Case 1. (b) Case 2. (c) Case 3. (d) Case 4.
Abdominal ultrasound at 5 weeks of age further defined her abdominal cyst as arising from the stomach wall and wrapping around the inferior vena cava. The cyst was resected and she had an unsuccessful attempt at umbilical hernia repair. She has had severe gastroesophageal disease and feeding difficulties since birth, requiring multiple high dose antireflux medications. Conductive hearing loss was detected at 6 months of age and required the insertion of myringotomy tubes. She underwent a right corneal transplant and left cataract excision at 4 months of age. With the use of a contact lens in the left eye, her visual acuity was 20/190 in the left eye and 20/960 in the right eye. A 2.7 × 2.8 cm subcutaneous mass was noted on the posterior right thigh. CT scan of the mass suggested that it was likely a hemangioma. No medical intervention was required. Growth parameters at 8 months of age revealed weight less than the 3rd centile, length at the 25th centile, and head circumference just below the 50th centile. Her physical features, including microphthalmia, were similar to her newborn exam. There was central hypotonia. At 18 months of age, the patient's first tooth erupted. Tooth shape was normal. A repeat attempt at surgical repair of the umbilical hernia and her extensive diastasis recti was successful at 20 months of age.
Developmental concerns were noted in the first year of life as she had significant hypotonia and visual impairment. She was smiling at 3 months of age, reaching and grasping at 8 months of age. She began rolling over at eight months. Following her surgery at 20 months, she began to sit independently, crawl and stand with support. She had a formal communication assessment at 15 months of age which indicated that her receptive language abilities were in the 7–9 month old range, and her expressive skills were in the 5 month old range. She was babbling at 23 months but did not yet have specific words. She receives occupational therapy, speech therapy, and is enrolled in an infant development program.
Case 2 (Edinburgh)
This girl was the first child of non-consanguineous parents. She was born by spontaneous vaginal delivery weighing 3,080g (12th percentile) at 41 weeks of gestation. An increased nuchal translucency was noted during the pregnancy but no invasive testing or detailed ultrasound examination was carried out. She was noted to be hypotonic soon after birth and was admitted to the neonatal intensive care unit. A cardiac ultrasound demonstrated an atrioventricular septal defect. She was thought to have facial dysmorphism compatible with a diagnosis of Down syndrome but chromosome analysis revealed a 46,XX apparently normal female karyotype. At this point she was reviewed by a clinical geneticist (DRF) who noted significant craniofacial dysmorphisms including upslanting palpebral fissures, large anterior fontanelle, brachycephaly, hypoplastic supraorbital ridges and a depressed nasal root (Fig. 1b). Her eye examination was remarkable with a left sided iris ‘coloboma’ caused by segmental aniridia with no evidence of an optic fissure closure defect. Her occipito-frontal cirumference at one week of age was 34 cm (25th percentile). She had minor digital dysmorphisms with 5th finger clinodactyly and mild syndactyly of the 2nd and 3rd toes on the left foot. She had unusual buttock folds. At this point she had further investigations including FISH for deletion 22q11.2 and Smith-Magenis syndrome, a full skeletal survey, a diasiallotransferrin assay for congenital disorders of glycosylation, and quantitative plasma amino acid urinary organic acids analysis, all of which were normal. She had an emergency admission for four different infective episodes during the first five months of life: bronchiolitis, adenovirus pneumonia, pneumococcal conjunctivitis and Clostridium difficile. At the age of seven months she had an elective repair of her AVSD and a secundum atrial septal defect. She had a prolonged recovery in intensive care and required continuous inotropic support for 38 days following the operation.
She had a Griffiths assessment at the age of 10 months and 24 days which showed global developmental delay with a developmental age equivalence for locomotor 2.75 mo., personal and social 3.5 mo., hearing and language 6.5 mo., eye and hand coordination 4.5 mo. and performance 3.5 mo. She was noted to have a mild ataxia and a brain MRI at the age of 4.3 years showed a small cerebellar vermis. When last reviewed at the age of 8.6 years her height was −3.3 SD, weight 21st percentile and OFC −3.2 SD. She is a very pleasant and friendly girl who was in good general health. She remains hypotonic and mildly ataxic. She speaks in sentences and has no behavioural problems. She attends a special educational establishment where she is making progress with all aspects of her development but she has significant global cognitive impairment.
The proband's mother and 19-year-old maternal half-sister were both healthy (See pedigree, Fig. 2b). The mother had a subsequent pregnancy that resulted in a termination for multiple fetal anomalies identified on antenatal ultrasound scanning. An autopsy on this fetus showed esophageal atresia with a tracheoesophageal fistula, a ventricular septal defect, truncus arteriosus, bilateral renal agenesis, bilateral radial aplasia, bilateral postaxial polydactyly of the feet and bilateral syndactyly of the 2nd/3rd and 3rd/4th toes. The mother's full sister, who is healthy, had a child who died as a result of a complex cardiac defect. The proband's maternal half uncle had been well until the age of 45 years when he was diagnosed with renal cell carcinoma.
Case 3 (Leiden)
This 16 year old girl was born at term with normal birth weight (Fig. 1c). There were no neonatal feeding problems or hypotonia. Motor development was slightly retarded. She was able to walk at two years of age. There was a more severe delay in speech development. At 10 years of age her vocabulary covered 40 words. MRI at 12 years of age showed no brain abnormalities. At 16 years of age the girl was not toilet-trained. When walking she would easily stumble over. She was obese and had a small, narrow forehead, straight eyebrows, narrow palpebral fissures, hypotelorism, open mouth appearance, crowding of the teeth and low posterior hairline. There was profound mental retardation.
Both brothers of this girl attended special schools because of learning difficulties (See pedigree, Fig. 2c). The father of the girl lives in an institution. His IQ is 64. He is unable to read or write. He has straight eyebrows, deep set eyes and narrow palpebral fissures.
Case 4 (Toronto)
The proband is an adopted male who was thirty years of age at the time of last examination. He was the product of first pregnancy for a then 16 year old mother, who gave him up for adoption soon after birth. He was born at full term via vaginal delivery in breech presentation with a birth weight of 2,150 g (below 3rd centile). He was noted at birth to have micrognathia, significant limb reduction defects of four extremities, congenital right hip dislocation, grade 1 hypospadias and left cryptorchidism. At 17 months of age his weight was 5.4 kg (well below the 3rd centile), head circumference 45.5 cm (–2 SD); he had mild dysmorphisms described as a hypoplastic mandible with overbite as well as mild developmental and significant speech delay. Cardiac evaluation revealed a grade 2/6 systolic murmur, but his EKG was normal. His hearing was tested at two years of age and was low-normal, with very mild conductive hearing loss in the left ear. ENT evaluation at 4 years of age (Fig. 1d) revealed a narrow, high vaulted palate with submucous cleft palate and very mild tongue coordination difficulties. He was assessed by ophthalmology at eight years of age and was found to have slight nystagmus, visual acuity of 20/20 and no structural eye defects. At age 11 (Fig. 1d), he was assessed by the craniofacial service because of severe class II malocclusion and underwent extensive orthodontic treatment and surgery including LeFort 1 to intrude the maxilla, mandibular sagittal split advancement and vertical reduction with advancement genioplasty. At 15 years of age he had left inguinal exploration that revealed an atrophic testis that was removed.
The patient has mild developmental delay and learning disabilities. His milestones were delayed and he did not sit by himself until 2 years of age. At 34 months of age he was performing at the level of a 20 month old, with prominent speech delay. The patient received therapy and was able to attend regular school with additional help due to learning disabilities affecting his reading comprehension. He finished high school, obtained a college degree, and now lives independently and works in customer care services.
At last examination at 30 years of age his head circumference was 58 cm (+ 2SD); his features include a broad nasal bridge, high arched palate; ears that are normally placed but have simple, pointed pinnae with a thin upper border. He has increased adipose tissue and has developed multiple stria in the torso and abdomen. His extremities show significant transverse reduction defects. His most well developed limb is his upper right arm which includes a normal humeral arm segment and a partly developed forearm that extends 20 cm below the elbow and ends on a blind stump. The left arm and both legs consist only of proximal segments. All extremities have dermatoglyphic patterns at the tips, suggesting at least partial development of the hands and feet. However, no digits or metacarpals are appreciated.
Materials and methods
Case 1 (Toronto)
PHA-stimulated lymphocytes from peripheral blood were cultured for 72 h with thymidine synchronization. GTG-banding analysis was performed on peripheral blood lymphocytes using standard cytogenetic techniques. G-banded karyotypes at 500 band resolution were prepared for the patient and both of her parents. The de novo change in our patient was further evaluated using fluorescent in situ hybridization (FISH). FISH was performed on cultured lymphocytes using the following probes: a chromosome 3q subtelomeric probe (Oncor, Gaithersburg, MD), and BAC clones RP11-159K3 and RP11-962B7, directly labeled with Spectrum Orange and Spectrum Green, respectively (Figs. 3 and 4). Hybridized metaphase spreads were analyzed using a Zeiss Axioplan 2 epifluorescence microscope. Images were captured by an Axiocam MRm Camera (Imaging Associates, Bicester, UK) and analyzed using an imaging system with MetaSystems Isis Software version 5.1.110 (Boston, MA).
Fig. 3.
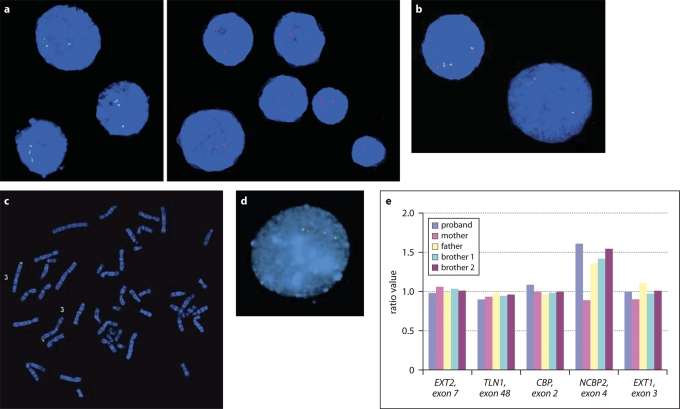
Confirmatory studies of duplications. (a) FISH confirmation in Case 1, showing duplication of BAC probes RP11-962B7 (green) and RP11-159K3 (red). (b) FISH in Case 1. Cohybridization of RP11-962B7 and RP11-159K3, demonstrating a tandem, directly oriented duplication. (c) FISH in Case 1, showing two copies of the 3q subtelomeric probe (green). (d) FISH confirmation in Case 3, showing duplication of probe CTC-196F4 (red) but not of control probe GS-1186B18 (green). (e) MLPA confirmation in Case 3 and family members. The MLPA probe in the NCBP2 gene shows a 3:2 ratio in the proband, the father and both brothers, while the control probes located elsewhere in the genome show a normal 2:2 ratio. The mother shows no duplication of the NCBP2 MLPA probe.
Fig. 4.
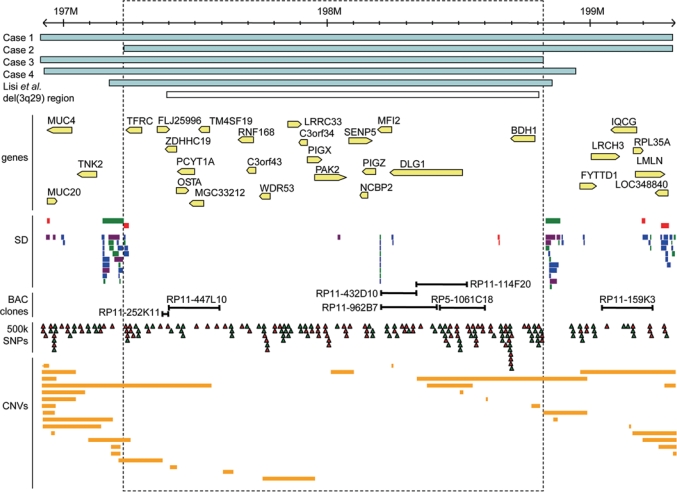
Microduplication region at 3q29. Scale at the top is in millions of base pairs (NCBI Build 35). The duplicated regions in four cases are shown (blue bars). The approximate location of the duplication described previously is shown (Lisi et al., 2008). The region of recurrent 3q29 microdeletion described in Willatt et al. (2005) is also indicated (white bar); this corresponds to the duplication reported in Ballif et al. (2008). Feature tracks from the Database of Genomic Variants are shown below: known genes (arrow indicating direction of transcription), segmental duplications (SD), SNPs on the Affymetrix 500K array set (red triangles: NspI array; green triangles: StyI array) and previously reported copy number variants (CNVs; orange bars). Locations of BAC clones used for FISH mapping (Case 1) or duplicated on BAC-CGH array (Cases 3, 4) are also indicated. Duplicated probe CTC-196F4 in Case 3 is located within BAC clone RP5-1061C18 (shown here; see also Willatt et al, 2005).
Case 2 (Edinburgh)
The 3q29 duplication in the proband was initially discovered with the BlueGnome CytoChip V1.1 1Mb BAC-CGH array (BlueGnome Ltd., Cambridge, UK), which has contig coverage of microdeletion regions. BAC array-CGH was performed on the proband and both parents where genomic DNA from each case was labeled by random priming. Hybridization and washes were performed on an HS 400™ Pro hybridization station (Tecan Ltd., UK). Each subarray was prehybridized for 45 min at 37°C with 1.5 μg of herring sperm DNA (Sigma-Aldrich, UK) in 75 μl of hybridization buffer (50% formamide, 7% dextran sulphate, 2× saline sodium citrate (SSC), 10 mM Tris-HCl pH 7.5, and 0.1% (v/v) Tween 20). Test and reference samples were mixed, co-precipitated, and resuspended in a 75 μl hybridization solution that also contained 2.5 μg/μl Cot-1 DNA (Invitrogen), denatured at 75°C for 15 min, incubated for 2 h at 37°C to block repetitive sequences, and hybridized for 21 h. Post-hybridization washes were performed using three wash cycles in each of PBS, 0.05% Tween at 37°C, 0.1× SSC at 54°C, 1× PBS at 37°C, and a final wash in PBS, 0.05% Tween at 23°C. Slides were dried using high purity nitrogen. Arrays were scanned using a GenePix Pro 5.0 array scanner (Axon Instruments, UK) and analysed using BlueFuse for Microarrays analysis software version 3.4 (BlueGnome Ltd, UK).
The proband, an unaffected sister (age 19) and mother, as well as an uncle who has renal cancer at the age of 49 and a maternal aunt and her child who died with complex congenital heart disease were also assayed by MLPA. Confirmatory MLPA was performed using both P036B and P070 human telomere assays (MRC Holland, Netherlands), which contain two independent probes for the 3q29 region. The P036B probe is situated in the BDH gene on 3q. The proximal probe sequence was GCCACCGGGAGGAACTGGGCCAT and the distal probe sequence TCTAACACCCGTTGCTACCATGCTGGCCACCCGCCTCTCCAGA. The second probe on 3q, P070, is located in KIAA0226 and has a proximal probe sequence 5′-CTCTTTCTCCAGGTCACTGCGCTGGAGGACAG and distal probe sequence 5′-ATGTGCCGTCTTGTCCTGCCTGTTTCACATCAGCATAGGATCA. MLPA products were processed using an ABI 3100 Genetic Analyzer with ABI GeneScan™ ROX500™ size standard. Quantitative data analysis was obtained using the SoftGenetics® Gene Marker® v1.4 software.
Case 3 (Leiden)
Conventional cytogenetic analysis on GTG-banded chromosomes from cultured lymphocytes of the index case was performed according to standard techniques. Array-CGH was performed on all five family members using the ∼1.0 Mb spaced whole genome large insert clone arrays, for which the clones were kindly made available by the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk). The clones were grown, PCR amplified and spotted as previously described (Fiegler et al., 2003; Knijnenburg et al., 2005). Genomic DNA of the patient was isolated using standard techniques, and 500 ng was labeled with Cy3-dCTP (GE Healthcare, Diegem, Belgium) using the BioPrime® DNA Labeling System (Invitrogen, Breda, the Netherlands). As a reference DNA, 500 ng female human genomic DNA (Promega, Leiden, the Netherlands) was labeled using Cy5-dCTP. Hybridization and slide washing was performed without prehybridization on an HS400 hybridization station (Tecan, Giessen, the Netherlands). Arrays were scanned with a GenePix 4100A scanner (Axon Instruments, Union City, CA) and images were processed with GenePix Pro 4.1 software. Final analysis of the intensity ratios of the hybridized DNA was as previously described (Knijnenburget al., 2005).
Confirmatory MLPA was performed on the index case as described (White et al., 2004). The selected probes were located in the NCBP2 gene. The proximal sequence was 5′-GGCCGCGGGAATTCGATTGGTGATGTTCTTCAGCAAATTCAACAGGCCAAAGGAGTGTTT and the distal sequence was 5′-GTCACTGACAGAGCTCTCACCACTCACACTAGTGAATTCGCGGC.
Quantitative readout was performed with an ABI 3730 DNA analyzer. The accompanying Genescan 3.5 software was used for peak analysis and further downstream normalization and calculations were performed as described (Whiteet al., 2004). Two-colour interphase FISH confirmation of the duplication in the proband was performed with clones CTC-196F4 at 3q29, partly overlapping the DLG1 gene (as in Willatt et al., 2005), and 3p subtelomeric clone GS-1186B18 as a control.
Case 4 (Toronto)
Routine cytogenetic workup was as for Case 1, above. The initial karyotype report of 46,XY was followed up with chromosomal microarray analysis (Kleberg Cytogenetics Laboratory, Baylor College of Medicine, Houston, TX, USA; CNV version 5.0).
Affymetrix genome-wide SNP array and copy number analyses
For CNV analysis, we adhered to recommended guidelines (Scherer et al., 2007). In order to maximize consistency between samples collected at the three sites (Toronto, Leiden and Edinburgh), all samples were characterized with the Affymetrix 500K array set at The Centre for Applied Genomics in Toronto. Each sample was genotyped with the GeneChip® Human Mapping NspI and StyI Arrays (Affymetrix, Inc., Santa Clara, CA) according to the manufacturer's instructions and as described previously (Kennedy et al., 2003). For copy number determination, we used three approaches: DNA Chip Analyzer (dChip) (Li and Wong, 2001; Lin et al., 2004; www.dchip.org), CNAG (Nannya et al., 2005) and GEMCA (Komura et al., 2006). The first two algorithms were applied separately to each 250K array, and GEMCA was applied to combined 500K array data. CNVs were scored if they were detected in the same individual either a) on both arrays, or b) by two of the algorithms. In our hands, these criteria result in high confidence CNV calls that are >95% likely to be confirmed by an independent technology such as qPCR (Pinto et al., 2007; Marshall et al., 2008). In the cases of copy number losses, SNP genotypes were examined in order to determine parent of origin.
Results
All phenotype and CNV data are entered in the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER; http://decipher.sanger.ac.uk/).
Case 1 (Toronto)
A subtle cytogenetically detectable differenceat 3q29 was detected at a G-band resolution of 500 bands in Case 1. This alteration was not observed in her parents’ chromosomes at the same resolution (data not shown). The chromosomal difference was determined to be interstitial as the chromosome 3q subtelomeric probe revealed two normal signals in the correct position in this patient (Fig. 3c). Further investigation using the Affymetrix 250K NspI Array revealed a 2.4 Mb duplication of 3q29 (Fig. 4; Table 1). The duplication was determined to be de novo, as neither parent revealed a CNV at this locus.
Table 1.
CNVs detected in patient families with Affymetrix 500K microarrays
| Cytoband |
Estimated size (bp) |
Type |
Status in childrena |
Gene(s) involved |
|---|---|---|---|---|
| Family of Case 1 | ||||
| Proband | ||||
| 3q29 | 2,399,433 | gain | de novo | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 6q16.1 | 60,058 | loss | inherited (paternal)b | no genes; overlaps known CNVs |
| 8p23.1 | 407,187 | gain | inherited (maternal) | FAM86B1, DEFB130, LOC44005; region of segmental duplications and known CNVs |
| Mother | ||||
| 8p23.1 | 202,167 | gain | – | no known genes; numerous cDNAs; region of segmental duplications and known CNVs |
| 10q11.1-q11.21 | 906,591 | gain | – | ZNF33B, BMS1L and numerous cDNAs; region of segmental duplications and known CNVs |
| 11q22.1 | 626,031 | loss | – | cDNA AK128111; overlaps known CNVs |
| 15q11.2 | 1,908,357 | loss | – | OR4N2, OR4M4, POTE15, LOC283755 and multiple cDNAs |
| 22q11.23 | 211,233 | gain | – | LRP5L and multiple cDNAs; region of segmental duplications and known CNVs |
| Father | ||||
| 6q16.1 | 60,058 | loss | – | no genes; overlaps known CNVs |
| 10q11.22 | 124,801 | gain | – | no genes; region of segmental duplications and known CNVs |
| 14q11.2 | 153,147 | gain | – | OR4N2, OR4K2, OR4K5, OR4K1 (odorant receptor gene cluster) |
| Family of Case 2 | ||||
| Mother | ||||
| 3q29 | 2,086,988 | gain | – | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 7q11.23 | 428,467 | gain | – | POMZP3, UPK3B, cDNA BC043544, intron of cDNA BC013192 |
| 14q21.1 | 357,718 | loss | – | cDNA BX248273; encompasses small CNV |
| Proband | ||||
| 3q29 | 2,086,988 | gain | inherited (apparently maternal) | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 7q11.23 | 422,126 | gain | inherited (apparently maternal) | POMZP3, most of UPK3B, cDNA BC043544, intron of cDNA BC013192 |
| 14q11.2 | 219,459 | gain | unknown | OR4Q3, OR4M1, OR4N2, OR4K2, OR4K5, OR4K1 (odorant receptor cluster) |
| 14q21.1 | 368,345 | loss | inherited (maternal)b | cDNA BX248273; encompasses small CNV |
| 15q11.2 | 1,662,281 | gain | unknown | OR4N2, OR4M4, POTE15, LOC283755 and multiple cDNAs |
| 17q21.31 | 183,068 | gain | unknown | 5′ end of KIAA1267, and cDNAs BC018467 and BC000924 |
| Family of Case 3 | ||||
| Proband | ||||
| 3q29 | 1,893,889 | gain | inherited (paternal) | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 8p23.2-p23.1 | 176,963 | gain | inherited (paternal) | 5′ end of MCPH1, and cDNAs including AK025595 |
| 15q11.2 | 1,378,020 | loss | de novoc | OR4N2, OR4M4, POTE15, LOC283755 and multiple cDNAs |
| 19q13.42 | 456,306 | gain | inherited (maternal) | cDNA BX248273; encompasses small CNV |
| Mother | ||||
| 12q24.31 | 114,901 | gain | – | 3′ ends of P2RX7 and CAMKK2, and all of P2RX4 |
| 19q13.42 | 996,692 | gain | – | multiple genes |
| brother 1 | ||||
| 1q31.3 | 142,667 | loss | inherited (paternal)d | CFHR3, CFHR1 and 5′ end of CFHR4 |
| 3q29 | 1,893,889 | gain | inherited (paternal) | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 8p23.2-p23.1 | 221,809 | gain | inherited (paternal) | 5′ end of MCPH1, and cDNAs including AK025595 |
| 10q11.22 | 1,087,629 | gain | inherited (paternal) | GPRIN2, PPYR1, ANXA8L1, ANXA8, and multiple other cDNAs |
| 12q24.31 | 79,614 | gain | inherited (maternal) | 3′ ends of P2RX7 and CAMKK2, all of P2RX4 |
| 14q11.2 | 229,920 | gain | inherited (paternal) | OR4Q3, OR4M1, OR4N2, OR4K2, OR4K5, OR4K1 (odorant receptor gene cluster) |
| 15q11.2 | 1,662,281 | gain | de novoc | OR4N2, OR4M4, POTE15, LOC283755 and multiple cDNAs |
| 19q13.42 | 857,878 | gain | inherited (maternal) | multiple genes |
| Brother 2 | ||||
| 3q29 | 1,893,889 | gain | inherited (paternal) | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 4q24 | 831,405 | gain | inherited (paternal) | TACR3 |
| 8p23.2-p23.1 | 198,636 | gain | inherited (paternal) | 5′ end of MCPH1, and cDNAs including AK025595 |
| 10q11.22 | 848,700 | gain | inherited (paternal) | SYT15, GPRIN2, PPYR1, ANXA8L1 |
| 15q11.2 | 1,662,281 | gain | de novoc | OR4N2, OR4M4, POTE15, LOC283755 and multiple cDNAs |
| 19q13.42 | 541,228 | gain | inherited (maternal) | multiple genes |
| Father | ||||
| 3q29 | 1,893,889 | gain | – | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 4q24 | 556,763 | gain | – | TACR3 |
| 8p23.2-p23.1 | 224,359 | gain | – | 5′ end of MCPH1, and cDNAs including AK025595 |
| 10q11.22 | 848,700 | gain | – | SYT15, GPRIN2, PPYR1, ANXA8L1 |
| 14q11.2 | 222,787 | gain | – | OR4Q3, OR4M1, OR4N2, OR4K2, OR4K5, OR4K1 (odorant receptor gene cluster) |
| Case 4 | ||||
| 2q27.3 | 81,134 | loss | unknown | multiple cDNAs; encompassed by known CNV |
| 3q29 | 2,041,109 | gain | unknown | multiple genes; overlapping with 3q29 microdeletion syndrome (Willatt et al., 2005) |
| 6q24.2-q24.3 | 2,637,073 | gain | unknown | 3′ end of UTRN, EPM2A, GRM1, RAB32, FLJ44955, FBX030, SHPRH, various cDNAs; encompasses several small CNVs |
| 14q11.2 | 249,343 | loss | unknown | OR4N2, OR4K2, OR4K5, OR4K1, OR4K13, OR4K14, OR4K15 (odorant receptor gene cluster; encompasses known CNV and segmental duplications) |
| Xp11.23 | 67,327 | loss | unknown | ZNF630, SSX6; encompasses known CNV and segmental duplications |
Parental origin is inferred assuming Mendelian inheritance of CNV events. In Case 2, these are apparently maternal; however, the father's sample was not available for analysis.
Parental origin of losses in Case 1 and Case 2 were confirmed by examination of SNP genotypes in these regions.
The 15q11.2 region is very complex and apparently de novo events may actually be inherited; in particular, the loss in Case 3 contains multiple heterozygous SNPs and is therefore not a simple hemizygous deletion.
SNP genotypes are consistent with paternal inheritance of the 1q31.3 loss in this individual. The corresponding CNV in the father was identified by only one algorithm and thus is not reported.
The patient also had a 60 kb loss at 6q16.1 and a 407 kb gain at 8p23.1 (Table 1). The 6q16.1 locus contains no known genes and overlaps numerous known segmental duplications and CNVs, and the 8p23.1 region is a locus of known copy-number polymorphisms and segmental duplications in the vicinity of the beta-defensin gene DEFB130. This CNV overlaps with the proximal end of the region of 8p23.1 duplication reported by Barber et al. (2008) (see Discussion). Interphase FISH analysis of the 3q29 region using BAC clone probes RP11-159K3 and RP11-962B7 revealed three signals for each probe, confirming the duplication (Fig. 3a). Clone RP11-962B7 is located approximately in the middle of the region identified as a duplication by microarray, while clone RP11-159K3 is located approximately 600 kb distal to RP11-962B7, also within the duplicated region in this patient (Fig. 3). Co-hybridization of the two BAC clone probes suggested that the structure of the rearrangement was a tandem, direct duplication (Fig. 3b). The parents of this patient had the normal two signals for each probe, confirming that the duplication occurred de novo in our patient (data not shown). These same FISH probes were hybridized to metaphase spreads which confirmed their localization to 3q29 only, in both the patient and her parents (data not shown).
In order to rule out non-paternity (and thus the possibility that the 3q29 microduplication was in fact inherited, rather than de novo), we used PedCheck (O’Connell and Weeks, 1998) to detect markers incompatible with the pedigree. Of the 262,264 SNPs on the NspI array, 271 autosomal and 20 X chromosome SNPs were inconsistent either between the father or mother and the proband. Of the autosomal SNPs, 117 were inconsistent between mother and child, 115 between father and child, and 39 were consistent between the child and each parent separately, but not as a pair. Since the proportion of incompatible markers is low (∼0.1%), the pedigree is consistent with the SNP data. Moreover, the rates of inconsistency between mother and child (43.2%: 117/271) versus father and child (42.4%: 115/271) are nearly identical, indicating that these are due to random genotyping errors and ruling out non-paternity in this family.
Case 2 (Edinburgh)
High resolution SNP array analysis confirmed a duplication at 3q29 of 2.08 Mb in size in the proband as detected originally using a 1 Mb BAC array platform. Notably, the distal boundary was identical to that of Case 1, although the proximal breakpoint was slightly farther distal, accounting for the difference in size (2.08 Mb vs. 2.4 Mb in Case 1; see Fig. 4). However, the unaffected mother and a maternal half-sister carry the apparently identical duplication, as does a maternal half-uncle who has renal cancer at the age of 45. No imbalance of the 3q29 region was evident in the proband's father as seen by BAC array-CGH or MLPA. The mother and proband were evaluated with Affymetrix 500K SNP arrays and their duplications were found to be identical in extent (Table 1). Unfortunately there was no material from the subsequent affected fetus of this mother to test for the duplication. A maternal aunt, and her child who died with complex congenital heart disease, do not have the duplication (as assayed by MLPA; data not shown; see pedigree, Fig. 2b) and thus this heart disease in the extended family does not appear to be related to the 3q29 duplication. CNVs detected in the proband (at 7q11.23, 14q11.2, 14q21.1, 15q11.2 and 17q21.31) and mother (7q11.23 and 14q21.1) are previously reported polymorphic CNVs found in apparently healthy individuals (Table 1) and are thus unlikely to contribute to the phenotype seen in this patient. The ∼360 kb loss at 14q21.1 and 420–430 kb gain at 14q21.1 are also present in the proband's unaffected mother.
Case 3 (Leiden)
BAC-CGH analysis confirmed that the proband, her father and two affected brothers carry duplications of BAC clones RP11-252K11 and RP11-114F20 (data not shown; see Fig. 4). SNP microarray analysis demonstrated an identical 1.9 Mb duplication at 3q29 in these individuals (Table 1). The proximal boundary was identical to that of Case 1, and the distal boundary was somewhat more proximal (Fig. 4). Other CNVs detected in this family include loci at 1q31.3, 4q24, 7q11.23, 10q11.22, 12q24.31, 14q11.2, 14q21.1, 15q11.2, 17q21.31, 19q13.42, all of which overlap known CNVs found in apparently healthy individuals (Table 1). An apparent de novo loss at 15q11.2 in the proband was not supported by examination of genotype data, as 16 SNPs in this region were heterozygous (data not shown); as this is in a complex region including multiple similar odorant receptor genes, this CNV may be a false positive, or may represent a loss to two copies as compared with higher copy number in other members of this family. One additional CNV in this family, at 8p23.2→p23.1, is also in a region of known CNVs and is proximal to the microduplication 8p23.1 region described by Barber et al. (2008) (see Discussion). The duplication in the proband was confirmed by two-colour interphase FISH (Fig. 3d). The MLPA probe in the NCBP2 gene also confirmed the duplication (3:2 ratio as compared with control) in the father and both brothers, while control probes located elsewhere in the genome show a normal 2:2 ratio. The mother showed no duplication of the NCBP2 MLPA probe (Fig. 3e).
Case 4 (Toronto)
BAC-CGH array analysis revealed a gain in copy number of two clones (RP11-447L10 and RP11-432D10; Fig. 4) located at cytogenetic band 3q29, with no other sites of copy number change detected. Microarray analysis demonstrated a duplication of 2.0 Mb at 3q29 (Table 1). The proximal boundary was similar to those of Cases 1 and 3, and the distal boundary just slightly distal to that of Case 3 but still within a cluster of segmental duplications (Fig. 4). Other CNVs detected in this individual that are also seen in healthy individuals were at 2q27.3, 14q11.2 and Xp11.23 (Table 1). One additional large (2.6 Mb) CNV at 6q24.2→q24.3 encompasses several small known CNVs but also results in a previously unreported copy number gain of a number of known genes including Utrophin (UTRN), the Lafora progressive myoclonus epilepsy gene EPM2A, a metabotropic glutamate receptor (GRM1), Ras oncogene family member RAB32, two genes apparently involved in protein ubiquitination (SHPRH and FBXO30) and an expressed repetitive element (FLJ44955).
Discussion
Duplication 3q syndrome (dup3q) has been described in the literature, consisting of dysmorphic features including microcephaly, low-set ears, downturned corners of the mouth, bushy eyebrows and long eyelashes, along with eye, palate, renal and cardiac anomalies (Steinbach et al., 1981; Aqua et al., 1995). The phenotype has been said to partially overlap that of Brachmann de Lange/Cornelia de Lange syndrome (OMIM#122470). Many groups have described the cytogenetic critical region associated with the 3q duplication syndrome as involving 3q26 (Aqua et al., 1995; Rizzu et al., 1997; Faas et al., 2002); however Battaglia et al. (2006) suggested that it was 3q29. The four index patients which we have presented do not have a phenotype consistent with Brachman de Lange syndrome, suggesting that 3q29 is unlikely to be involved in the previously described ‘Duplication 3q syndrome’.
Prior to the past year, there were few cases described with pure duplications of chromosome 3q. Faas et al. (2002) described three patients with cytogenetically visible chromosome 3q duplications that extended to include 3q29. The duplications in these patients included more proximal cytogenetic bands in addition to 3q29, whereas our reported cases had small duplications localized within 3q29; however, features common to both groups included mental retardation and ocular anomalies.
As molecular cytogenetic techniques have advanced over the recent years, we have been able to better detect and more precisely define microdeletions and microduplications in this region. The presence of microdeletions (Willatt et al., 2005) and the abundance of segmental duplications in this region (in particular at approximately 196.6 and 199 Mb) suggested that the reciprocal microduplications might exist, and two recent reports (Ballif et al., 2008; Lisi et al., 2008) and the cases reported here support this hypothesis.
As demonstrated by our cases, there appears to be a variable clinical phenotype associated with this duplication (see Table 2), characterized by ocular and cardiac anomalies, hypotonia, developmental and speech delay. However, the phenotype is variable with reduced penetrance; the mother and maternal half sister of Case 2 are unaffected, and the two brothers of Case 3 have milder phenotypes. Case 4 demonstrates the wide spectrum of phenotypic differences, although it is possible that his tetramelia may be unrelated to the duplication (see below). The previously reported family (Lisi et al., 2008) also included individuals with some similar features, notably frequent developmental delay and some with palpebral fissure anomalies. The cases reported by Ballif et al. (2008) had duplications varying in size from 0.2 to 2.4 Mb, but only five of these had the apparent reciprocal duplication of the previously reported deletion (Willatt et al., 2005). Of these, clinical information was reported for three, with mild to moderate developmental delay as the only common feature, and craniosynostosis, high palate, seizures and a ventricular septal defect occurring in two cases each (Ballif et al., 2008).
Table 2.
Comparison of clinical features in four patients with duplication of 3q29
| Clinical feature |
Case 1 (Toronto; 23 months) |
Case 2 (Edinburgh; 8 years) |
Case 3 (Leiden; 16 years) |
Case 4 (Toronto; 30 years) |
|---|---|---|---|---|
| Birth weight | 2,850g (50–75th centile) | 3,080g (12th centile) | 3,200g (25th centile) | 2,150g (<3rd centile) |
| Microcephaly | – | + | not reported | – |
| Generalized obesity | – | – | + | + |
| Ocular anomalies | + | + | not reported | – |
| Palpebral fissure anomalies | + | + | + | – |
| Cleft palate | + | – | – | + |
| Dental anomalies | + | – | + | + |
| Conductive hearing loss | + | – | not reported | + |
| Structural brain anomaly | + | + | – | – |
| Developmental delay | + | + | + | + |
| Hypotonia | + | + | – | – |
| Congenital heart disease | + | + | – | – |
| Musculoskeletal anomalies | + | + | not reported | + |
Notably, the duplication events in all four of the families presented here overlap, and those of Cases 2, 3 and 4 each share one of the boundaries of Case 1, the largest of the duplications. Together, these define a minimum critical region of approximately 1.58 Mb in size, encompassing the region from the TFRC to BDH1 genes. The proximal and distal ends of this minimal region correspond to clusters of segmental duplications (Fig. 4), indicating a possible recombination-mediated mechanism for the formation of these duplications. The region is similar in size to the 3q29 microdeletions previously reported (Willatt et al., 2005), which extend from BAC clone RP11-252K11 (proximal end at 197.40 Mb) to RP11-535N19 (distal end at 198.81 Mb) (see Fig. 4), further supporting the hypothesis that these are reciprocal products of a deletion/duplication event mediated by non-allelic homologous recombination at segmental duplications (Lupski, 2004; Ballif et al., 2008; Lisi et al., 2008). This region contains 20 known genes (Table 3).
Table 3.
Genes in minimal region of 3q29 duplication
| Gene |
Function |
Known syndromes/diseases |
MIM |
|---|---|---|---|
| BDH1 | 3-hydroxybutyrate dehydrogenase, type 1; interconversion of acetoacetate and (R)-3-hydroxybutyrate during fatty acid catabolism | none | – |
| C3orf43 | unknown | none | – |
| C3orf34 | unknown | none | – |
| DLG1 | homologue of Drosophila tumour suppressor gene; putative lymphocyte-specific TSG | homologous to X-linked mental retardation gene DLG3 (Willatt et al., 2005); eye anomalies in knockout mouse (Nguyen et al., 2003) | 601014 |
| FLJ25996 | unknown | none | – |
| LRRC33 | leucine rich repeat containing 33 | none | – |
| MFI2 | melanoma cell surface glycoprotein; similar to transferring | none | 155750 |
| MGC33212 | TCTEX1D2, Tctex1 domain containing 2 | none | – |
| NCBP2 | nuclear cap-binding protein 2 | none | 605133 |
| OSTA | organic solute transporter alpha subunit; basolateral bile acid and steroid transporter | none | – |
| PAK2 | p21 activated kinase | homologous to X-linked mental retardation gene PAK3 (Willatt et al., 2005) | 605022 |
| PCYT1A | phosphate cytidylyltransferase 1, choline, alpha isoform; phosphatidyl choline synthesis | none (essential for survival in cultured cells) | 123695 |
| PIGX | phosphatidyl inositol glycan, class X; synthesis of glycosyl- phosphatidylinositol in the endoplasmic reticulum | none | 610276 |
| PIGZ | phosphatidyl inositol glycan, class Z; synthesis of glycosyl- phosphatidylinositol | none | 611671 |
| RNF168 | ring finger protein 168 | none | – |
| SENP5 | SUMO1/sentrin specific peptidase 5; required for mitosis and/or cytokinesis | none | – |
| TFRC | transferrin receptor 1 | cellular receptor for New World hemorrhagic fever arenaviruses; knockout results in severe anemia and neurologic abnormalities in mouse model | 190010 |
| TM4SF19 | transmembrane 4 L six family member 19 | none | – |
| WDR53 | WD repeat domain 53 | none | – |
| ZDHHC19 | homologue of palmitoyltransferase of NRAS and HRAS (ZDHHC9) | homologous to ZDHHC9 causing X-linked mental retardation with Marfanoid habitus | – |
DLG1 seems to be a good candidate for the ocular aspects of the dup(3)(q29) phenotype (microphthalmia in Case 1, partial aniridia in Case 2). DLG1 is expressed in the developing lens and retinal pigment epithelium, and a DLG1 gene-trap homozygous mouse has overgrowth of the lens epithelium as one part of the phenotype (Nguyen et al., 2003). With regards to developmental delay, Willatt et al. (2005) in their description of microdeletions of 3q29 in this region (see Fig. 3) point out that two of the genes in this region, PAK2 and DLG1, are homologues of the X-linked mental retardation genes PAK3 and DLG3. We also note that this region contains the ZDHHC19 gene, a homologue of the ZDHHC9 palmitoyltransferase, mutations of which have been shown to cause X-linked mental retardation with Marfanoid habitus (OMIM 300646). Importantly, all of these are increased in copy number in the 3q29 duplication patients described here, whereas they are reduced to haploidy in 3q29 microdeletion patients. Possibly, these or other genes in the region are sensitive to both increases and decreases in gene dosage, either of which might disrupt normal development.
The other CNVs detected in these patients overlap previously reported polymorphic CNVs present in the general population (reflected by their presence in the Database of Genomic Variants; Iafrate et al., 2004), with two exceptions. The most obvious potentially pathogenic CNV is at 8p23.1 in Case 1, overlapping the proximal end of the region seen in 8p23.1 duplication syndrome patients (Barber et al., 2008). This patient shares some clinical features with the patients of Barber et al., notably a high forehead, cardiac malformations, low-set ears and partial 2/3 toe syndactyly. The Family 1 proband of Barber et al. also had a high arched palate which could be compared to the cleft palate of our Case 1. We cannot rule out that some of these features seen in our Case 1 may be due to the overlap of the 8p23.1 CNV with the proximal end of the 8p23.1 microduplication region; however, there is a relatively small region of overlap (407 kb of ∼3.75 Mb total) containing only the beta-defensin gene DEFB130 and two genes of unknown function (FAM86B1, and LOC440053 which has homology to zinc finger protein genes). Furthermore, our Case 1 and Case 2 probands share common anomalies and phenotypic features, and Case 2 was not found to have an 8p23.1 CNV, suggesting that their similarities are more likely a result of their common 3q29 duplication. In contrast, the 8p23.2→ p23.1 CNV seen in the family of our Case 3 is distinct, being located distal to this microduplication region. Although Barber et al. ran a custom Agilent 44K genome-wide oligonucleotide CGH array on their patients (http://www.ngrl.org.uk/Wessex/array.htm), they report no results indicating whether changes in the 3q29 region (or anywhere else in the genome other than 8p23.1) were observed.
The second potentially pathogenic CNV is the gain at 6q24.2→q24.3 seen in Case 4. This 2.6 Mb region might contribute to the tetramelia seen only in this patient, possibly due to the involvement of the Utrophin/Dystrophin-like protein (UTRN) gene, or the putative protein-ubiquitin ligases SHPRH and FBXO30. However, there is currently no experimental evidence that gains of activity or genomic copy number of these or the other genes in this region (GRM1, EPM2A, RAB32 and FLJ44955) can affect limb development, although extensive work characterizing UTRN knockouts has been performed. Given its role in normal muscle development, further characterization of overexpression of Utrophin in limb development may shed some light on this patient's phenotype.
In summary, we have described four new cases with 3q29 duplications, with a minimum region of overlap of 1.6 Mb corresponding in location to the previously reported 3q29 microdeletions. The phenotype in these families reveals variable expressivity and reduced penetrance. Global developmental delay was the most consistent feature in our cases and two other studies (Ballif et al., 2008; Lisi et al., 2008). Other features common to some of the patients included ocular anomalies, congenital heart defects, structural brain anomalies and hypotonia. Further phenotypic characterization of these patients, in combination with improved molecular understanding of the 3q29 duplicated region, will better delineate potential dosage sensitive genes in this genomic interval and their possible roles in cognitive and ocular development.
Management and anticipatory care
Management guidelines for duplication of chromosome 3q29 have not been previously published. Our recommendations are based on the phenotypes described in our newly reported cases and those of the cases recently described by Lisi et al. 2008 and Ballif et al. in 2008. Importantly, as we discuss in this paper, the CNV content at other sites should also be considered in performing genotype and phenotype correlations.
Infancy and initial diagnosis:
1) Ophthalmologic evaluation
2) Echocardiogram
3) Brain imaging
4) Developmental assessment by 6 months of age, and continuing every 1–3 years as needed
5) Hearing evaluation
6) Skeletal survey
7) Early intervention services
8) Offer cytogenetic/molecular testing to parents to determine if the duplication of 3q29 is de novo or familial. Refer parents for genetic counseling if contemplating future pregnancies.
9) Family support
Childhood:
1) Ongoing developmental services and therapy. Individualized educational plan if appropriate. Children will likely benefit from speech therapy, occupational therapy, and physical therapy.
2) Referral to paediatric dentistry
3) Encourage physical activity and balanced diet, given reported obesity in some children/adults with duplication 3q29
Adolescence and adulthood:
1) Annual medical examination as per standard medical practice
2) Ongoing developmental services, individualized educational plan, and counseling regarding work placement appropriate for level of development
3) Education regarding sexual development and recurrence risk in offspring if appropriate for level of development.
These recommendations will be updated as we learn more about the natural history and variable phenotype of this condition, as well as the impact of CNVs at different sites in the genome.
Web resources
The Database of Genomic Variants (DGV): http://projects.tcag.ca/variation/
Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER): http://decipher.sanger.ac.uk/
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM
Acknowledgements
We thank the TCAG microarray facility, Dr. Ann George for helpful discussions concerning the manuscript, Dr. Marjolein Kriek and Emmelien Aten for help with MLPA analysis and Marlies Hoogenboom for confirmation FISH (Leiden group), and Pingzhao Hu (TCAG Statistical Analysis Facility) for paternity analysis of Case 1.
Footnotes
Work at The Centre for Applied Genomics was supported by Genome Canada/Ontario Genomics Institute, the McLaughlin Centre for Molecular Medicine, the Canadian Institutes of Health Research (CIHR), the Canadian Institute for Advanced Research, the Canada Foundation for Innovation, the Ontario Ministry of Research and Innovation, and The Hospital for Sick Children (SickKids) Foundation. C.R.M. is supported by the SickKids Foundation and the National Alliance for Research on Schizophrenia and Depression (NARSAD). S.W.S. is the GlaxoSmithKline-CIHR Pathfinder Chair in Genetics and Genomics at SickKids and a Fellow of the CIHR at the University of Toronto. This collaboration arose as a result of interaction at the Third International DECIPHER Symposium (Wellcome Trust Sanger Centre, Hinxton, UK; May 16–18, 2007).
References
- Aqua MS, Rizzu P, Lindsay EA, Shaffer LG, Zackai EH, et al. Duplication 3q syndrome: molecular delineation of the critical region. Am J Med Genet. 1995;55:33–37. doi: 10.1002/ajmg.1320550111. [DOI] [PubMed] [Google Scholar]
- Autism Genome Project Consortium, Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JCK, Maloney VK, Huang S, Bunyan DJ, Cresswell L, et al. 8p23.1 duplication syndrome; a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet. 2008;16:18–27. doi: 10.1038/sj.ejhg.5201932. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Novelli A, Ceccarini C, Carey JC. Familial complex 3q;10q rearrangement unraveled by subtelomeric FISH analysis. Am J Med Genet A. 2006;140:144–150. doi: 10.1002/ajmg.a.31042. [DOI] [PubMed] [Google Scholar]
- Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39(7 Suppl):S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitropoulos A, Schultz RT. Autistic-like symptomatology in Prader-Willi syndrome: a review of recent findings. Curr Psychiatry Rep. 2007;9:159–164. doi: 10.1007/s11920-007-0086-7. [DOI] [PubMed] [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas BH, De Vries BB, Van Es-Van Gaal J, Merkx G, Draaisma JM, Smeets DF. A new case of dup(3q) syndrome due to a pure duplication of 3qter. Clin Genet. 2002;62:315–320. doi: 10.1034/j.1399-0004.2002.620411.x. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, et al. Large-scale genotyping of complex DNA. Nat Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- Knijnenburg J, Szuhai K, Giltay J, Molenaar L, Sloos W, et al. Insights from genomic microarrays into structural chromosome rearrangements. Am J Med Genet A. 2005;132:36–40. doi: 10.1002/ajmg.a.30378. [DOI] [PubMed] [Google Scholar]
- Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- Lisi EC, Hamosh A, Doheny KF, Squibb E, Jackson B, et al. 3q29 interstitial microduplication: a new syndrome in a three-generation family. Am J Med Genet A. 2008;146:601–609. doi: 10.1002/ajmg.a.32190. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Hotspots of homologous recombination in the human genome: not all homologous sequences are equal. Genome Biol. 2004;5:242. doi: 10.1186/gb-2004-5-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt JL, Lindor NM. Further clinical description of duplication of Williams-Beuren region presenting with congenital glaucoma and brachycephaly. Am J Med Genet A. 2008;146A:1055–1058. doi: 10.1002/ajmg.a.32235. [DOI] [PubMed] [Google Scholar]
- Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Nguyen MM, Nguyen ML, Caruana G, Bernstein A, Lambert PF, Griep AE. Requirement of PDZ-containing proteins for cell cycle regulation and differentiation in the mouse lens epithelium. Mol Cell Biol. 2003;23:8970–8981. doi: 10.1128/MCB.23.24.8970-8981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana C, Bernabeu J, Monfort S, Roselló M, Oltra S, et al. Duplication of the Williams-Beuren critical region: case report and further delineation of the phenotypic spectrum. J Med Genet. 2008;45:187–189. doi: 10.1136/jmg.2007.054064. [DOI] [PubMed] [Google Scholar]
- Osborne LR, Martindale D, Scherer SW, Shi XM, Huizenga J, et al. Identification of genes from a 500-kb region at 7q11.23 that is commonly deleted in Williams syndrome patients. Genomics. 1996;36:328–336. doi: 10.1006/geno.1996.0469. [DOI] [PubMed] [Google Scholar]
- Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;16(Spec No. 2):R168–173. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, et al. Molecular mechanism for duplication 17p11.2 – the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzu P, Haddad BR, Vallcorba I, Alonso A, Ferro MT, et al. Delineation of a duplication map of chromosome 3q: a new case confirms the exclusion of 3q25–q26.2 from the duplication 3q syndrome critical region. Am J Med Genet. 1997;68:428–432. [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39(7 Suppl):S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach P, Adkins WN, Jr, Caspar H, Dumars KW, Gebauer J, et al. The dup(3q) syndrome: report of eight cases and review of the literature. Am J Med Genet. 1981;10:159–177. doi: 10.1002/ajmg.1320100210. [DOI] [PubMed] [Google Scholar]
- Torniero C, Dalla Bernardina B, Novara F, Cerini R, Bonaglia C, et al. Dysmorphic features, simplified gyral pattern and 7q11.23 duplication reciprocal to the Williams-Beuren deletion. Eur J Hum Genet. 2008;16:880–887. doi: 10.1038/ejhg.2008.42. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- White SJ, Vink GR, Kriek M, Wuyts W, Schouten J, et al. Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat. 2004;24:86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- Willatt L, Cox J, Barber J, Cabanas ED, Collins A, et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GN, Hieber VC, Schmickel RD. The association of chromosome 3 duplication and the Cornelia de Lange syndrome. J Pediatr. 1978;93:783–788. doi: 10.1016/s0022-3476(78)81077-4. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]