Abstract
Recent large-scale genomic studies within human populations have identified numerous genomic regions as copy number variant (CNV). As these CNV regions often overlap coding regions of the genome, large lists of potentially copy number polymorphic genes have been produced that are candidates for disease association. Most of the current data regarding normal genic variation, however, has been generated using BAC or SNP microarrays, which lack precision especially with respect to exons. To address this, we assessed 2,790 candidate CNV genes defined from available studies in nine well-characterized HapMap individuals by designing a customized oligonucleotide microarray targeted specifically to exons. Using exon array comparative genomic hybridization (aCGH), we detected 255 (9%) of the candidates as true CNVs including 134 with evidence of variation over the entire gene. Individuals differed in copy number from the control by an average of 100 gene loci. Both partial- and whole-gene CNVs were strongly associated with segmental duplications (55 and 71%, respectively) as well as regions of positive selection. We confirmed 37% of the whole-gene CNVs using the fosmid end sequence pair (ESP) structural variation map for these same individuals. If we modify the end sequence pair mapping strategy to include low-sequence identity ESPs (98–99.5%) and ESPs with an everted orientation, we can capture 82% of the missed genes leading to more complete ascertainment of structural variation within duplicated genes. Our results indicate that segmental duplications are the source of the majority of full-length copy number polymorphic genes, most of the variant genes are organized as tandem duplications, and a significant fraction of these genes will represent paralogs with levels of sequence diversity beyond thresholds of allelic variation. In addition, these data provide a targeted set of CNV genes enriched for regions likely to be associated with human phenotypic differences due to copy number changes and present a source of copy number responsive oligonucleotide probes for future association studies.
Copy number variation within genes has long been recognized as an important force in human evolution and disease. Prior to the advent of whole-genome based approaches, many examples of gene CNVs had been described within the human population due to their association with phenotype and disease, including RHD (Colin et al., 1991), opsins (Nathans et al., 1986), olfactory receptors (Trask et al., 1998), CYP2D6 (Heim and Meyer, 1992), amylase (Groot et al., 1989), complement C4 (Schneider et al., 1986) and HLA-DR loci (Zhang et al., 1990). Many of these genes are postulated to have played important roles in human adaptation to changing environmental conditions and infectious pathogens. Early FISH experiments also demonstrated the presence of large-scale CNV regions near telomeres at clusters of olfactory receptors (Trask et al., 1998) and within pericentromeric regions (Ritchie et al., 1998; Barber et al., 1999). Targeted studies of individual euchromatic regions also provided insight into the complexity of gene copy number variation including CCL3L1-CCL4L1 (Townson et al., 2002) and beta defensin loci (Hollox et al., 2003). Additional examples of genetic copy number variation were detected as sequence misassemblies during the Human Genome Project but were initially triaged or flagged as gaps in an effort to generate a single, consistent reference genome (Eichler et al., 2004).
Two initial genome-wide studies (Iafrate et al., 2004; Sebat et al., 2004), followed by a series of large-scale scans for CNVs (de Vries et al., 2005; Sharp et al., 2005; Tuzun et al., 2005; Conrad et al., 2006; Hinds et al., 2006; Locke et al., 2006; McCarroll et al., 2006) reignited interest in structural variation of our genome. As no single detection method is currently capable of capturing the full spectrum of copy number variation these studies have been complementary – each adding to our understanding and contributing to an ever-growing catalog of CNV regions and potential CNV genes. Currently, the Database of Genomic Variants catalogs 29% (863 Mb) of genomic sequence as involved in CNVs >1 kb (Iafrate et al., 2004). Not surprisingly, it has been predicted that many of the thousands of genes within this sizable portion of the genome may themselves be CNV and that these gene CNVs may explain a significant fraction of human phenotypic variation. The significance of copy number variation in human disease is supported by the growing number of disease associations detected through targeted assessments of individual genes (Gonzalez et al., 2005; Hollox et al., 2008) and whole-genome association studies (Marshall et al., 2008; Walsh et al., 2008).
Nonetheless, our current view of copy number variation within genes lacks precision. The vast majority of CNV regions remain ill-defined at the sequence level (Cooper et al., 2007; Kidd et al., 2008). Both sequence- and microarray-based techniques have limitations. End sequence pair (ESP) approaches are dependent upon detecting length and orientation discrepancies after alignment of end sequences against the reference genome (Tuzun et al., 2005; Korbel et al., 2007; Kidd et al., 2008). Such approaches have reduced power to detect copy number variation in duplicated regions of near perfect identity because end sequences cannot be unambiguously placed. In addition, the insert size of the clone library introduces theoretical size limitations for the detection of insertions that can be partially overcome with modified analysis techniques (Cooper et al., 2008). Similarly, aCGH-based approaches are dependent upon the spacing of probes (i.e. oligonucleotides) and/or the nature of the molecules used on the microarray. CNV regions detected by BAC-based aCGH, for example, may be due to a partial loss or gain of sequence represented by the clone and therefore the span of the clone does not necessarily reflect the boundaries of the CNV or the involvement of an underlying gene. CNV regions detected by widely spaced oligonucleotide probes do not necessarily imply that genes within the intervening regions are truly CNVs. Although sequence-based approaches will ultimately dominate, it is currently impossible to rapidly ascertain the full spectrum of CNVs at the nucleotide level on a genome-wide scale for thousands of individuals. With a catalog of CNV regions covering almost one-third of the genome and the prediction that many of these boundaries are inflated (4–10 fold) (Kidd et al., 2008), it is important to determine how many underlying genes are truly CNV. CNV regions appear to be enriched for genes within segmental duplications (SDs) and under positive selection, but it is important to confirm these biases on a validated set of CNV genes (Nguyen et al., 2006; Cooper et al., 2007). Such an assessment will allow us to focus on the variants most likely to have biological implications through changes in gene dosage or function, and through future studies judge their relative contribution to human phenotypic variation and disease. Towards this end we have systematically identified candidate genes underlying CNVs in a small number of normal individuals and experimentally validated gene CNVs at the resolution of the individual exon using an oligonucleotide aCGH platform. In addition, based on these cross-platform comparisons, we further refine ESP-based approaches to capture a greater fraction of copy number polymorphic genes within duplicated regions of the genome.
Methods
Individuals assayed
We assessed nine individuals from the HapMap project (GM12878, GM12156, GM18502, GM18507, GM18517, GM18555, GM18956, GM19129 and GM19240) that have been extensively analyzed for CNVs (Conrad et al., 2006; Locke et al., 2006; McCarroll et al., 2006; Redon et al., 2006). All but GM18502 have undergone fosmid ESP sequencing and analysis (Kidd et al., 2008). The control genome for comparative hybridization (G248, aka NA15510) represents a female individual for which CNVs detected by fosmid ESP mapping have been published (Tuzun et al., 2005).
CNV candidate regions
We collected all available copy number variation data (as of February 2007) mapped against build35 of the human genome (http://genome.ucsc.edu). When possible, results were collected and analyzed at the clone and individual levels by examining both the published primary and supplemental data. In total we cataloged 4,083 overlapping CNV positive regions spanning 289,437,161 bases of the genome. We limited our analysis to CNV regions ≥1 kb and <1000 kb as these were large enough to avoid most retroelements and affect entire exons. CNV sites originated primarily from BAC aCGH and SNP-based methods and included:
708 redundant regions at the individual level from whole-genome BAC tiling path aCGH and 921 redundant regions detected by loss of heterozygosity within SNP arrays; we included only regions found within our study individuals (Redon et al., 2006)
927 BACs detected by whole-genome BAC tiling path aCGH; we included only BACs with CNV detected in >2 individuals to reduce false positives (Wong et al., 2007)
599 redundant regions detected by Mendelian inconsistencies within SNP arrays (Conrad et al., 2006)
538 regions detected by Mendelian failures and null phenotypes within SNP arrays (McCarroll et al., 2006)
410 BACs detected by duplication-targeted BAC aCGH (Sharp et al., 2005; Locke et al., 2006)
253 non-redundant BAC regions detected by discontinuous BAC tiling path aCGH (Iafrate et al., 2004)
236 non-redundant regions detected by fosmid end sequence pairs within our control individual G248 as compared to human reference genome (Tuzun et al., 2004)
73 non-redundant regions detected by discontinuous ROMA tiling (Sebat et al., 2004)
63 regions detected by SNP-based methodology and confirmed by targeted high-density oligonucleotide array (Hinds et al., 2006)
47 non-redundant regions detected by whole-genome BAC aCGH (de Vries et al., 2005).
Reference gene set
To compile a non-redundant set of transcribed loci, we started with the curated NCBI RefSeq mRNAs (build21: Jan 6, 2007) as placed on the human reference genome (build35: hg17) within the UCSC browser (http://genome.ucsc.edu). The NCBI RefSeq dataset is a highly curated set of reference mRNAs that contains splicing isoforms and is redundantly placed at allelic levels of variation upon the genome in the UCSC browser. The total 24,345 unique RefSeq IDs had 24,744 placements and represented 18,397 genes. Rather than discard alternatively spliced exons, we identified and collapsed the sequence of overlapping isoforms into a non-redundant set of genomic coordinates incorporating all possible exons. Among the 18,666 remaining gene structures were instances of overlapping genes, particularly in duplicated regions. To remove these redundancies, we checked for gene overlap at the exon level and, if found, we kept only a single gene based on the greater percent genomic identity, size of transcript, and level of transcript validation. In the event of equally best placements ≥99.5% identity, a gene was assigned to multiple locations as long as another gene did not overlap. This process resulted in a final, non-overlapping set of 18,373 gene loci.
CNV gene set
Our goal was to maximize the inclusion of putative CNV genes that would be present in our nine HapMap individuals or G248 control. Putative CNV genes were identified by a two-pronged approach by including genes overlapping frequent CNVs – detected multiple times within one study or across studies – or CNVs specific to our study individuals. We also included any gene if a variant had been specifically validated. This approach allowed us to maximize the inclusion of common gene CNVs within our individuals, while minimizing the number of genes studied, and thereby, our false positive rates. Our control set consisted of 80 unique genes (928 exons) that lacked evidence for copy number variation based on high-density targeted oligonucleotide CNV (Wong et al., 2007). Initial positive controls were collected from known variant genes that had multiple methodologies and literature validation. The expected individual copy number differences relative to control were determined through analyses of the combined fosmid ESP data (Tuzun et al., 2004; Kidd et al., 2008).
Exon microarray design
From our identified list of candidate CNV genes and controls, we targeted an approximate equal number of probes to each exon by including nearly equivalent amounts of sequence for probe design (Fig. 1). This scheme essentially increased the weight of small exons with the inclusion of flanking sequence and decreased the weight of large exons by only sampling a limited portion. The inclusion of flanking non-transcribed sequence also helped to limit the detection of processed pseudogenes. Overall each of the exons for the candidate genes included in our design was represented by between 203–600 bases of sequence. These probe design regions were merged into a non-overlapping set of sequence from which NimbleGen Systems algorithms selected appropriate isothermal oligonucleotides (without regard to probe copy number). Probes were spaced on average every 20 bases at NimbleGen Systems resulting in an array containing 380,002 probes (averaging 12 probes per targeted exon). Chips manufactured from this design were hybridized with given individuals and control using standard conditions (Selzer et al., 2005). Analysis was performed on the resulting normalized hybridization datasets provided by NimbleGen. All hybridization data is available at the supplemental web site (http://humanparalogy.gs.washington.edu/geneCNVs).
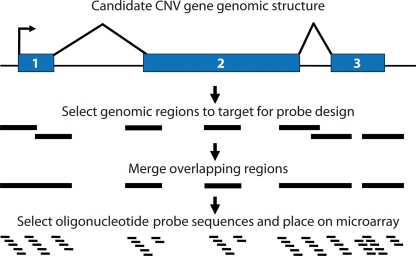
Fig. 1.
Exon-targeted oligonucleotide array CGH design. From our identified list of candidate CNV genes and controls, we targeted an equal number of probes to each exon by including nearly equivalent amounts of sequence for probe design. For each exon, we identified two regions for probe design: 200 bp centered at the beginning and 200 bp centered at the end of the exon. For small exons (<200 bp) this amounted to 100 bp flanking either side plus the length of the exon since these regions overlapped. For medium size exons (200–999 bp) this amounted to 400 bp with equivalent amounts of flanking and exonic sequence. For large exons (≥1 kb), we added an additional 200 bp directly in the center of the exon to provide a measure of continuity in these larger regions. This scheme essentially increased the weight of small exons with the inclusion of flanking sequence and decreased the weight of large exons by only sampling a limited portion. The inclusion of flanking non-transcribed sequence also limited the detection of processed pseudogenes. Overall each of the exons for the candidate genes were represented by 203–600 bases of sequence. These probe design regions were merged into a non-overlapping set of sequence from which NimbleGen algorithms choose appropriate oligonucleotide sequences for array synthesis.
Detection algorithm
Our detection strategy was developed as a simple threshold-per-exon test with chaining of exons to predict the extent of the variation. The thresholds for each individual were calculated based on multiples of the standard deviation of the normalized relative signal intensity for probes outside of duplicated and CNV regions. For each exon, the normalized relative hybridization intensities of its oligonucleotide probes were averaged after trimming the upper and lower deciles (minimum of one each). Exons lacking the requisite number of probes were not scored. Detection thresholds and chaining rules were developed based on the mean relative hybridization intensity of the 80 negative control genes. For each gene, the exons within a given individual were chained to identify the overall size of the CNV based on the described rules (Supplemental Fig. 1; see www.karger.com/doi/10.1159/000184713 for all supplemental material). The average relative hybridization intensity of the entire gene was measured and only CNVs with the same relative gain or loss compared to control were detected. Thus, complex patterns of both loss and gain could not be discerned.
Initial analysis of our 80 negative control genes showed that one sample (GM12828) with the highest standard deviation demonstrated an increased false positive rate compared to all other samples (5 false positive CNVs each 2–3 exons in length). Rather than entirely discard this sample, we empirically corrected for this increased experimental noise by raising the detection thresholds by 30%. This limited GM12828 to only one false positive within all of the control genes. While this decreased our sensitivity in this sample, we opted to limit the number of false positives. Similarly we conservatively avoided analysis of the X chromosome in the single male sample due to high standard deviation likely reflecting the variation of paralogous sequences known to exist on the Y. With these adjustments, the rates of false positives were as described in the results. Additionally the individual rates of negative control exons passing the high (1.3 SD), medium (1.0 SD) and low (0.5 SD) thresholds were 0.7, 2.4 and 22%, respectively over all individuals.
Segmental duplication analysis
SDs were downloaded from the Segmental Duplication Database (http://humanparalogy.gs.washington.edu/build35/build35.htm) representing regions ≥1 kb and ≥90% nucleotide identity (Bailey et al., 2001).
Removal of cross hybridization signals in clustered regions
Cross hybridization signals were first removed in an automated fashion. Gene CNVs were clustered based on SDs with ≥95% identity. Within a cluster, individual genes were compared if they both had evidence for copy number variation. Signal intensities for all exons implicated as CNV were compared. The differences in relative hybridization intensities for each putative exon were averaged. A gene was removed if it demonstrated a consistent pattern of decreased relative intensity (an absolute difference ≥0.2 SD based on known control clusters) to all gene CNVs within a cluster. Gene CNVs were not removed if they were encompassed within the span of a highly similar duplicon (>99%) suggesting that the genes may have been duplicated together, and therefore, vary in concert. To estimate remaining cross hybridization signals, we conservatively assumed that all duplication clusters were the result of variation at a single locus unless there was evidence of an encompassing duplication. We retained the CNV gene within a given cluster that showed the most extensive evidence of variation. This stringent removal provides a lower bound for the number of CNV genes within the genome.
GO and Panther analyses
GO and Panther biologic and functional analyses were performed similarly by comparing the complete set of 357 CNV gene transcripts to the overall gene starting set of 18,373 RefSeq placements. The GOStat web service was used to analyze GO terms for biologic process and molecular function (Beissbarth and Speed, 2004). Significance was adjusted for multiple comparisons using the default method of Benjamini and Hochberg (1990).
The Panther database was analyzed using the web service for both biologic process and molecular function using default parameter (Thomas et al., 2006).
Fosmid ESP analysis
We compared whole-gene CNVs to published fosmid ESP mapping and validation (Kidd et al., 2008), everted ESPs (Cooper et al., 2008) and low similarity ESPs (Kidd, unpublished). As GM18502 has not undergone fosmid ESP analysis, we excluded three whole-gene CNVs that had detectable variation only within this individual. All fosmid ESP mapping data is available at http://hgsv.washington.edu/.
Supplementary web site
Data sets for all major steps in the analysis, including graphical browser representation of probe intensities, raw hybridization data, complete detection results, and gene annotation, are located at http://humanparalogy.gs.washington.edu/geneCNVs.
Results
Identification of candidate CNV genes and exon-targeted aCGH
We targeted a subset of CNV regions identified from eleven different studies that assayed CNVs using a variety of methodologies (Iafrate et al., 2004; Sebat et al., 2004; Tuzun et al., 2004; de Vries et al., 2005; Sharp et al., 2005; Conrad et al., 2006; Hinds et al., 2006; Locke et al., 2006; McCarroll et al., 2006; Redon et al., 2006; Wong et al., 2007). Among these, BAC aCGH predominated followed by SNP-based algorithms that leverage loss of heterozygosity, null phenotypes, or Mendelian inconsistencies in adjacent SNPs to detect copy loss. Other methods contributed to our collection of CNV regions, including oligonucleotide aCGH and fosmid ESP mapping. We identified candidate genes from non-redundant placements of the curated NCBI RefSeq mRNAs (see Methods). In this study, we focused on the detailed characterization of nine HapMap DNA samples for which sequence and clone resources exist but did not include sites detected by the fosmid structural variation project (Kidd et al., 2008). Genes were tagged as CNV candidates if the overlapping CNV regions demonstrated variation within our study individuals or if the frequency suggested a common variant (see Methods). These criteria aimed to maximize the detection of CNV genes within our study individuals while minimizing false positives and rare variants unlikely to be present in our individuals. In total, we identified 2,796 candidate CNV gene transcripts (30,203 exons), which represented 15.2% of all genomic transcripts.
For the 2,796 CNV candidates and 80 negative control genes, we designed a customized oligonucleotide microarray (NimbleGen) targeted specifically to the 30,203 exons. We sought to assess each candidate exon with an equivalent number of probes regardless of exon size. As the aCGH probe design algorithm was limited to select equally spaced probes across a defined set of regions, we compensated for the wide variation in exon size (3–21,694 bases) by restricting probe design to equivalently sized regions at the boundaries of each exon (Fig. 1, Methods). The regions were targeted with overlapping oligonucleotide probes synthesized onto a microarray. Each of our nine individuals was hybridized on an individual microarray in comparison to a control individual (G248) (Methods). Visual representation and raw data are available at the Supplemental Web Site (http://humanparalogy.gs.washington.edu/geneCNVs). In total, greater than 99.3% of all exons had six or more probes assessed on the microarray. Only 21 genes were missing coverage of more than one exon at this level and only six genes were considered to have failed design and hybridization (<50% of exons represented).
Detection algorithm and estimation of false positive and negative rates
While a number of CNV detection algorithms are now available, none are designed to deal with the overlapping and clustered nature of our probes in these experiments. Thus, we developed a simple algorithm that chained together exons with similar deviations in relative hybridization intensity beyond a series of thresholds indicative of a CNV (Supplemental Fig. 1, Methods). The algorithm was calibrated to maximize CNV detection within duplicated regions while maintaining a low false positive rate based on 80 negative control genes that demonstrated no evidence for copy number variation based on targeted aCGH (Methods). Within control genes, we detected only three small (two-exon long) false positive CNVs representing a false positive rate of 0.6% per exon assayed across all individuals. We estimated the number of false positive CNV genes at approximately 10 single- and 100 multi-exon genes. For multi-exon genes, the vast majority of false positives were expected to be small (two exons in length) and located in larger genes.
Positive controls were examined to determine if the detected pattern in a given individual was consistent with their known CNV content (Supplemental Table 1). All 11 control genes known to be variant within these individuals were appropriately determined to be CNV gene transcripts. For each of the individuals compared to the control, we identified 93% (51/55) of expected detectable variants and of these 87% of the whole-gene CNVs were correctly recognized as involving all exons. Two of the four missed CNVs appeared to be subtle changes in copy number in extensive highly similar tandem duplications, where the ratio in copy number led to minimal change in relative hybridization intensity. The other two missed variants occurred in sample GM12878 for which we raised the threshold to decrease the false positive rate indicating that this sample's sensitivity has been decreased (Methods). We also subsequently examined a set of control CNVs defined at the base pair level by complete sequencing of fosmid clones in specific study individuals (Kidd et al., 2008). All nine CNVs were detected within the specific individuals from whom the sequenced fosmid clones originated. The CNV exons were accurately defined in all but one case involving a region of tandem gene duplication in which 15 rather than 13 exons were detected as variant (Supplemental Table 2). Overall, these results indicate that our algorithm is able to properly detect and delineate CNVs even within regions of segmental duplications.
Detection of CNV genes
Using our detection algorithm, we initially identified 470 genes with evidence for partial- or whole-gene transcript CNVs (see Fig. 2for examples). We observed that highly similar gene paralogs (≥95% identity at the exon level) often demonstrated similar patterns of relative hybridization intensity but the strongest signal intensity difference often corresponded to the known variant locus. For example, we detected signal intensity differences at both of the RHD and RHCE genes, which represent paralogs with 98% genomic identity (Supplemental Fig. 2). The consistently stronger signal within RHD, known to be deleted, compared to RHCE suggested that cross hybridization was resulting in a false CNV signal on RHCE.
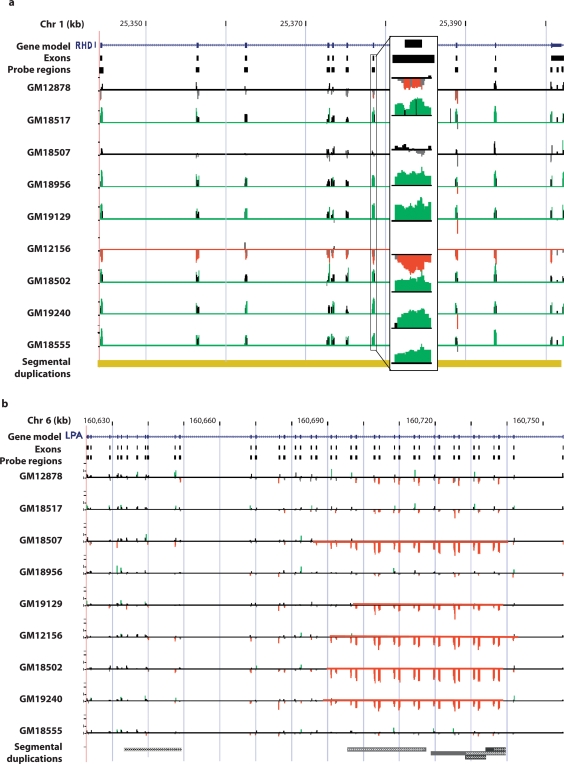
Fig. 2.
Examples of detected CNV transcripts. The observed relative signal intensities and results of the chaining algorithm are depicted for (a) the complete deletion of the RhD Blood group antigen gene (RHD) and (b) the partial-gene CNV of the lipoprotein Lp(a) precursor (LPA). Each gene is depicted (blue), the regions used for probe selection, and the relative signal intensities of the probes for each individual assayed. Individual probe signals with absolute relative deviations >1.0 SD are colored green for gain and red for loss. For RHD, an expanded area shows the probes for exon 6 in detail. The results of our detection algorithm are depicted by a red or green line indicating a region of loss or gain. In the case of RHD, these represent detection of gains and losses of the entire transcript. For LPA, the detected regions demonstrate partial transcript loss relative to the control. The region identified in LPA represents a series of variously-sized tandem deletions and duplications based on a 2-exon module containing Kringle domains. Vertical scales represent the natural log of the normalized relative hybridization intensities.
To identify and remove these false CNV signals due to cross hybridization, we clustered genes involved in SDs with ≥95% identity and operationally assigned copy number variation to the locus where signal intensity difference was consistently greater for all exons (Methods). This allowed us to remove 73 putative gene CNVs as regions of paralogous cross hybridization and provided a more accurate estimate of 397 CNV genes. Of these, 166 were classified as whole-gene CNVs, showing evidence for variation across all exons in at least one individual (Table 1). Thirty percent (50/166) of the whole-gene CNVs are represented by single exon genes. The majority of gene CNVs (205/397) occur more than once within our nine individuals; however, our study from the outset sought to enrich for such common variants and thus does not provide an unbiased estimate of their frequency.
Table 1.
Exon-targeted detection of gene CNVs
| Locia |
Total count (%) |
Conservative/stringentb count (%) |
|
|---|---|---|---|
| All gene CNVs | 397 (14) | 255 (9) | |
| Signal: | gain only | 170 (43) | 97 (38) |
| loss only | 123 (31) | 80 (31) | |
| gain & loss | 104 (26) | 78 (31) | |
| Within SDs | 239 (60) | 163 (64) | |
| Common | 205 (52) | 164 (64) | |
| Whole gene CNV | 166 (6) | 134 (5) | |
| Signal: | gain only | 61 (37) | 50 (37) |
| loss only | 47 (28) | 41 (31) | |
| gain & loss | 58 (35) | 40 (30) | |
| Within SDs | 136 (82) | 96 (72) | |
| Common | 127 (77) | 85 (63) | |
| Single exon | 50 (30) | 30 (22) | |
| Partial gene CNV only | 231 (8) | 121 (4) | |
| Signal: | gain only | 109 (47) | 47 (39) |
| loss only | 76 (33) | 39 (32) | |
| gain & loss | 46 (20) | 35 (29) | |
| Within SDs | 111 (48) | 67 (55) | |
| Common | 95 (41) | 79 (65) | |
| Genes negative for CNV | 2393 (86) | 2535 (91) | |
| Within SDs | 333 (14) | 409 (16) | |
| Total candidates | 2790 | 2790 | |
| Within SDs | 572 (21) | 572 (21) | |
Within SDs: =50% of exonic sequence fall within segmental duplications (SD). Common: Detected in >1 individual.
Estimate based on retaining one locus per duplication cluster and removing two exon CNVs in large genes (>5 exons) appearing in a single individual as likely false positives; thereby providing a minimal estimate of CNV within our studied individuals.
Among the identified 63 duplicated gene clusters, there remained potential cases of cross hybridization due to the presence of multiple, highly similar paralogous gene transcripts. The majority appeared to represent tandem duplications with near allelic levels of sequence similarity (>99% identity). Completely disentangling such regions to determine the variant paralog(s) will most certainly require direct sequencing in multiple individuals. In these cases, we conservatively corrected each duplication cluster for cross hybridization by selecting the single paralogous locus with the most extensive CNV pattern (Methods). As an additional measure of stringency, we also removed possible false positive signals similar to those observed in our control regions – two-exon partial CNVs from large genes (>5 exons) detected in only a single individual. This left 255 CNV genes comprising 134 whole and 121 partial gene transcript CNVs and provided a lower boundary for gene CNVs within our assessed individuals (Table 1, Supplemental Table 3 for complete list). We determined that the individuals in our study differed from the control by an average 104 gene CNVs (63 whole and 41 partial) (Supplemental Table 4). Of our initial 2,796 candidate genes, 14% showed exon copy number variation. This percentage dropped to 9% if we applied the most stringent criteria. Of the 702 candidates identified by full-tiling path BAC aCGH within these same nine individuals (Redon et al., 2006), we detected 23–32% of the genes as CNV (Table 2). Of the 177 candidates that would have been implicated by fosmid ESP analysis, we detected 37–50% at the genic level using this exon-targeted aCGH approach.
Table 2.
Validation of candidate genes implicated from studies of the same individuals
| Study | Gene set | Genes |
|||
|---|---|---|---|---|---|
| Implicated candidatesa |
CNP partial |
CNP whole |
CNP total (%) |
||
| Full-tiling path BAC aCGH | total | 702 | 108 | 119 | 227 (32) |
| stringent | 702 | 63 | 98 | 161 (23) | |
| Fosmid ESPb | total | 177 | 37 | 52 | 89 (50) |
| stringent | 177 | 25 | 40 | 65 (37) | |
Candidate genes with CNV regions specifically within our assayed individuals.
Fosmid ESP sites were overlaid after initial candidate gene discovery.
Previous analyses have detected significant associations between SD and CNV regions (Sebat et al., 2004; Locke et al., 2006; Redon et al., 2006; Cooper et al., 2007). We determined if SDs overlapped ≥50% of total exon bases of each candidate gene (Methods). By this criterion, 21% of candidate genes were located within SDs. Compared to candidate genes, stringent gene CNVs showed a significant enrichment with 64% located within SDs (P = 6.1 × 10–45, Fisher's exact test). The SD enrichment for whole-gene CNVs was significantly greater than partial-gene CNVs (72 and 55%, respectively, P = 0.0031, Fisher's exact). More stringent and relaxed definitions of SD content did not significantly alter the remarkable association for whole-gene CNVs and suggests that these as a class are particularly ensconced within SDs.
To better understand the nature of the detected CNV transcripts, we investigated their biologic and functional associations using both the GO and Panther annotation databases (Supplemental Tables 5, 6, 7, 8, Methods). Both analyses demonstrated considerable enrichment in CNV genes related to pathogen defense (bacterial defense, IgG domains and receptors, humoral immunity, complement activation), reproductive biology (egg-sperm binding for fertilization, cell-cell interaction), and environmental interaction (olfaction, digestion, steroid/toxin hormone metabolism) (Supplemental Tables 5 and 7). Molecular functions mirrored biologic processes with additional specific enrichments for IgG binding, amylase, immunoglobulin receptors, and galactosidase (Supplemental Tables 6 and 8). These enrichments are similar to previous observations for genes underlying CNV regions (Cooper et al., 2007) as well as genes within SD (Bailey et al., 2002) or undergoing positive selection (Vallender and Lahn, 2004).
We carefully examined whole-gene CNVs for previous evidence of copy number variation outside of large-scale studies. It is notable that many detected whole-gene CNVs have been confirmed previously (Supplemental Table 3). These included pepsinogen (Taggart et al., 1985), amylase (Groot et al., 1989), natural killer cell receptors (Wilson et al., 2000), complement C4 (Schneider et al., 1986), HLA Class II proteins (Zhang et al., 1990), telomeric olfactory receptors (Trask et al., 1998), and low affinity Fc receptor III (Koene et al., 1998). If all detected olfactory receptor genes are included as confirmed, approximately half of all whole-gene CNVs showed evidence for copy number variation from experiments outside of large-scale studies (∼30% when excluding olfactory receptors). It also becomes apparent that many examples within our gene CNVs represent known genes or gene families with evidence for recent positive selection (Vallender and Lahn, 2004). This includes the defensins (Boniotto et al., 2003), the MHC region (Zhang et al., 1990), and KIRs (Hughes, 2002). Many genes from families known to be under recent positive selection but lacking previous confirmation of copy number variation CNV were detected (Supplemental Table 3). These include genes involved in reproduction such as SPANX and GAGE family members as well as genes thought to be important in microbial defense, such as the late cornified envelope proteins important for skin integrity. The CNV genes detected in this study are enriched for duplications and regions of positive selection – regions that have been shown to be important in human evolution and disease.
An integrated map of gene copy number variation
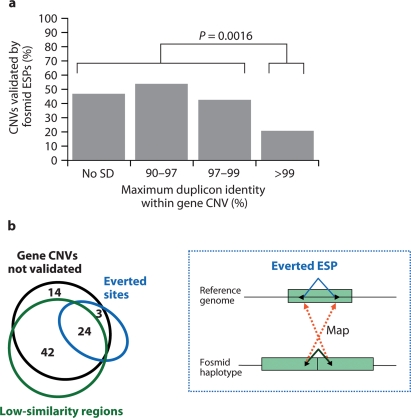
We compared our CNV genes with the sequence-based map that had been constructed using fosmid end sequence pairs (Kidd et al., 2008). Focusing on the 131 whole-gene CNVs detected within at least one of the eight individuals analyzed by fosmid ESP mapping, we found that only 37% (48/131) of these overlapped with the set of validated deletions or insertions reported in Kidd et al. (2008). This low rate of overlap may be partly explained by the observation that 70% (92/131) of these CNVs are located in SDs, which are genomic regions where the fosmid ESP approach is known to have limited detection power due to the difficulty in obtaining unique ESP map positions. As expected, we find that the fosmid ESP validation rate was the least at the highest levels of sequence identity (Fig. 3a). Only 20% (10/51) of CNVs mapping within segmental duplications >99% were validated indicating that sequence identity of duplicates is the most significant source of false negatives (Fisher's exact; P = 0.0016).
Fig. 3.
Whole-gene CNVs compared to fosmid ESP analysis. (a) Validation rates categorized by the percent identity of the most similar duplicon within each whole-gene CNV region. There is a significant decrease in the validated fraction for regions containing duplicons >99% identity. (b) Venn diagram showing the association of the 83 whole-gene CNVs with best-placed fosmid ESPs of low-similarity (98–99.5%) suggesting more divergent unrepresented CNV paralogs and/or with best-placed everted ESPs (>99.5%) suggesting highly similar tandem duplications. Interesting regions containing everted and low-similarity regions overlap suggesting a more complex nature for these CNV genes. The inset depicts the basis for the formation of everted fosmid ESPs, where a clone that traverses the boundary of a tandem duplication can only map to the single copy contained within the reference genome (Cooper et al., 2008).
We explored in more detail the 83 CNV genes that were not validated by the fosmid ESP analysis (Fig. 3b) in an effort to enhance our power to detect these variant duplicated genes. We recently developed a modification to the fosmid ESP approach that allows large tandem duplication events to be identified based on a unique pattern of ESP placements (Cooper et al., 2008). Tandem duplications of sequences in fosmid haplotypes that are not tandemly duplicated in the reference assembly can be identified by characteristic clusters of ESPs whose ends map in an everted orientation (that is, the end sequences map onto opposite strands but are pointing outward) (Fig. 3b inset). We found that 33% (27/83) of the non-validated CNVs intersect with everted sites identified by two or more clones from a single fosmid library. This indicates that a substantial fraction of the whole-gene CNVs are tandem duplication events not represented in the reference genome.
The fosmid ESP mapping algorithm also requires high sequence identity alignments (>99.5%) for both end sequence placements against the reference genome (Tuzun et al., 2005). Duplicated sequences not represented within the human reference genome may, however, have greater sequence divergence. We, therefore, examined all ESPs of high quality (Q > 30) that mapped to a best location but with less sequence identity. We determined that 80% (66/83) of these intervals overlap with best-placements with alignment identity between 98 and 99.5%. Interestingly, this set includes 89% (24/27) of the everted placements. However, the fraction of everted ESPs >99.5% compared to all everted clones (98–100%) was greater than 0.5 for 89% (24/27) of the above everted regions consistent with unrepresented highly similar paralogs (>99.5% identity to our assayed locus). These results suggest that such unrepresented regions may be the result of more divergent tandem duplications within these genomes. We predict that a consideration of both everted placements and high-quality, but low-sequence identity placements, will allow ∼90% of CNVs to be accurately predicted.
Discussion
While thousands of genes have been implicated as underlying CNV regions, the imprecision of current large-scale detection methods leaves the true extent of gene copy number variation poorly defined. To date, no genome-wide analyses have been published with sufficient resolution to determine the exact boundaries for all detected CNVs, which would provide an accurate determination of gene copy number variation. Here, we presented an assessment of 2,790 of the most promising candidate genes as identified by previous copy number variation studies. We used a novel approach targeting multiple oligonucleotide probes to each candidate exon, which provided the power to detect variation even in highly duplicated regions. In total, we conservatively detected 255 gene loci as CNV in transcribed sequence and estimated that any two humans differ by a minimum of 100 gene CNVs. Our analysis has several important biological and practical implications.
First, the number of CNV and copy number polymorphic genes is not as extensive as anticipated. We detected copy number variation in only 14% of our candidate genes despite enriching for probable common variants within our individuals. This is not simply an effect of our small sample size, since comparisons within the same individuals found only one-third of the BAC aCGH implicated candidate genes confirmed by our assay. While false positives in large-scale aCGH account for a proportion of this low confirmation rate, the majority of CNV-negative genes are probably due to the imprecise determination of the variant CNV regions leading to implication of genes that lie nearby the true CNV. This conclusion is supported by the fact that we observed greater confirmation rates (up to 50%) for more precise fosmid-based methods as well as the general trend within studies that more precise methods yield lower-fractions of CNVs overlapping genes (Hinds et al., 2006; McCarroll et al., 2006; Redon et al., 2006; Wong et al., 2007; Kidd et al., 2008). Given that many CNV detection methods appeared to perform poorly in highly tandemly duplicated regions, our estimate of an average of 100 variant loci in individuals compared to a control is consistent with an average of 65 CNVs detected in two recently sequenced and assembled individual genomes (Levy et al., 2007; Wheeler et al., 2008).
From a biological perspective, the amount of common gene CNVs is consistent with thoroughly characterized genetic systems, such as the human blood group antigens, where SNPs currently account for the majority of clinically relevant phenotypic differences. Gene CNVs account for only a few notable common antigenic differences within the blood groups but do account for numerous rare null alleles (Daniels, 2002). Recent experiments examining levels of gene expression, a phenotype that can result directly from changes in gene dosage, found that SNPs still accounted for the vast majority of associated variation, capturing 84% of expression variation compared to 18% for CNVs (Stranger et al., 2007).
Another important finding is that gene CNVs appear to be more tightly associated with regions of SDs than previously appreciated. Sixty-four percent of detected gene CNVs and 72% of whole-gene CNVs fell within regions of segmental duplication. For whole-gene CNVs, we estimate that 84% (113/134) of whole-gene CNVs map to high-identity duplications. Given that our study was enriched for common variants, this suggests that the vast majority of common CNV genes are the result of the gain or loss of whole-gene SDs. This only heightens the need to resolve this aspect of human genetic diversity at the sequence level. While we were able to detect and remove a proportion of false positive loci due to cross hybridization, we could not always determine which gene copies were copy number variant – particularly between paralogs with allelic levels of sequence identity. Developing further experimental and computational methods to identify cross hybridization will be of increasing importance as higher-resolution arrays begin to assess variant tandem duplications with a growing number of probes.
Furthermore, we demonstrate the value of cross-platform studies on the same individuals in obtaining a comprehensive map of structural variation. For example, we show that a large fraction of our whole-gene CNVs were missed by standard application of fosmid ESP, but that with some additional modification to the end-sequence pair mapping algorithm, most could be recovered. We found that 83% (69/83) of exon-aCGH-detected CNVs that were missed could be captured if everted ESP sites and/or low-similarity (98–99.5%) best-mapped ESPs were considered. The vast majority of everted clones appear to represent tandem duplications at near allelic levels of identity not present in the reference sequence. The regions containing low-similarity (98–99.5%) fosmids suggest unrepresented paralogs below allelic levels of variation. This picture is complicated as almost all gene CNVs with everted clones also contain low-similarity ESPs suggesting the presence of complex rearrangements and/or multiple unrepresented variant paralogs. These regions will require detailed sequence analysis of clone inserts to resolve the structural complexity of these regions (Eichler et al., 2007) and this study has highlighted dozens of these regions for further analyses.
Finally, our study provides a targeted set of common CNV transcripts that can provide the basis for association studies. These CNV gene transcripts demonstrate an enrichment of functions that are associated with processes undergoing adaptive evolution including response to environmental stimuli, reproductive biology and response to infectious disease. Many of the loci that we detected have already been associated with human phenotypes and regions of selective pressure, the genes within our set are likely to yield further associations with phenotype and disease. Utilizing expanding genotyping data for these regions (Kidd et al., 2008) as well as further probe screening (Sharp et al., 2007) should soon enable the selection of specific subsets of oligonucleotide probes that are the most responsive to the dosage of each gene CNV. Such next generation microarrays will provide a targeted platform to robustly examine these transcript CNVs in relation to important human diseases.
Supplementary Material
CNV detection algorithm
Depicted is a hypothetical example of CNV detection illustrating the rules governing detection and chaining. For each exon, the average hybridization intensity was calculated after trimming the extreme probes (upper and lower deciles). For single exon genes, a threshold of ≥ 1.3 STD was indicative of CNV. For multi-exon genes, a seeding region was required consisting of two contiguous exons that met the threshold of ≥1.3 STD and ≥1.0 STD, respectively. Further chaining was less stringent to allow for variation in the copy-number responsiveness of an exon's probes. Chain extension continued in three ways: rules #1–3. Rule#1 is simple extension at a high threshold. Rule #2 allowed for dips in relative hybridization intensity (0.5–1.0 SD) as long as the subsequent exons showed strong (≥1 SD) relative hybridization intensity. Rule #3 allowed for the detection of apparent whole-gene CNVs where the terminal exon showed reduced intensity (0.5–1.0 SD). Chaining is terminated if rules #1–3 could not be applied. In this hypothetical example, the seed occurs in exons 2 (≥1.0 SD) and 3 (≥1.3 SD). Continued chain extension occurs to the left by rule #2 for exon 4 and then rule #1 for exon 5. Termination occurs before the inclusion of exon number 1 where the mean relative hybridization intensity is in the opposite direction. Thus, a partial-gene copy number gain of exons 2 through 5 is identified in this example.
False CNV signatures at RHCE due to paralogous CNV RHD deletion
Depicted is the 110 kb region containing the RHD and RHCE genes along with the observed relative signal intensities of the oligonucleotide probes for all nine individuals. Probes are labeled green and red if the respective relative gain or loss in intensity exceeds a standard deviation for a given individual. The paralogous genes are related by a 50 kb inverted tandem duplication that has 98% nucleotide identity. RHD contains a null mutation due to a deletion of the entire gene that renders 15% of the Caucasian population negative for the protein. The G248 control is heterozygous for this deletion. Within RHCE there are no known common CNVs. Interestingly, RHCE shows a similar albeit weaker pattern of variation relative to RHD in all individuals. These signals on RHCE are due to cross hybridization of RHD DNA onto the highly similar RHCE probes. Gene structures are depicted in blue with RHD and RHCE in transcribed opposite orientations. Probes are labeled red or green if they exceed. Probe regions delineate the regions from which the probes were chosen from vertical bars represent a scale of 5 kb. The SD containing the genes and its inverted orientation is in yellow.
CNV Detection Results for Positive Control Genes
Detection of known gene CNVs defined by complete fosmid clone sequencing
Stringent list of detected gene CNVs
Detected gene CNVs in each individual relative to control
Gene Ontology biologic processes within CNV genes demonstrating statistically significant deviations compared to all RefSeq transcripts
Gene Ontology molecular functions within CNV genes showing statistically significant deviations compared to all RefSeq transcripts
Panther biologic process classification of detected CNV genes compared to all RefSeq transcripts
Panther molecular function classification of detected CNV genes compared to all RefSeq transcripts
Intersection of whole-gene CNV sites with fosmid ESP validated sites and additional signatures of CNV∗
Footnotes
J.A.B. was supported by an ARC Transfusion Medicine Fellowship. J.M.K. was supported by a NSF Graduate Research Fellowship. This work was supported in part by NIH grant HG004120 to E.E.E.
References
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Barber JC, Reed CJ, Dahoun SP, Joyce CA. Amplification of a pseudogene cassette underlies euchromatic variation of 16p at the cytogenetic level. Hum Genet. 1999;104:211–218. doi: 10.1007/s004390050938. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Boniotto M, Tossi A, DelPero M, Sgubin S, Antcheva N, et al. Evolution of the beta defensin 2 gene in primates. Genes Immun. 2003;4:251–257. doi: 10.1038/sj.gene.6363958. [DOI] [PubMed] [Google Scholar]
- Colin Y, Cherif-Zahar B, Le Van Kim C, Raynal V, Van Huffel V, Cartron JP. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39:S22–29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy-number variant detection via genome-wide single nucleotide polymorphism genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. Human Blood Groups. 2nd ed. Blackwell Science: Malden; 2002. [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat Rev Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Groot PC, Bleeker MJ, Pronk JC, Arwert F, Mager WH, et al. The human alpha-amylase multigene family consists of haplotypes with variable numbers of genes. Genomics. 1989;5:29–42. doi: 10.1016/0888-7543(89)90083-9. [DOI] [PubMed] [Google Scholar]
- Heim MH, Meyer UA. Evolution of a highly polymorphic human cytochrome P450 gene cluster: CYP2D6. Genomics. 1992;14:49–58. doi: 10.1016/s0888-7543(05)80282-4. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JA, Barber JC. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Evolution of the human killer cell inhibitory receptor family. Mol Phylogenet Evol. 2002;25:330–340. doi: 10.1016/s1055-7903(02)00255-5. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koene HR, Kleijer M, Roos D, de Haas M, Von dem Borne AE. Fc gamma RIIIB gene duplication: evidence for presence and expression of three distinct Fc gamma RIIIB genes in NA(1+,2+) SH(+) individuals. Blood. 1998;91:673–679. [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness D. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Nguyen DQ, Webber C, Ponting CP. Bias of selection on human copy-number variants. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie RJ, Mattei MG, Lalande M. A large polymorphic repeat in the pericentromeric region of human chromosome 15q contains three partial gene duplications. Hum Mol Genet. 1998;7:1253–1260. doi: 10.1093/hmg/7.8.1253. [DOI] [PubMed] [Google Scholar]
- Schneider PM, Carroll MC, Alper CA, Rittner C, Whitehead AS, et al. Polymorphism of the human complement C4 and steroid 21-hydroxylase genes. Restriction fragment length polymorphisms revealing structural deletions, homoduplications, and size variants. J Clin Invest. 1986;78:650–657. doi: 10.1172/JCI112623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Itsara A, Cheng Z, Alkan C, Schwartz S, Eichler EE. Optimal design of oligonucleotide microarrays for measurement of DNA copy-number. Hum Mol Genet. 2007;16:2770–2779. doi: 10.1093/hmg/ddm234. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart RT, Mohandas TK, Shows TB, Bell GI. Variable numbers of pepsinogen genes are located in the centromeric region of human chromosome 11 and determine the high-frequency electrophoretic polymorphism. Proc Natl Acad Sci USA. 1985;82:6240–6244. doi: 10.1073/pnas.82.18.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–3026. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Trask B, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Tuzun E, Bailey JA, Eichler EE. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 2004;14:493–506. doi: 10.1101/gr.1907504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Vallender EJ, Lahn BT. Positive selection on the human genome. Hum Mol Genet. 2004;13:R245–254. doi: 10.1093/hmg/ddh253. Spec No 2: [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Torkar M, Haude A, Milne S, Jones T, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Degli-Esposti MA, Cobain TJ, Cameron PU, Christiansen FT, Dawkins RL. Differences in gene copy number carried by different MHC ancestral haplotypes. Quantitation after physical separation of haplotypes by pulsed field gel electrophoresis. J Exp Med. 1990;171:2101–2114. doi: 10.1084/jem.171.6.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CNV detection algorithm
Depicted is a hypothetical example of CNV detection illustrating the rules governing detection and chaining. For each exon, the average hybridization intensity was calculated after trimming the extreme probes (upper and lower deciles). For single exon genes, a threshold of ≥ 1.3 STD was indicative of CNV. For multi-exon genes, a seeding region was required consisting of two contiguous exons that met the threshold of ≥1.3 STD and ≥1.0 STD, respectively. Further chaining was less stringent to allow for variation in the copy-number responsiveness of an exon's probes. Chain extension continued in three ways: rules #1–3. Rule#1 is simple extension at a high threshold. Rule #2 allowed for dips in relative hybridization intensity (0.5–1.0 SD) as long as the subsequent exons showed strong (≥1 SD) relative hybridization intensity. Rule #3 allowed for the detection of apparent whole-gene CNVs where the terminal exon showed reduced intensity (0.5–1.0 SD). Chaining is terminated if rules #1–3 could not be applied. In this hypothetical example, the seed occurs in exons 2 (≥1.0 SD) and 3 (≥1.3 SD). Continued chain extension occurs to the left by rule #2 for exon 4 and then rule #1 for exon 5. Termination occurs before the inclusion of exon number 1 where the mean relative hybridization intensity is in the opposite direction. Thus, a partial-gene copy number gain of exons 2 through 5 is identified in this example.
False CNV signatures at RHCE due to paralogous CNV RHD deletion
Depicted is the 110 kb region containing the RHD and RHCE genes along with the observed relative signal intensities of the oligonucleotide probes for all nine individuals. Probes are labeled green and red if the respective relative gain or loss in intensity exceeds a standard deviation for a given individual. The paralogous genes are related by a 50 kb inverted tandem duplication that has 98% nucleotide identity. RHD contains a null mutation due to a deletion of the entire gene that renders 15% of the Caucasian population negative for the protein. The G248 control is heterozygous for this deletion. Within RHCE there are no known common CNVs. Interestingly, RHCE shows a similar albeit weaker pattern of variation relative to RHD in all individuals. These signals on RHCE are due to cross hybridization of RHD DNA onto the highly similar RHCE probes. Gene structures are depicted in blue with RHD and RHCE in transcribed opposite orientations. Probes are labeled red or green if they exceed. Probe regions delineate the regions from which the probes were chosen from vertical bars represent a scale of 5 kb. The SD containing the genes and its inverted orientation is in yellow.
CNV Detection Results for Positive Control Genes
Detection of known gene CNVs defined by complete fosmid clone sequencing
Stringent list of detected gene CNVs
Detected gene CNVs in each individual relative to control
Gene Ontology biologic processes within CNV genes demonstrating statistically significant deviations compared to all RefSeq transcripts
Gene Ontology molecular functions within CNV genes showing statistically significant deviations compared to all RefSeq transcripts
Panther biologic process classification of detected CNV genes compared to all RefSeq transcripts
Panther molecular function classification of detected CNV genes compared to all RefSeq transcripts
Intersection of whole-gene CNV sites with fosmid ESP validated sites and additional signatures of CNV∗