Abstract
Pharmacogenetics is the study of the role of inheritance in variation to drug response. Drug response phenotypes can vary from adverse drug reactions at one end of the spectrum to equally serious lack of the desired effect of drug therapy at the other. Many of the current important examples of pharmacogenetics involve inherited variation in drug metabolism. Sulfate conjugation catalyzed by cytosolic sulfotransferase (SULT) enzymes, particularly SULT1A1, is a major pathway for drug metabolism in humans. Pharmacogenetic studies of SULT1A1 began over a quarter of a century ago and have advanced from biochemical genetic experiments to include cDNA and gene cloning, gene resequencing, and functional studies of the effects of single nucleotide polymorphisms (SNPs). SNP genotyping, in turn, led to the discovery of functionally important copy number variations (CNVs) in the SULT1A1 gene. This review will briefly describe the evolution of our understanding of SULT1A1 pharmacogenetics and CNV, as well as challenges involved in utilizing both SNP and CNV data in an attempt to predict SULT1A1 function. SULT1A1 represents one example of the potential importance of CNV for the evolving disciplines of pharmacogenetics and pharmacogenomics.
Introduction
Human cytosolic sulfotransferase (SULT) enzymes catalyze the sulfate conjugation of many drugs, other xenobiotics, neurotransmitters, and hormones – especially steroid hormones. 3′-Phosphoadenosine 5′-phosphosulfate (PAPS) is the sulfate donor for reactions catalyzed by SULTs (Weinshilboum and Otterness, 1994; Falany, 1997). These reactions typically result in increased water solubility and enhanced urinary excretion of the conjugated substrate (Glatt and Meinl, 2004). In addition to the biological inactivation of substrates, the SO3– transferred by SULTs can also serve as a leaving group, resulting in the formation of an active, potentially toxic or carcinogenic metabolite (Miller and Surh, 1994; Glatt, 2000a; Glatt and Meinl, 2004). Therefore, SULT enzymes play an important role in drug metabolism, in toxicology, and in chemical carcinogenesis. As a result, variation in the ability to catalyze these reactions could potentially result in altered drug efficacy as well as risk for adverse drug reactions or carcinogenesis.
Drug metabolism is often separated into Phase I (oxidation, reduction, and hydrolysis) and Phase II (conjugation) reactions (Weinshilboum, 2003). The role of copy number variations (CNVs) in the pharmacogenetics of a major class of enzymes that catalyze Phase I reactions, the cytochromes P450 (CYPs), is addressed elsewhere in this volume (Johansson and Ingelman-Sundberg, 2008). The SULT family of Phase II conjugating enzymes has, like the CYPs, been the subject of intense pharmacogenetic studies (Glatt, 2000b; Glatt et al., 2001; Coughtrie, 2002; Glatt and Meinl, 2004; Weinshilboum and Adjei, 2005; Nowell and Falany, 2006). They were designed to determine the contribution of inheritance to individual differences in response to drugs that undergo sulfation as well as the possible role of inherited variation in sulfate conjugation in disease risk. As described in subsequent paragraphs, sulfation pharmacogenetics initially involved studies of single nucleotide polymorphisms (SNPs), but we now know that we must also take copy number variations into consideration (Hebbring et al., 2007). The evolution of our understanding of SULT pharmacogenetics – particularly pharmacogenetics of the important drug- and hormone-metabolizing enzyme SULT1A1 – illustrates the importance of including CNVs as a potential pharmacogenetic mechanism. This review will briefly describe the human SULT gene family, including the SULT1A subfamily that includes SULT1A1. The focus will then be placed on SULT1A1, a widely expressed isoform that plays an important role in the biotransformation of a large number of drugs and steroid hormones, especially estrogens. SULT1A1 represents an excellent example of the challenges and the importance for pharmacogenetics of unifying SNP data, CNV data, and function.
The human SULT gene family
The human cytosolic SULT gene family includes 13 members (Table 1). These genes map to clusters on chromosomes 2, 4, 16, and 19, plus two single genes on chromosomes 2 and 22 (Blanchard et al., 2004; Freimuth et al., 2004). The chromosome 2 cluster includes SULT1C2, 1C3, and 1C4, all located within a 150-kb region at 2q12.3. Proteins encoded by SULT1C family members have been shown to catalyze the metabolic ‘activation’ of procarcinogens such as N-hydroxyl-2-acetylaminofluorene to form potent carcinogens (Glatt et al., 2000). The chromosome 4 SULT genes include SULT1B1, 1E1, and 1D1. These genes map to a 110-kb cluster at 4q13.3. SULT1E1 was previously referred to as estrogen sulfotransferase since this isoform has the highest affinity for estrogens of any member of the SULT family (Adjei and Weinshilboum, 2002). However, SULT1A1 may play a more important role in overall estrogen sulfate conjugation than does SULT1E1 because of its ubiquitous expression. SULT1B1 has been reported to contribute to thyroid hormone sulfate conjugation (Wang et al., 1998), although there is evidence that other SULT isoforms can also catalyze that reaction (Glatt and Meinl, 2004). Genes encoding the two members of the human SULT2 subfamily map to chromosome 19. These genes encode enzymes that metabolize steroids such as dehydroepiandrosterone, pregnenolone, and cholesterol (Falany, 1997). Because of their possible pharmacogenetic importance, many of the human SULT genes have been subjected to comprehensive gene resequencing, followed by functional genomic studies (Her et al., 1996; Raftogianis et al., 1997, 1999; Freimuth et al., 2001; Thomae et al., 2002, 2003; Adjei et al., 2003; Hildebrandt et al., 2004; Ji et al., 2007) (Table 1). The most intensively studied SULT genes are those in the SULT1A subfamily within a gene cluster at chromosome 16p11.2. SULT1A1, 1A2, 1A3, and 1A4 map to an area on the short arm of chromosome 16 in a region rich in repetitive sequences and segmental duplications (Hildebrandt et al., 2004; Bradley and Benner, 2005). The four enzymes encoded by these genes are highly homologous in their amino acid sequences. For example, SULT1A3 and 1A4 are identical in sequence, SULT1A1 and 1A2 are 96% identical, and SULT1A1 and 1A3/1A4 are 93% identical in amino acid sequence (Raftogianis et al., 1996; Hildebrandt et al., 2004). SULT1A1 is widely expressed in a variety of human tissues and plays an important role in the biotransformation of a large number of drugs and of estrogen hormones (Adjei and Weinshilboum, 2002; Coughtrie, 2002; Glatt and Meinl, 2004), but with lower affinity than SULT1E1 (Adjei et al., 2003). SULT1A2 is not widely expressed (Coughtrie, 2002; Nowell et al., 2005), while SULT1A3 and 1A4, both of which encode and transcribe active proteins (Wood et al., 1994; Hildebrandt et al., 2004), catalyze the biotransformation of catecholamine neurotransmitters such as dopamine and norepinephrine (Glatt and Meinl, 2004). The structures of SULT1A3 and 1A4 illustrate the fact that large segmental duplications occur in this area of chromosome 16. These genes, with virtually identical sequences, are contained within two 146-kb duplicated segments, separated by 728 kb of DNA (Hildebrandt et al., 2004). Because it is so widely expressed and because it plays such a critical role in drug metabolism, SULT1A1 has been the most intensively studied of the SULT isoforms, both from a biochemical and a pharmacogenetic perspective.
Table 1.
List of the 13 human SULT genes with chromosomal band and physical locations indicated
| Gene | Chromosome band | Physical location |
|---|---|---|
| SULT1C2a | 2q12.3 | 108271527–108292803 |
| SULT1C3 | 2q12.3 | 108230083–108248239 |
| SULT1C4 | 2q12.3 | 108360853–108370702 |
| SULT6B1 | 2p22.2 | 37248467–37269 194 |
| SULT1B1 | 4q13.3 | 70627275–70661019 |
| SULT1E1a | 4q13.3 | 70741519–70760459 |
| SULT1A1a | 16p11.2 | 28524419–28528858 |
| SULT1A2a | 16p11.2 | 28510767–28515302 |
| SULT1A3a | 16p11.2 | 29378695–29383802 |
| SULT1A4a | 16p11.2 | 30118037–30123121 |
| SULT2A1a | 19q13.32 | 53065682–53081405 |
| SULT2B1a | 19q13.32 | 53747241–53794496 |
| SULT4A1 | 22q13.2 | 42551720–42589711 |
Gene has undergone resequencing studies designed to identify and functionally characterize common polymorphisms (Her et al., 1996; Raftogianis et al., 1997, 1999; Freimuth et al, 2001; Thomae et al, 2002; Adjei et al, 2003; Thomae et al, 2003; Hildebrandt et al., 2004; Ji et al., 2007).
SULT1A1 pharmacogenetics
Pharmacogenetic studies of SULT1A1 focused initially on SNPs, both non-synonymous and functional promoter SNPs of the gene that influence transcription (Prondzinski et al., 2003; Buckland et al., 2005; Hebbring et al., 2007). The association of those SNPs with function, although clearly defined during in vitrostudies (Prondzinski et al., 2003), did not always correlate well with in vivofunction (Raftogianis et al., 1997; Glatt and Meinl, 2004). Therefore, the recent discovery of functionally important SULT1A1 CNVs (Hebbring et al., 2007) – when combined with SNP data – may enhance our understanding of SULT1A1 pharmacogenetics. Pharmacogenetic studies of SULT1A1 also illustrate, as described subsequently, the practical difficulty of interpreting SNP genotypes in conjunction with CNV data. Subsequent paragraphs will briefly describe the discovery and characterization of SULT1A1 SNPs and their role in the ensuing discovery of CNVs for this important gene, followed by a description of attempts to interpret the functional effects of SULT1A1 SNPs together with CNVs to help make it possible to correlate genotypic data with function.
Studies of the genetic regulation of human SULT1A1 began long before either the cDNA or gene encoding this enzyme had been cloned and characterized. Biochemical genetic studies performed in the 1980s using human blood platelets (an easily obtained tissue in which the enzyme is expressed) determined that inheritance played a major role in the variation of two phenotypes, level of enzyme activity and enzyme thermal stability (Van Loon and Weinshilboum, 1984; Campbell et al., 1987; Anderson et al., 1988; Price et al., 1989). The decade of the 1990s witnessed the cloning and characterization of the human SULT1A1 cDNA and gene (Wood et al., 1994; Aksoy and Weinshilboum, 1995; Raftogianis et al., 1996), followed quickly by resequencing of the gene's coding region using 150 DNA samples from Caucasian subjects (Raftogianis et al., 1997). Two common non-synonymous SNPs were observed, SULT1A1∗2(rs9282861, G638A, Arg213His) and SULT1A1∗3 (rs1801030, A667G, Met223Val). The SULT1A1∗2 variant was the most common of the two, with a minor allele frequency (MAF) of 31–33% in Caucasian subjects while SULT1A1∗3 had a MAF of ∼1% in these subjects (Carlini et al., 2001). As we have now learned to expect, there were striking differences among ethnic groups in allele frequencies, with SULT1A1∗2 having a frequency of only 8% and SULT1A1∗3 afrequency of 0.6% in Han Chinese subjects. However, SULT1A1∗3 had a frequency of 23% in African-American subjects (Carlini et al., 2001). We will see subsequently that ethnic variation in frequency also occurs with SULT1A1 CNVs. The SULT1A1∗2 allele was associated with a substantial decrease in levels of both enzyme activity and thermostability (Raftogianis et al., 1997) as a result, in part, of altered degradation of that variant allozyme (Raftogianis et al., 1999). Subsequently, two SNPs in the promoter of SULT1A1 were reported to influence transcription (Prondzinski et al., 2003; Hebbring et al., 2007). Therefore, three SNPs, 1A1∗2 (rs9282861) and two promoter SNPs (rs3760091 and rs750155), have been the subject of a series of molecular epidemiology studies that often resulted in equivocal or inconclusive results (Glatt and Meinl, 2004). However, it was observations made in the course of genotyping those three SNPs which led to the discovery that SULT1A1 also displayed functionally significant CNVs.
SULT1A1 CNVs
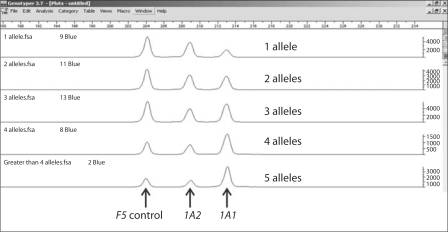
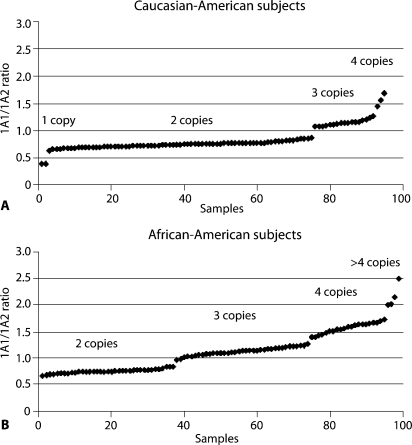
When SULT1A1 genotyping was performed using the semi-quantitative pyrosequencing assay, an unusual peak distribution for the SULT1A1∗2 polymorphism was observed. After confounding artifacts were eliminated as the cause, it seemed possible that these observations might be due to CNVs. Therefore, a quantitative fluorescent-based PCR assay was designed in which both SULT1A2 and the Coagulation Factor V (F5) genes were used as controls to which SULT1A1 gene amplification could be compared. When that assay was applied, the results were compatible with the conclusion that one to five copies of SULT1A1 were present in individual DNA samples (Fig. 1). This PCR-based assay was then used to study a series of DNA samples obtained from human liver surgical biopsy samples as well as from the Coriell Cell Repository. The results of those studies were also compatible with the existence of SULT1A1 CNVs, with striking frequency differences between Caucasian-American (CA) and African-American (AA) subjects (Fig. 2). For example, 4.7% of CA subjects appeared to have only one copy of SULT1A1 in their genome, while 25.7% had three or more copies. However, none of the 100 AA subjects studied had only a single copy of the gene, but 62.6% had three or more.
Fig. 1.
Quantitative fluorescent-based PCR assay. The figure shows peaks for the controls, Coagulation Factor V (F5) and SULT1A2, as well as peaks for 1–5 copies of SULT1A1 (Hebbring et al., 2007, with the permission of Human Molecular Genetics).
Fig. 2.
Peak height ratios of SULT1A1 to SULT1A2 for 100 DNA samples from both (A) Caucasian-American and (B) African-American subjects graphed to indicate SULT1A1 copy numbers (Hebbring et al., 2007, with the permission of Human Molecular Genetics).
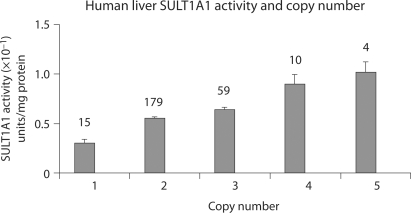
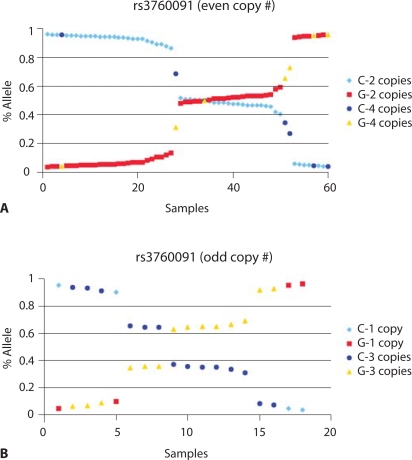
Furthermore, when SULT1A1 copy number was correlated with the results of a SULT1A1-specific radiochemical enzymatic assay performed using cell lysates from 267 human liver samples, CNV correlated well with enzyme activity (Fig. 3). However, the functional SNPs that had been assayed in a large number of molecular epidemiology studies over the past decade (over 100 such studies were listed by PubMed after search with the term ‘SULT1A1 polymorphism’) showed little correlation when CNV was taken into consideration (Hebbring et al., 2007). In summary, this series of experiments suggested that both SNP genotype and CNV need to be determined in order to correlate SULT1A1 genotype with clinically and biologically relevant phenotypes. However, assessing SULT1A1 SNP genotype presents a challenge in the presence of CNVs. That is true because many SNP genotyping methods can not reliably detect an imbalance between two SNP alleles. When those techniques are used, it can be determined whether a sample is homozygous or heterozygous, but when more than two alleles are present, the number of each allele can not be resolved. In order to assay for allele ‘dosage’, techniques such as pyrosequencing are required to determine the ratio between signals from the two alleles. For example, when pyrosequencing is utilized, allele ratios can be used, together with copy number genotype, to determine how many copies of each allele are present. That can be accomplished by plotting ‘percent signal’ for each allele, separated into groups on the basis of copy number. Plots of the data can then be analyzed to assess the number of copies of each allele present. For example, if a sample has a copy number of three, and the pyrosequencing peak height ratio is 66.6% G and 33.4% C, that sample would have two alleles with G and one allele with C at the locus of interest. In our experience, it is easiest to assess SNP allele dosage if the SNP data are grouped by odd or even copy numbers, as shown in Fig. 4. In that figure, the ‘even copy number’ plot shows that alleles with either two or four copies are identical with regard to SNP genotype when half of the alleles are of one type and half of the other, i.e., ∼0.5 on the ‘% allele’ axis (1 of 2 or 2 of 4). Furthermore, compound heterozygotes (1 of 4) for subjects carrying four copies can be easily identified with this type of plot, i.e., they are all at either 0.25 or 0.75 on the Y-axis. Perhaps even more striking, compound heterozygotes can be easily separated in plots for samples that carry an odd number of SULT1A1 copies (one or three copies). This type of visual depiction makes it possible to easily add SNP allele ‘calls’ to copy number data (Fig. 4B). When we used this approach to genotype DNA samples from 761 randomly selected Caucasian women for the three common SULT1A1 functional SNPs (two in the gene promoter and the SULT1A1∗2 non-synonymous SNP), we observed every possible combination of SNP genotypes for the duplicated alleles (Table 2). This observation contrasts with the situation for the functionally and clinically relevant cytochrome P450 gene CYP2D6, a gene that has also undergone deletion and duplication in the course of evolution (Johansson et al., 1993; Dalen et al., 1998; Johansson and Ingelman-Sundberg, 2008). When multiple copies of CYP2D6 are present, they generally involve only a single duplicated allele, not the variety of combinations that we observed for SULT1A1 (Table 2). This difference probably results from different mechanisms by which CNVs evolved.
Fig. 3.
Plot of SULT1A1 enzymatic activity in 267 human liver biopsy samples as a function of SULT1A1 copy number. The values above the bar represent the number of samples tested in each group (Hebbring et al., 2007, with the permission of Human Molecular Genetics).
Fig. 4.
Plot of SULT1A1 SNP data sorted by ‘% allele’ from pyrosequencing data for each sample. (A) Samples having an even number of copies of SULT1A1 (2 and 4). (B) Samples having an odd number of copies (1 and 3).
Table 2.
The table lists genotype frequencies (in %) for the two SULT1A1 promoter SNPs (rs3760091 and rs750155), and the non-synonymous SNP (Arg213His, rs9282861) with respect to copy number in 761 randomly selected Caucasian women. The ‘A’ allele represents C for all SNPs, while ‘B’ represents the Gallele in rs3760091 and T in rs750155 and rs9282861.
| Genotype |
SULT1A1 promoter SNPs |
SULT1A1∗2 |
|
|---|---|---|---|
| rs3760091 | rs750155 | rs9282861 | |
| A | 1.8 | 2.1 | 2.7 |
| B | 1.8 | 1.6 | 1.2 |
| AA | 24.9 | 16.0 | 22.3 |
| AB | 31.9 | 32.3 | 31.0 |
| BB | 7.7 | 15.8 | 11.0 |
| AAA | 5.4 | 3.6 | 12.7 |
| AAB | 7.0 | 9.2 | 9.4 |
| BAA | 8.8 | 8.8 | 2.8 |
| BBB | 4.2 | 3.9 | 0.4 |
| AAAA | 0.9 | 0.7 | 3.6 |
| AAAB | 1.2 | 1.7 | 1.3 |
| AABB | 1.8 | 1.9 | 1.3 |
| BAAA | 1.6 | 1.5 | 0.0 |
| BBBB | 0.9 | 1.1 | 0.3 |
Conclusion
Pharmacogenetics has been heralded as one of the major applications of genomics to help ‘individualize’ 21st century medicine. The initial focus of pharmacogenetics has understandably been on SNPs. However, the examples provided by SULT1A1 and CYP2D6 (Johansson and Ingelman-Sundberg, 2008) make it clear that both SNPs and CNVs may need to be taken into account to successfully link genotypic variation to variation in drug response phenotypes. Once techniques are developed that make this process efficient, accurate, and cost effective, there can be little doubt that both the ‘discovery phase’ of pharmacogenetics and pharmacogenetic clinical assays will – when appropriate – routinely obtain both CNV data and SNP genotyping, as described here for SULT1A1, to help make it possible to better individualize drug therapy.
Footnotes
This work is supported, in part, by NIH grants R01 GM28157, R01 GM35720, R01 CA132780, U01 GM61388 (The Pharmacogenetics Research Network), and a PhRMA Foundation ‘Center of Excellence in Clinical Pharmacology’ Award.
References
- Adjei AA, Weinshilboum RM. Catecholestrogen sulfation: possible role in carcinogenesis. Biochem Biophys Res Commun. 2002;292:402–408. doi: 10.1006/bbrc.2002.6658. [DOI] [PubMed] [Google Scholar]
- Adjei AA, Thomae BA, Prondzinski JL, Eckloff BW, Wieben ED, Weinshilboum RM. Human estrogen sulfotransferase (SULT1E1) pharmacogenomics: gene resequencing and functional genomics. Br J Pharmacol. 2003;139:1373–1382. doi: 10.1038/sj.bjp.0705369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy IA, Weinshilboum RM. Human thermolabile phenol sulfotransferase gene (STM): molecular cloning and structural characterization. Biochem Biophys Res Commun. 1995;208:786–795. doi: 10.1006/bbrc.1995.1406. [DOI] [PubMed] [Google Scholar]
- Anderson RJ, Jackson BL, Liebentritt DK. Human platelet thermostable phenol sulfotransferase from blacks and whites: biochemical properties and variations in thermal stability. J Lab Clin Med. 1988;112:773–783. [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- Bradley ME, Benner SA. Phylogenomic approaches to common problems encountered in the analysis of low copy repeats: the sulfotransferase 1a gene family example. BMC Evol Biol. 2005;5:22. doi: 10.1186/1471-2148-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O'Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- Campbell NR, Van Loon JA, Weinshilboum RM. Human liver phenol sulfotransferase: assay conditions, biochemical properties and partial purification of isozymes of the thermostable form. Biochem Pharmacol. 1987;36:1435–1446. doi: 10.1016/0006-2952(87)90108-0. [DOI] [PubMed] [Google Scholar]
- Carlini EJ, Raftogianis RB, Wood TC, Jin F, Zheng W, et al. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics. 2001;11:57–68. doi: 10.1097/00008571-200102000-00007. [DOI] [PubMed] [Google Scholar]
- Coughtrie MW. Sulfation through the looking glass – recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- Dalen P, Dahl ML, Bernal Ruiz ML, Nordin J, Bertilsson L. 10-hydroxylation of nortriptyline in white persons with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin Pharmacol Ther. 1998;63:444–452. doi: 10.1016/S0009-9236(98)90040-6. [DOI] [PubMed] [Google Scholar]
- Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- Freimuth RR, Eckloff B, Wieben ED, Weinshilboum RM. Human sulfotransferase SULT1C1 pharmacogenetics: gene resequencing and functional genomic studies. Pharmacogenetics. 2001;11:747–756. doi: 10.1097/00008571-200112000-00002. [DOI] [PubMed] [Google Scholar]
- Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J. 2004;4:54–65. doi: 10.1038/sj.tpj.6500223. [DOI] [PubMed] [Google Scholar]
- Glatt HR. An overview of bioactivation of chemical carcinogens. Biochem Soc Trans. 2000a;28:1–6. doi: 10.1042/bst0280001. [DOI] [PubMed] [Google Scholar]
- Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000b;129:141–170. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- Glatt H, Meinl W. Pharmacogenetics of soluble sulfotransferases (SULTS) Naunyn Schmiedebergs Arch Pharmacol. 2004;369:55–68. doi: 10.1007/s00210-003-0826-0. [DOI] [PubMed] [Google Scholar]
- Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, et al. Sulfotransferases: genetics and role in toxicology. Toxicol Lett. 2000;112–113:341–348. doi: 10.1016/s0378-4274(99)00214-3. [DOI] [PubMed] [Google Scholar]
- Glatt H, Boeing H, Engelke CE, Ma L, Kuhlow A, et al. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat Res. 2001;482:27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- Hebbring SJ, Adjei AA, Baer JL, Jenkins GD, Zhang J, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16:463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- Her C, Raftogianis R, Weinshilboum RM. Human phenol sulfotransferase STP2 gene: molecular cloning, structural characterization, and chromosomal localization. Genomics. 1996;33:409–420. doi: 10.1006/geno.1996.0216. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Salavaggione OE, Martin YN, Flynn HC, Jalal S, et al. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. doi: 10.1016/j.bbrc.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Ji Y, Moon I, Zlatkovic J, Salavaggione OE, Thomae BA, et al. Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. J Pharmacol Exp Ther. 2007;322:529–540. doi: 10.1124/jpet.107.122895. [DOI] [PubMed] [Google Scholar]
- Johansson I, Ingelman-Sundberg M. CNVs of human genes and their implication in pharmacogenetics. Cytogenet Genome Res. 2008;123:195–204. doi: 10.1159/000184709. [DOI] [PubMed] [Google Scholar]
- Johansson I, Lundqvist E, Bertilsson L, Dahl ML, Sjoqvist F, Ingelman-Sundberg M. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci USA. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Surh YJ. Sulfonation in chemical carcinogenesis. In: Kauffman FC, editor. Handbook of Pharmacology Vol. 112, Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. New York: Springer; 1994. pp. 429–458. [Google Scholar]
- Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- Nowell S, Green B, Tang YM, Wiese R, Kadlubar FF. Examination of human tissue cytosols for expression of sulfotransferase isoform 1A2 (SULT1A2) using a SULT1A2-specific antibody. Mol Pharmacol. 2005;67:394–399. doi: 10.1124/mol.104.006171. [DOI] [PubMed] [Google Scholar]
- Price RA, Spielman RS, Lucena AL, Van Loon JA, Maidak BL, Weinshilboum RM. Genetic polymorphism for human platelet thermostable phenol sulfotransferase (TS PST) activity. Genetics. 1989;122:905–914. doi: 10.1093/genetics/122.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prondzinski J, Thomae B, Wang L, Eckloff B, Wieben E, Weinshilboum R. Sulfotransferase (SULT) 1A1 pharmacogenetics: functional 5′-flanking region (5′-FR) polymorphisms. Clin Pharmacol Ther. 2003;73:P77. [Google Scholar]
- Raftogianis RB, Her C, Weinshilboum RM. Human phenol sulfotransferase pharmacogenetics: STP1 gene cloning and structural characterization. Pharmacogenetics. 1996;6:473–487. doi: 10.1097/00008571-199612000-00001. [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun. 1997;239:298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem Pharmacol. 1999;58:605–616. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- Thomae BA, Eckloff BW, Freimuth RR, Wieben ED, Weinshilboum RM. Human sulfotransferase SULT2A1 pharmacogenetics: genotype-to-phenotype studies. Pharmacogenomics J. 2002;2:48–56. doi: 10.1038/sj.tpj.6500089. [DOI] [PubMed] [Google Scholar]
- Thomae BA, Rifki OF, Theobald MA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catecholamine sulfotransferase (SULT1A3) pharmacogenetics: functional genetic polymorphism. J Neurochem. 2003;87:809–819. doi: 10.1046/j.1471-4159.2003.02027.x. [DOI] [PubMed] [Google Scholar]
- Van Loon J, Weinshilboum RM. Human platelet phenol sulfotransferase: familial variation in thermal stability of the TS form. Biochem Genet. 1984;22:997–1014. doi: 10.1007/BF00499627. [DOI] [PubMed] [Google Scholar]
- Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol Pharmacol. 1998;53:274–282. doi: 10.1124/mol.53.2.274. [DOI] [PubMed] [Google Scholar]
- Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Adjei AA. Sulfate conjugation: pharmacogenetics and pharmacogenomics. In: Pacific GM, Coughtrie MWH, editors. Human Cytosolic Sulfotransferases. Boca Raton: CRC Press; 2005. pp. 61–78. [Google Scholar]
- Weinshilboum RM, Otterness DM. Conjugation-deconjugation reactions in drug metabolism and toxicity. In: Kauffman FC, editor. Handbook of Experimental Pharmacology Series. New York: Springer; 1994. p. 33. [Google Scholar]
- Wood TC, Aksoy IA, Aksoy S, Weinshilboum RM. Human liver thermolabile phenol sulfotransferase: cDNA cloning, expression and characterization. Biochem Biophys Res Commun. 1994;198:1119–1127. doi: 10.1006/bbrc.1994.1159. [DOI] [PubMed] [Google Scholar]