Abstract
Background
Expressed Sequence Tag (EST) has been a cost-effective tool in molecular biology and represents an abundant valuable resource for genome annotation, gene expression, and comparative genomics in plants.
Results
In this study, we constructed a cDNA library of Prunus mume flower and fruit, sequenced 10,123 clones of the library, and obtained 8,656 expressed sequence tag (EST) sequences with high quality. The ESTs were assembled into 4,473 unigenes composed of 1,492 contigs and 2,981 singletons and that have been deposited in NCBI (accession IDs: GW868575 - GW873047), among which 1,294 unique ESTs were with known or putative functions. Furthermore, we found 1,233 putative simple sequence repeats (SSRs) in the P. mume unigene dataset. We randomly tested 42 pairs of PCR primers flanking potential SSRs, and 14 pairs were identified as true-to-type SSR loci and could amplify polymorphic bands from 20 individual plants of P. mume. We further used the 14 EST-SSR primer pairs to test the transferability on peach and plum. The result showed that nearly 89% of the primer pairs produced target PCR bands in the two species. A high level of marker polymorphism was observed in the plum species (65%) and low in the peach (46%), and the clustering analysis of the three species indicated that these SSR markers were useful in the evaluation of genetic relationships and diversity between and within the Prunus species.
Conclusions
We have constructed the first cDNA library of P. mume flower and fruit, and our data provide sets of molecular biology resources for P. mume and other Prunus species. These resources will be useful for further study such as genome annotation, new gene discovery, gene functional analysis, molecular breeding, evolution and comparative genomics between Prunus species.
Background
Mei (Prunus mume) originated in China and belongs to the sub-family Prunoideae within the Rosaceae family and is widely cultivated in East Asian countries. Mei trees are classified into two groups, fruiting mei and flowering mei, based on their uses [1]. The fruiting mei has been cultivated in China for more than 7000 years according to the historical records, and now it is also widely cultivated in Japan [2]. The processed products of mei include salted mei, mei wine and juice have high nutritional and medicinal value and are consumed in some countries, including China, Japan, and Korea. Chu [3] reported that mei flower and young fruit can tolerate relatively low temperatures (-4°C ~ -2°C) in early spring. These attributes make it an economically important and excellent ornamental tree in south China. Mei has long played an important role in human diet and health, but there is dearth of information about its flower and fruit development, physiology and biochemistry. The scarcity of knowledge contributes to difficulties in breeding, production, and storage of fruits. Mei cDNA library construction and EST analysis could be employed for the study of important genes responsible for flower and fruit development and their evolution in the plant kingdom.
Expressed sequence tags (ESTs) are partial sequences of expressed genes prepared by reverse transcribing mRNA and cloning the cDNA fragments into a plasmid and they are also gained by large scale sequencing at one instance [4]. They represent a snapshot of genes expressed in a given tissue and/or at a given developmental stage [5]. In plants, this method was initially applied to the model plant Arabidopsis thaliana [6]. Recently, many EST libraries of a wide range of plant species have been constructed for the genes involved in plant growth and differentiation [7], secondary metabolism [8] and biochemical pathways [9,10] as well as responses to pathogen attack and environmental stresses [11]. ESTs are proven cost-effective tools and represent abundant valuable resources for genome annotation, gene expression, and comparative genomics in non-model plants [12,13]. EST has also played an important role in functional genomics research on new functional gene discovery other than the whole genome [14-16]. By April 23, 2010, a total of 65,389,085 ESTs had been submitted to the National Center for Biotechnology Information (NCBI) from 1,989 species. The numbers of ESTs submitted are increasing rapidly at an approximate monthly rate of one million.
EST libraries for fruit trees such as grape, citrus, almond, strawberry, apricot, apple, blackberry and peach [12,13,17-22] have been constructed, sequenced, analyzed and deposited in databases. However, for P. mume, only 116 hits were found in the dbEST of the GenBank http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html in May 2010, a much lower figure than those of other main Prunus plants. In this study, a suitable high quality cDNA library from flowers and fruits of mei was constructed for the analysis of expressed sequence tags (ESTs), and to generate an EST resource for EST-SSR development of P. mume, the cDNA-based design of microarrays needed for understanding gene expression, and provide a platform for mei functional genomic studies on growth, development, metabolic regulation mechanisms, and so on. In this study, 1,294 unique sequences generated from 4,473 unigenes have homology with other plants, and 1,130 could be annotated with known and putative functions.
EST-SSR has been applied in molecular biology studies of fruiting trees as a new technology, especially on its application in some research fields of pomology, including genetic diversity analysis [23], genetic linkage map construction [24,25], comparative mapping [26,27], molecular phylogeny and cultivar identification [28]. Based on these EST resources, types and frequency of simple sequence repeats (EST-SSRs) with a motif length of 2-6 bp were searched by MISA software. The EST-SSRs generated in this study will be an efficient tool for additional genetic and genomic research in mei and even in Prunus species.
Results
Quality inspection of the cDNA library
The cDNA library of mei was constructed based on the pDNR-LIB vector (Invitrogen Biotechnology Co., Ltd). The results showed that the primary titer of the constructed cDNA library was 1.4 × 106 pfu/mL, while the recombination rate was about 97.5%, and the distribution of insert sizes of the library was about 1.0-3.0 kb based on random PCR analysis of 30 clones (Table 1). These results indicated that the constructed mei cDNA library was of a high quality (high titer, high recombinant rate and large inserted fragments) and it is also the first cDNA library of mei flower and fruit.
Table 1.
Quality inspection and summaries of EST sequence analysis of the cDNA library of P. mume.
| Group | Records | |
|---|---|---|
| Primary titer | 1.4 × 106 pfu/mL | |
| Recombination rate | 97.5% | |
| Insert sizes | 1.0-3.0 kb | |
| Number of raw sequences | 10,123 | |
| Number of high quality sequences | 8,656 | |
| Average EST length | 464 bp | |
| Number of unigenes | 4,473 | |
| Singletons | 2,981 | |
| Contigs | 1,492 | |
| Average unigene length | 560 bp | |
| Average number of ESTs per unigene | 1.9 | |
| Average number of ESTs per contig | 3.8 | |
| Maximum number of ESTs per contig | 84 | |
| Minimum unigene length | 101 bp | |
| Maximum unigene length | 1471 bp | |
| Minimum ORF length | 102 bp | |
| Maximum ORF length | 1323 bp | |
| Average ORF length | 383 bp |
ESTs obtained and assembled
A total of 10,123 clones were randomly picked and subjected to sequencing for the flower and fruit cDNA library. The leading vector, tailing of the sequence, and poor-quality sequences were excluded first, and 8,656 non-redundant EST sequences were obtained, showing a high success rate of sequencing. The average length of ESTs without vector was 464 bp. 4,473 non-redundant unigenes were assembled and most of them (3,672 unigenes, 82.1%) have sequence lengths between 400 and 800 bp, and only 2.4% (110) were shorter than 200 bp (see Additional file 1). The average length of unigene sequences was 560 bp, which was longer than that in other fruit trees such as persimmon [29], grapevine [12,30], apricot [20] and citrus [31], but shorter than almond [18] and peach [22]. The minimum and maximum length of these unigenes were 101 bp and 1,471 bp, and the average number of ESTs per unigene was 1.9. These unigenes were composed of 1,492 (33.36%) contigs and 2,981 (66.64%) singletons, with the number of average and maximum ESTs per contig being 3.8 and 84, respectively (Table 1). Only 11.6% of the contigs were composed of more than 4 ESTs (Additional file 2), suggesting that the redundancy rate is relatively low in this normalized library. In addition, the sequences data were submitted to the EST database of NCBI and publicly available (GW868575-GW873047).
EST annotation and functional classification
We used BLASTX to annotate our P. mume unigene sequences. 1,294 (28.9%) of the unigenes matched genes in other species with an expectation value of 1e-10 or better in a search against the NCBI nr protein database (released in October 2009), which is lower than other Prunus species, such as apricot [20], and peach [22]. That may because mei is different from other stone fruit species or the total number of ESTs sequenced were less than in the other two species, or for other reasons. The unigenes which showed similarity with sequences in public databases were classified into 23 groups based on their similarities as in Table 2. The results revealed that 1,130 (87.33%) of matched unigenes were known or predicted functional genes, 164 (12.67%) with unknown functional. Those unigenes with unknown functions could be considered as novel or specific genes in mei. Based on the known or predicted annotation, a large number of unigenes (353, 31.24%) were involved in transport and metabolism including carbohydrate, amino acid, lipid, inorganic, coenzyme and nucleotide. There were also 56 (4.95%) unigenes involved in DNA/RNA processing and transcription. Only 8 were seen to be involved in the defense mechanisms covering 0.62% of the functional genes.
Table 2.
Grouping of unigenes from flower and fruit cDNA library of P. mume.
| Category | Number of ESTs in Category | Percentage (%) |
|---|---|---|
| General function prediction only | 237 | 18.32 |
| Function unknown | 164 | 12.67 |
| Posttranslational modification, protein turnover, chaperones | 146 | 11.28 |
| Carbohydrate transport and metabolism | 113 | 8.73 |
| Amino acid transport and metabolism | 95 | 7.34 |
| Energy production and conversion | 94 | 7.26 |
| Translation, ribosomal structure and biogenesis | 86 | 6.65 |
| Lipid transport and metabolism | 58 | 4.48 |
| Inorganic ion transport and metabolism | 42 | 3.25 |
| Cell wall/membrane/envelope biogenesis | 37 | 2.86 |
| Coenzyme transport and metabolism | 37 | 2.86 |
| Signal transduction mechanisms | 36 | 2.78 |
| Replication, recombination and repair | 26 | 2.01 |
| Transcription | 26 | 2.01 |
| Secondary metabolites biosynthesis, transport and matabolism | 25 | 1.93 |
| Cytoskeleton | 17 | 1.31 |
| Intracellular trafficking, secretion, and vesicular transport | 16 | 1.24 |
| Cell cycle control, cell division, chromosome partitioning | 16 | 1.24 |
| Nucleotide transport and metabolism | 8 | 0.62 |
| Defense mechanisms | 8 | 0.62 |
| RNA processing and modification | 4 | 0.31 |
| Chromatin structure and dynamics | 2 | 0.15 |
| Cell motility | 1 | 0.08 |
| Total | 1294 | 100 |
Alignment against EST database of Prunus species
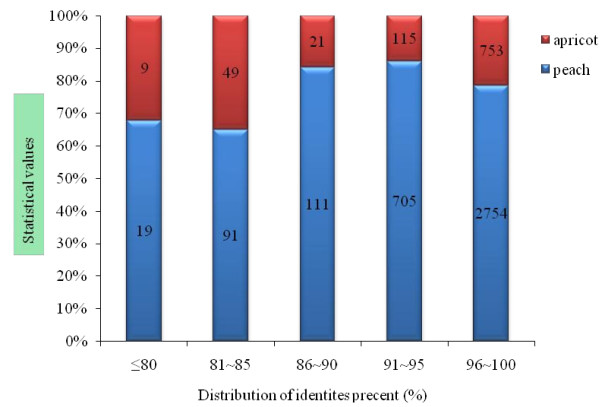
By April 28, 2010, 100,071 ESTs from all Prunus species had been deposited in NCBI. However, only 116 ESTs were those of P. mume and this data was much lower than those Prunus persica (peach; 79,567) and Prunus armeniaca (apricot; 15,105). It is evident that the ESTs deposited from this work will provide vital data for P. mume genomic and genetic study. Here, we first carried out alignment between the unigenes (4,473) generated from P. mume and the EST database of two other Prunus species (P. persica, P. armeniaca) openly accessible in NCBI (by April 23, 2010). The results showed that 948 unigenes of mei aligned with P. armeniaca, at an identity level of 79%~100% (the average level was 96.2%), while 3,680 were aligned with P. persica and the average identity level was 96.53%. The numbers and distribution of identity level of mei unigenes which have aligned with two other species are shown in Figure 1. It was clearly discernible that most of the identity levels were over 96%, and 753 and 2,754 mei ESTs have homology with apricot and peach, respectively. The description above indicated that most of the P. mume ESTs had predicted functions and could be a great help for further research on functional genomics and genetic analysis of P. mume.
Figure 1.
Identities distribution of alignment results against the EST database of two other Prunus species. The data in red and blue column were the numbers of mei ESTs which have homology with apricot and peach EST database in different identities.
Characterization of microsatellite sequences
Microsatellites or Simple Sequence Repeats (SSRs) have become one of the most useful molecular marker systems in plant breeding [32]. They are widely used in cultivar fingerprinting, genetic diversity assessment, molecular mapping, and marker assisted breeding. The development of SSR markers from genomic libraries is expensive and at times inefficient [33]. However, with the availability of large numbers of expressed sequence tags (ESTs), development of SSR markers from ESTs through data mining has become an efficient and low cost option for many plant species. In this study, MISA http://pgrc.ipk-gatersleben.de/misa was used to identify SSR markers in the unique sequences, and the results showed that 4,352 sequences were examined, with a total size of 2,455,352 bp, including 1,233 SSRs distributed in 935 sequences. 225 sequences were examined in more than 1 SSR loci and 179 SSR loci were present in compound formation. The SSRs found are summarized in Table 3. There were a total of 1,233 SSR sites comprising of 438 (35.52%) dinucleotides, 510 (41.36%) trinucleotides, 159 (12.9%) tetranucleotides, 53 (4.3%) pentanucleotides, and 73 (5.92%) hexanucleotides in the EST datasets. 41.36% (510) of these are the tri-nucleotide repeats (Table 3), which is in agreement with previous studies of other plant species [34-36]. The main type of dinucleotides was AG/CT with a frequency of 93.15% (408) and the second was AC/GT (16, 3.65%). The most frequent tri-nucleotide repeat was AAG/CTT (195, 38.24%) described in additional file 3.
Table 3.
Distribution to different repeat type classes of EST library in mei.
| Unit size | Number of SSRs | Percent (%) | Frequency (%) | Average distance(Kb) |
|---|---|---|---|---|
| 2 | 438 | 35.52 | 10.06 | 5.61 |
| 3 | 510 | 41.36 | 11.72 | 4.81 |
| 4 | 159 | 12.9 | 3.65 | 15.44 |
| 5 | 53 | 4.3 | 1.22 | 46.33 |
| 6 | 73 | 5.92 | 1.68 | 33.63 |
| Total | 1233 | 100 | 28.33 | 1.99 |
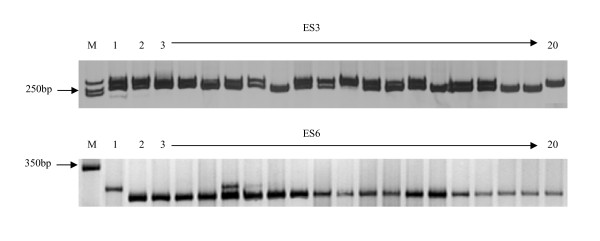
To investigate whether the potential SSR loci mined are the true-to-type one and can be used for genetic analysis, 42 EST-SSR primer pairs were designed and verified in 20 mei cultivars. The PCR amplified results showed that only 14 (33.3%) could generate clear DNA banding patterns with the expected size. Two polyacrylamide gel electrophoresis patterns (amplified with ES3 and ES6 primers) are shown as examples in Figure 2. The information of these SSRs primers are shown in Table 4.
Figure 2.
Polyacrylamide gel electrophoresis patterns of microsatellite alleles amplified in P. mume using ES3 and ES6 SSR markers. M: Maker DL2000; 1-20: Accession numbers of P. mume, representing cultivar names were listed in Table 5.
Table 4.
Summaries of EST-SSR primers and repeat motif.
| Primer | Repeat motif | Primer sequence (5'→3')* | Tm(°C) | Expected product (bp) |
|---|---|---|---|---|
| ES1 | (AG)10 | F: CTCTTGTTTCTCCTCCTACT | 57.6 | 253 |
| R: GACGAGTTTATCGTTGACT | ||||
| ES2 | (TG)13+(TC)20 | F: TTGAGGTTGAGCCTACAC | 56 | 306 |
| R: GCAAGGTCACTATCTTTTTC | ||||
| ES3 | (TGG)7+(TGGTGT)3 | F: GCTATACACGCTGCTAATA | 54.6 | 259 |
| R: AAGGCATCACATCAACAC | ||||
| ES4 | (AGC)7 | F: CCGTAATAACCACCGTCC | 58.2 | 240 |
| R: CCGCCTTCATCATCCTCT | ||||
| ES5 | (CAA)8 | F: CCAGATCCACTATTTCTTC | 55.9 | 267 |
| R: GTGTTAGAGCCAGAAACC | ||||
| ES6 | (AGTG)5 | F: GCACTCTTCTCTCTCTCTCT | 56.5 | 312 |
| R: GAGACCTTATGGAAGAAAAC | ||||
| ES7 | (TTTTG)4 | F: GAGAGAGACAAACAAGTGAA | 55.5 | 253 |
| R: CTTGAGGAGTGATTTCCTA | ||||
| ES8 | (TC)23 | F: ACAGTTTCAGAATCTCACAG | 57.6 | 284 |
| R: GATGGGACTTAAGAAGAGTC | ||||
| ES9 | (AG)15 | F: CCCTCTTATTCTCTCTCACT | 56.5 | 207 |
| R: CTTCAATATACTTGGTGAGC | ||||
| ES10 | (GA)22 | F: CTACGTACTTCCTGAGTGAG | 56.5 | 220 |
| R: CTAAAGATCGTTCAGACTGT | ||||
| ES11 | (GTCT)3(GA)13 | F: GGGTGTTGTGTCTGTTGGA | 58.7 | 287 |
| R: ACGAGGAAGATGAGGAGGG | ||||
| ES12 | (TTG)7 | F: TTCCTGCTATCTGCTCCAAT | 58.7 | 344 |
| R: GTGACGATGCTGTGCTCTGT | ||||
| ES13 | (AT)10 | F: GAGGAAATATTCCTGCATCA | 55.5 | 188 |
| R: CTGTTTCGTCATCTTTTTCC | ||||
| ES14 | (AGA)7(GGA)4 | F: ACATATCCACCACCACCAAC | 58.6 | 254 |
| R: AAAACAGAACACGACCCAGA |
* Every two primers belong to one pair. F is the forward and R is the reverse primer.
Transferability of P. mume EST microsatellite markers
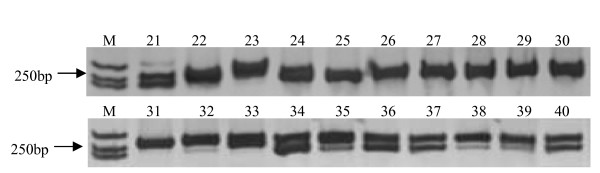
The application of a PCR-based marker system for comparative genomics would be highly desirable, because such a marker system can increase the efficiency of transferring genetic information across species. This transferability can also make the EST-SSR markers have good potential application in comparative genomics between various plants and in their genetic analysis [34,37,38]. In this study, 14 EST-SSR markers developed from P. mume ESTs were used to test the transferability on other two main Prunus species (peach and plum). Clear DNA fingerprints of PCR amplification using mei SSR primer could be obtained, such as the polyacrylamide gels of SSR bands amplified by primer pairs 3 (Figure 3). The results showed that 13 of the primer pairs could produce anticipated simple sequence repeat (SSR) bands in plum and 12 in peach, and the rate of transferability was about 92% and 86%, respectively. This result indicated that the EST-SSR primers generated from mei had a higher generality among Prunus species.
Figure 3.
Polyacrylamide gel images of SSR bands amplified in 20 cultivars of P. persica (peach) and P. salicina (plum) by primer pairs 3. M: Maker DL2000; 21-30, 31-40: Accession numbers of P. persica and P. salicina, the cultivar names were listed in Table 5.
Analysis of genetic diversity
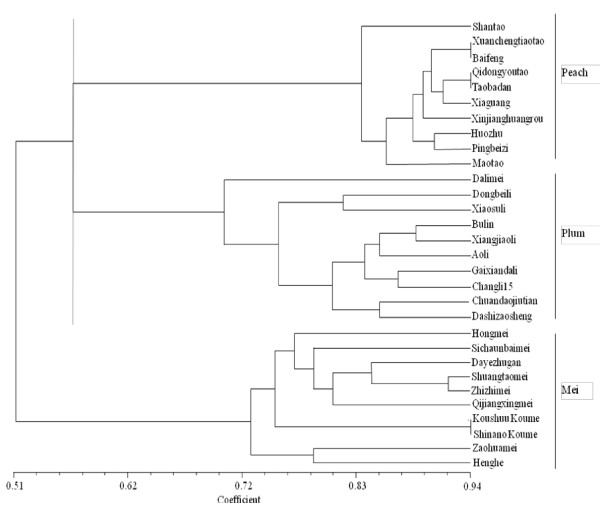
The genetic diversity among 30 varieties of peach, plum, and mei was analyzed with the new SSR primer pairs developed. 54 bands were totally amplified and high level of band polymorphism rate was observed in the mei species (70%), lower in plum (65%) and lest in peach (46%). A clustering diagram (Figure 4) showed that cultivars of these three species were divided into three groups, and the genetic difference among them was obvious, which was consistent with the fact that they are three different species. In the peach group, varieties belong to P. persica (L.) Batsch, such as 'Xuanchengtiantao' and 'Baifeng', 'Qidongyoutao' and 'Taobadan', 'Huozhu' and 'Pingbeizi' were clustered into three subgroups, with high similarity coefficients of 0.94, 0.94, 0.90, respectively. In the plum group, cultivars that had same geographical distribution were clustered together, such as 'Bulin' and 'Xiangjiaoli', with a genetic similarity coefficient of 0.88. In the mei group, all the ten cultivars were separated well, with 'Koushuu Koume' and 'Shinano Koume' introduced from Japan being clustered together. These results indicated that these P. mume SSR markers were useful in the evaluation of genetic relationships between and within the Prunus species.
Figure 4.
A dendrogram constructed using polymorphisms at 14 SSR loci with a total of 54 alleles. All these varieties of the three species were analyzed into three groups. The ten cultivars above were belong to P. persica marked as peach and middle were P. salicina marked as plum, down were P. mume marked as mei.
Discussion
Mei is an important fruit crop and famous ornamental tree in China, with processed mei products playing an important role in the economy and in people's daily lives. Expressed sequence tags (ESTs) are sequenced portions of messenger RNA, the projects of EST have been initiated for numerous plant and animal species, and generated large amount of sequence information that can be used for gene discovery, functional genetic studies, and marker development [39]. As we know, much more molecular researches have been carried out on mei, but the studies on flower and fruit development are poor. In this study, a high quality cDNA library of mei containing highly variable EST sequences was constructed, using a mixture of RNAs from plant materials at various stages, so as to understand the molecular mechanisms of growth, development and metabolic regulation at different development stages. The sequencing of mei ESTs can definitely provide an important platform for the studies of functional genomics, and can greatly accelerate the new gene discovery and study on EST-SSR development of P. mume.
In this study, the results of EST annotation and functional classification show that 1,130 unigenes have similarities to Vitis vinifera, Populus trichocarpa and P. persica. In the classification of known or putative functions (Additional file 4), the largest proportion of functionally assigned ESTs fell into three categories: primary transport and metabolism, protein synthesis and energy. Alignment of present ESTs was useful for comparative genomics. In this paper, we first compared the ESTs of P. mume with other Prunus plants, the results showed that these ESTs sequences were conserved also had high homology with other Prunus, which providing an important information for comparative genomics in Prunus species.
The scope of EST-SSR marker development is limited to the species for which sequence databases already exist. The SSR marker development for plant species lacking a sequence database can still be expensive and time consuming. An alternative approach for SSR marker development in those species could be the utilization of SSR markers from related species [32]. As the EST-SSR markers are derived from transcribed regions of DNA, they are expected to be more conserved and have a higher rate of transferability and polymorphism than genomic SSR markers [40]. By virtue of the sequence conservation of transcribed regions of the genome, a significant portion of the primer pairs designed from EST-SSRs is expected to function in distantly related species [36]. Transferability of EST-derived markers over different taxonomic levels has been demonstrated earlier [27,41]. In our study, the majority of our 14 EST-derived SSR loci from P. mume revealed cross-species amplification with alleles of comparable sizes in peach and plum. As expected, the transferability rate of the markers was high, which suggests that the flanking regions of these SSR loci are sufficiently conserved, and can be used for comparative analyses of genetic diversity, population structures and so on. It also indicated that these SSRs generated from P. mume can be useful for the development of conserved markers for linkage mapping and QTL analyses in different Prunus species.
In addition, the polymorphism rate in peach (46%) was lower than in plum (65%) and mei (70%). This may be due to the fact that most of the peach cultivars were from the same breeding program and thus have similar genetic background. The plum and mei cultivars have wider geographical distribution and bigger genetic differences. A dendrogram was constructed using the polymorphic bands in 30 cultivars of P. mume, P. persica and P. salicina, the relationships and diversity among these three species were analyzed accurately and can be roughly and visually reflected from the dendrogram. These results further suggest the applicability of mei EST-SSR markers in genetic study of Prunus species.
Conclusion
This paper provides the first P. mume flower and fruit cDNA library. In addition, large scale and valuable EST information which can be great help to some further research about gene cloning, gene function and expression analysis of P. mume were generated. Our analysis further highlights the efficient identification of SSR from mei ESTs and the high transferability of mei EST-SSR markers between peach and plum.
Methods
Sample collection and DNA isolation
Fruits and flowers of P. mume 'Xiyeqing' at different development stages were collected from the fruiting mei resource nursery of Nanjing Agricultural University, Najing PRC. The materials were collected from February 23, 2009 to May 6, 2009 and the collected tissues were frozen immediately in liquid nitrogen and stored at -40°C awaiting RNA extraction. The flowers were collected at four growing stages including small bud, medium bud, half-unfolding flower and completely unfolding flower, while the fruits were collected for seven weeks until the stone hardening stage, with the average size of 0.3, 0.6, 0.9, 1.2, 1.8, 2.6, 3.6 cm diameter.
All the samples of P. mume consisting of six introduced from Japan and used for EST-SSR analysis were collected on April 4, 2009 in this resource nursery and their names are described in Table 5. Cultivars of P. persica and P. salicina were obtained from National Germplasm Resources Garden of Peach in Jiangsu Academy of Agricultural Sciences, Najing PRC, the name of cultivars are also listed in Table 5. Genomic DNA was isolated from leaves using the DNeasy Plant Mini Kit (Qiagen, City, CA, USA) according to the manufacturer's recommendations.
Table 5.
List of the cultivars used in this study.
| No. | Cultivar name | Type | No. | Cultivar name | Type |
|---|---|---|---|---|---|
| 1 | 'Koushuu Koume' | P. mume | 21 | 'Shantao' | P. persica |
| 2 | 'Zaohuamei' | P. mume | 22 | 'Xuanchengtiantao' | P. persica |
| 3 | 'Lv' e mei' | P. mume | 23 | 'Maotao' | P. persica |
| 4 | 'Qijiangxingmei' | P. mume e | 24 | 'Qidongyoutao' | P. persica |
| 5 | 'Shuangtaomei' | P. mume | 25 | 'Huozhu' | P. persica |
| 6 | 'Sichaunbaimei' | P. mume | 26 | 'Taobadan' | P. persica |
| 7 | 'Zhizhimei' | P. mume | 27 | 'Xiaguang' | P. persica |
| 8 | 'Dayezhugan' | P. mume | 28 | 'Baifeng' | P. persica |
| 9 | 'Gyokuei' | P. mume | 29 | 'Xinjianghuangrou' | P. persica |
| 10 | 'Wanhong' | P. mume | 30 | 'Pingbeizi' | P. persica |
| 11 | 'Hongmei' | P. mume | 31 | 'Dalimei' | P. salicina |
| 12 | 'Oushuku' | P. mume | 32 | 'Dongbeili' | P. salicina |
| 13 | 'Henghe' | P. mume | 33 | 'Xiaosuli' | P. salicina |
| 14 | 'Xianmimei' | P. mume | 34 | 'Bulin' | P. salicina |
| 15 | 'Xiao'ou gongfen' | P. mume | 35 | 'Chuandaojiutian' | P. salicina |
| 16 | 'Shinano Koume' | P. mume | 36 | 'Dashizaosheng' | P. salicina |
| 17 | 'Taoxingmei' | P. mume | 37 | 'Aoli' | P. salicina |
| 18 | 'Momei' | P. mume | 38 | 'Xiangjiaoli' | P. salicina |
| 19 | 'Koushuu Saisyou' | P. mume | 39 | 'Gaixiandali' | P. salicina |
| 20 | 'Gojirou' | P. mume | 40 | 'Changli15' | P. salicina |
Note: The detail characteristic of P. mume cultivars are described in China fruit records-Mei and reference 2; the detail characteristic of P. persica cultivars are described in China fruit records-Peach and P. salicina are described in China fruit records-Plum.
cDNA library construction
Total RNA from the selected flowers and fruits of each stage was extracted by using Trizol (Takara Biotechnology, Dalian Co. Ltd.) respectively, and then 1 μg from each stage were mixed, while cDNA library construction was carried out according to the Creator SMART cDNA Construction Kit protocol (Clontech, USA, Cat.No.634903). The quantity and integrity of RNA was detected by BioPhotometer (Eppendorf, Gene Company Limited) while electrophoresis was done on agarose with Ethidium Bromide. The first strand and double strand cDNA were synthesized according to the protocol of Creator SMART cDNA Construction Kit (Clontech, USA, Cat.No.634903).
For first strand cDNA synthesis: 3 μl RNA sample, 1 μl SMART IV oligonucleotide (5'-AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG-3') and 1 μl CDS-3 M adapter (10 μM, 5'-AAGCAGTGGTATCAACGCAGAGTGGCCGAGGCGGCC (T)20VN-3') were mixed and incubated at 72°C for 2 min and placed in ice for another 2 min. 2.0 μl 5 × First-Strand Buffer, 1.0 μl DDT (20 mM), 1.0 μl dNTP Mix (10 mM), 1.0 μl PowerScript™ Reverse Transcriptase (Invitrogen Biotechnology Co., Ltd) were added to a total volume of 10 μl, mixed well and incubated at 42°C for 1 hour.
Double strand cDNA synthesis: The single strand cDNA was amplified for double strand cDNA synthesis by long-distance polymerase chain reaction (LD-PCR), in the following steps, 2 μl single-strand cDNA were added to 80 μl ddH2O, 10 μl 10 × Advantage 2 PCR Buffer, 2 μl 50 × dNTP Mix, 4 μl M1 PCR Primer (10 μM, 5'-AAGCAGTGGTATCAACGCAGAGT-3'), and 2 μl 50 × Advantage 2 Polymerase Mix then topped with ddH2O to make a total volume of 100 μl. The mixture was centrifuged then amplified as follows: 95°C for 1 min; 20 cycles of 95°C for 15 sec and 68°C for 6 min. Five microliters of the centrifuged PCR product were pipetted onto a 1.2% agarose/EtBr gel to check the double strand (ds)-cDNA quality. The concentration of the ds-cDNA was estimated by comparing with a suitable DNA marker.
The remaining double-strand cDNA was digested by SfiI Enzyme then sized, purified by a QIA quick PCR Purification Kit (Qiagen Cat.No.2810) and packaged in order to filter the fragments shorter than 400 bp. Double strand cDNA with cohesive ends was finally ligated into pDNR-LIB vector by T4 ligase, the recombinant vector was transformed into Escherichia coli DH10B by electroporation at 2.1 KV, cultured overnight in 37°C after directly applying onto a media plate with chloramphenicol.
Sequencing for the clones
After the cDNA library was plated onto LB media plates, the white clones were picked into a 96-well block containing 800 μl LB culture media (supplied with chloramphenicol 50 mg/ml) and incubated at 230 rpm and 37°C overnight. Before large scale sequencing, PCR reaction was initially conducted to check the size of inserted fragments using random selection of 30 clones [42]. Finally, about 10,000 fresh clones were sent to Beijing Huada Gene Company (Beijing, P.R. China) for sequencing.
EST processing
Among the vector sequences, low quality and redundant sequences were rejected with Chromas, Phred [43] and phd2fasta. Sequences longer than 100 bp were outputted after converting to FASTA forms. Uniform sequences were assembled using CAP3 [44] software for splicing into contigs with two or more ESTs.
EST annotation and function
For further functional annotation, comparative and classification analysis, the non-redundant sequences (edited ESTs sequences) considered as valid were subjected to BlastX analysis against the non-redundant protein database (nr) of National Center of Biotechnology Institute (NCBI) to search for similarity. Search results were imported into Microsoft Office Excel where sequence matches with E-value scores ≤10-10 were considered significant and used to categorize the ESTs based on their putative or known functions of plant genes.
EST alignment against Prunus EST database
The software package of blast was download from the FTP server ftp://ftp.ncbi.nlm.nih.gov/blast/executables/ of NCBI, then compiled and installed according to the instructions [45]. Alignment of nucleic acids were carried on among the ESTs of P. persica, P. armeniaca submitted in NCBI (by the end of April 1, 2010) and unigenes generated from P.mume cDNA library by using blastn with e-10 as the parameter.
Identification of EST-SSRs
The EST library was searched for sequences containing SSRs using MISA [37], a Perl script able to detect perfect as well as compound microsatellites in nucleotide sequences. Compound microsatellites were defined as repeats interrupted by a non-repetitive sequence of a maximum 100 nucleotides. MISA was set with the following minimum length criteria for the extraction of repeated units (unit size/minimum number of repeats): at least six dinucleotides (2/6); at least five trinucleotides (3/5); three tetranucleotides (4/3), three pentanucleotides (5/3) and three hexanucleotides (6/3). Sequences contained corresponding repeat units were selected for marker development.
EST-SSR primers designed and PCR amplication
Primer pairs flanking repeats with a minimum length 20 bp were designed using by Primer3.0 program http://frodo.wi.mit.edu/primer3/ and the parameters are as follows: Primer GC%: 40% ~ 60%, and the optimum value of 50%; Primer size: 18 ~ 22 bp; Primer TM: 50 ~ 60°C; Product size ranges: 150-350 bp. The primers designed were also identified by Oligo6 software in order to avoid the primer dimers, hairpin structure and the occurrence of mismatch and so on. Primers were synthesized by Invitrogen, Shanghai PR China.
The PCR amplification reactions were performed using an Eppendorf Authorized Thermal Cycler each and conducted in a total volume of 25 μl containing 50 -100 ng template DNA, 20 pmol each primer, 2 Units Taq DNA polymerase (purchased from Takara Biotechnology, Dalian Co. Ltd.), 2.5 μl 10 × PCR buffer, 2 μl MgCl2 (2.5 mM), 1.5 μl dNTPs (2.5 mM), and added ddH2O to the total volume. The PCR conditions were: 94°C for 5 min, followed by 35 cycles of 94°C for 30 sec, 50 ~ 60°C for 40 sec, 72°C for 1 min, and a final extension step at 72°C for 10 min. To check for PCR product quality, 8 μl of the PCR reaction was visualized on a 1.5% agarose gel and the remaining reaction was then electrophoresis on 5% polyacrylamide gel in 1 × TBE buffer at 80 W for 100 to 120 min. Gels were stained with silver nitrate following the protocol detailed by Bassam [46] with few modifications and photographed in white light.
Cross-species amplification
To assess the transferability of our EST-SSR markers, we tested their amplification in other Prunus species containing 10 cultivars of P. persica (peach) and 10 of P. salicina (plum). The PCR amplification reactions were performed as the same as above.
Data Analysis
According to presence or absence of bands on the polyacrylamide gel, the same electrophoretic bands were recorded as 1, no bands were recorded as 0. The polymorphisms data were inputted into Excel for the cluster analysis using NYSYS-pc software and a dendrogram was generated for the analysis of genetic relationship and diversity among the varieties of peach, plum and mei.
Authors' contributions
LX carried out the laboratory work and participated in manuscript draft writing. SL performed data analyses. SC, WC, GZ, and YH participated in the design and coordination the study. FJ conceived, designed the study and revised this paper. All authors read and approved the final manuscript.
Supplementary Material
Length distribution of uineges in the flower and fruit cDNA library of fruiting mei. This graph shows the description of the length distribution of uineges in the flower and fruit cDNA library of fruiting mei.
Composition of contigs. This table shows the composition of contigs in this study.
Types and frequency of SSRs in this study. Tables show the details of classified repeat types (considering sequence complementary).
Classification of P. mume unigenes with known or putative functions. This graph shows the detail classification of P. mume unigenes with known or putative functions.
Contributor Information
Xiaoying Li, Email: 2009204001@njau.edu.cn.
Lingfei Shangguan, Email: 2008104020@njau.edu.cn.
Changnian Song, Email: 2008204002@njau.edu.cn.
Chen Wang, Email: 2008104022@njau.edu.cn.
Zhihong Gao, Email: gaozhihong@njau.edu.cn.
Huaping Yu, Email: 2008104024@njau.edu.cn.
Jinggui Fang, Email: fanggg@njau.edu.cn.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (No. KYJ200909) and the Program of NCET (No. NCET96).
References
- Lu Y. Discussion on translation of Prunus mume name into English. Journal of Beijing Forestry University (The International Symposium on Mei and Wintersweets' Culture, Special Issue) 2000. pp. 17–18.
- Fang JG, Twito T, Zhang Z, Chao CT. Genetic relationships among fruiting-mei (Prunus mume Sieb.et Zucc.) cutivars evaluated with AFLP and SNP markers. Genome. 2006;49:1256–1264. doi: 10.1139/G06-097. [DOI] [PubMed] [Google Scholar]
- Chu MY. China Fruit Records - Mei (in Chinese) China Forestry Press, Beijing; 1999. [Google Scholar]
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert H. Genomic approaches to plant stress tolerance. Curr Opin Plant Biol. 2000;3:117–124. doi: 10.1016/S1369-5266(99)00052-7. [DOI] [PubMed] [Google Scholar]
- Höfte H, Desprez T, Amselem J, Chiapello H, Rouzé P, Caboche M, Moisan A, Jourjon MF, Charpenteau JL, Berthomieu P, Guerrier D, Giraudat J, Quigley F, Thomas F, Yu DY, Raynal M, Cooke R, Grellet F, Marcillac P, Gigot C, Fleck J, Philipps G, Axelos M, Bardet C, Tremousaygue D, Lescure B. An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J. 1994;4:1051–1061. doi: 10.1046/j.1365-313X.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Demura T, Galis I, Horiguchi T, Sasaki M, Tashiro G, Fukuda HA. A Comprehensive Gene Expression Analysis Towards the Understanding of Growth and Differentiation of Tobacco BY-2 Cells. Plant Cell Physiol. 2004;45:1280–1289. doi: 10.1093/pcp/pch155. [DOI] [PubMed] [Google Scholar]
- Park JS, Kim JB, Haha BS, Kim KH, Ha SH, Kim JB, Kim YH. EST analysis of genes involved in secondary metabolism in Camellia sinensis (tea) using suppression subtractive hybridization. Plant Sci. 2004;166:953–961. doi: 10.1016/j.plantsci.2003.12.010. [DOI] [Google Scholar]
- Remy I, Michnick SW. A cDNA library functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods. 2004;32:381–388. doi: 10.1016/j.ymeth.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Sumner LW. MedicCyc: a Biochemical pathway database for Medicago truncatula. Bioinformatics. 2007;23:1418–1423. doi: 10.1093/bioinformatics/btm040. [DOI] [PubMed] [Google Scholar]
- Sugui JA, Deising HB. Isolation of infection-specific sequence tags expressed during early stages of maize anthracnose disease development. Mol Plant Patho. 2002;3:197–203. doi: 10.1046/j.1364-3703.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- Moser C, Segala C, Fontana P, Salakhudtinov I, Gatto P, Pindo M, Zyprian E, Toepfer R, Grando MS, Velasco R. Comparative analysis of expressed sequence tags from different organs of Vitis vinifera L. Funct Integr Genomics. 2005;5:208–217. doi: 10.1007/s10142-005-0143-4. [DOI] [PubMed] [Google Scholar]
- Park S, Sugimoto N, Larson MD, Beaudry R, Nocker S. Identification of genes with potential roles in apple fruit development and biochemistry through large-scale statistical analysis of expressed sequence tags. Plant Physiol. 2006;141:811–824. doi: 10.1104/pp.106.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada-Akiyama H, Akiyama Y, Ebinaa M, Xu Q, Tsuruta S, Yazaki J, Kishimoto N, Kikuchi S, Takahara M, Takamizo T, Sugita S, Nakagawa H. Analysis of expressed sequence tags in apomictic guineagrass (Panicum maximum) J Plant Physiol. 2009;166:750–761. doi: 10.1016/j.jplph.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhao L, Gao Q. Generation and analysis of expressed sequence tags from the tender shoots cDNA library of tea plant (Camellia sinensis) Plant Sci. 2005;168:359–363. doi: 10.1016/j.plantsci.2004.08.009. [DOI] [Google Scholar]
- Zhao L, Ma C, Chen L. Construction and Expressed Sequence Tags Analysis of Young Roots cDNA Library of Tea Plant. Mol Plant Breeding. 2008;6:893–898. [Google Scholar]
- Forment J, Gadea J, Huerta L, Abizanda L, Agusti J, Alamar S, Alos E, Andres F, Arribas R, Beltran JP. Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol Biol. 2005;57:375–391. doi: 10.1007/s11103-004-7926-1. [DOI] [PubMed] [Google Scholar]
- Jiang YQ, Ma RC. Generation and analysis of expressed sequence tags from almond (Prunus dulcis Mill.) pistils. Sex Plant Reprod. 2003;16:197–207. doi: 10.1007/s00497-003-0190-8. [DOI] [Google Scholar]
- Folta KM, Staton M, Stewart PJ, Jung S, Bies DH, Jesdurai C, Main D. Expressed sequence tags (ESTs) and simple sequence repeats (SSR) markers from octoploid strawberry (Fragaria × ananassa) BMC Plant Biol. 2005;5:12. doi: 10.1186/1471-2229-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Romieu C, Audergon JM, Marty I, Albagnac G, Lambert P, Bouchet JP, Terrier N. Transcriptomic study of apricot fruit (Prunus armeniaca) ripening among 13,006 expressed sequence tags. Physiol Plant. 2005;125:281–292. doi: 10.1111/j.1399-3054.2005.00563.x. [DOI] [Google Scholar]
- Lewers KS, Saski CA, Cuthbertson BJ, Henry DC, Staton ME, Main DS, Dhanaraj AL, Rowland LJ, Tomkins JP. A blackberry (Rubus L.) expressed sequence tag library for the development of simple sequence repeat markers. BMC Plant Biol. 2008;8:69. doi: 10.1186/1471-2229-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchietti A, Lazzari B, Ortugno C, Bianchi F, Malinverni R, Caprera A, Mignani I, Pozzi C. Comparative analysis of expressed sequence tags from tissues in ripening stages of peach (Prunus persica L. Batsch) Tree Genet Genomes. 2009;5:377–391. doi: 10.1007/s11295-008-0193-6. [DOI] [Google Scholar]
- Xie H, Sui Y, Chang FQ, Xu Y, Ma RC. SSR allelic variation in almond (Prunus dulcis Mill) Theor Appl Genet. 2006;112:366–372. doi: 10.1007/s00122-005-0138-5. [DOI] [PubMed] [Google Scholar]
- Chen C, Bowman KD, Choi YA, Dang PM, Rao MN, Huang S, Soneji JR, Mccollum TG, Gmitter JR. EST-SSR genetic maps for Citrus sinensis and Poncirus trifoliate. Tree Genet Genomes. 2008;4:1–10. doi: 10.1007/s11295-007-0083-3. [DOI] [Google Scholar]
- Silfverberg-Dilworth E, Matasci CL, Vandeweg WE, Kaauwenm PW, Walser M, Kodde LP, Sogliov V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A. Mierosatellite markers spanning the apple (Malus domestica Borkh.) Tree Genet Genomes. 2006;2:202–224. doi: 10.1007/s11295-006-0045-1. [DOI] [Google Scholar]
- Bassil NV, Njuguna W, Slovin JP. EST-SSR markers from Fragaria vesca L. cv. Yellow Wonder. Mol Ecol Notes. 2006;6:806–809. doi: 10.1111/j.1471-8286.2006.01351.x. [DOI] [Google Scholar]
- Vendramin E, Deitori MT, Giovlnazzi J, Micali S, Quarta R, Verde I. A set of EST-SSRs isolated from peach fruit transcriptome and their transportability across Prunus species. Mol Ecol Notes. 2007;7:307–310. doi: 10.1111/j.1471-8286.2006.01590.x. [DOI] [Google Scholar]
- Rossetto M, Mcnally J, Henry RJ. Evaluating the potential of SSR flanking regions for examining taxonomic relationships in the vitaceae. Theor Appl Genet. 2002;104:61–66. doi: 10.1007/s001220200007. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nakatsuka A, Yano K, Yasugahira S, Nakamura R, Sun N, Itai A, Suzuki T, Itamura H. Expressed sequence tags from persimmon at different developmental stages. Plant Cell Rep. 2008;27:931–938. doi: 10.1007/s00299-008-0518-9. [DOI] [PubMed] [Google Scholar]
- Da Silva FG, Iandolino A, Al-Kayal F, Bohlmann MC, Cushman MA, Lim H, Ergul A, Figueroa R, Kabuloglu EK, Osborne C, Rowe J, Tattersall E, Leslie A, Xu J, Baek J, Cramer GR, Cushman JC, Cook DR. Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol. 2005;139:574–597. doi: 10.1104/pp.105.065748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausher M, Shatters R, Chaparro J, Dang P, Hunter W, Niedz R. An expressed sequence tag (EST) set from Citrus sinensis L. Osbeck whole seedlings and the implications of further perennial source investigations. Plant Sci. 2003;165:415–422. doi: 10.1016/S0168-9452(03)00202-4. [DOI] [Google Scholar]
- Saha MC, Mian MA, Eujayl I, Zwonitzer JC, Wang L, May GD. Tall fescue EST-SSR markers with transferability across several grass species. Theor Appl Genet. 2004;109:783–791. doi: 10.1007/s00122-004-1681-1. [DOI] [PubMed] [Google Scholar]
- Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W. How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol. 2003;12:1339–1348. doi: 10.1046/j.1365-294X.2003.01825.x. [DOI] [PubMed] [Google Scholar]
- Kantety RV, La Rota M, Matthews DE, Sorrells ME. Data mining for simple sequence repeats in expressed sequence tags from bar ley, maize, rice, sorghum and wheat. Plant Molecular Biology. 2002;48:501–510. doi: 10.1023/A:1014875206165. [DOI] [PubMed] [Google Scholar]
- Liang X, Chen X, Hong Y, Liu H, Zhou G, Li S, Guo B. Utility of EST derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol. 2009;9:35. doi: 10.1186/1471-2229-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia M, Oger-Desfeux C, Marais G, Widmer A. A White Campion (Silene latifolia) floral expressed sequence tag (EST) librar y: annotation, EST-SSR characterization, transferability, and utility for comparative mapping. BMC genomics. 2009;10:243. doi: 10.1186/1471-2164-10-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Eujayl I, Sledge MK, Wang L, May GD, Chekhovskiy K, Zwonitzer JC, Mian MA. Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theor Appl Genet. 2004;108:414–422. doi: 10.1007/s00122-003-1450-6. [DOI] [PubMed] [Google Scholar]
- Pashley CH, Ellis JR, McCauley DE, Burke JM. EST databases as a source for molecular markers: lessons from Helianthus. Journal of Heredity. 2006;97:381–388. doi: 10.1093/jhered/esl013. [DOI] [PubMed] [Google Scholar]
- Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ. Analysis of SSRs derived from grape ESTs. Theor Appl Genet. 2000;100:723–726. doi: 10.1007/s001220051344. [DOI] [Google Scholar]
- Decroocq V, Fave MG, Hagen L, Bordenave L, Decroocq S. Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet. 2003;106:912–922. doi: 10.1007/s00122-002-1158-z. [DOI] [PubMed] [Google Scholar]
- Gao QK, Hu C. Construction of a cDNA library of host recognition kairomone for telenomus theophilae. Entomol Sin. 2001;9:35–39. [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Huan X, Madan A. A DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- README for stand alone. Blast [EB/OL] 2002. pp. 4–23.ftp://ftp.ncbi.nih.gov/blast/executables/blast+/LATEST/user_manual.pdf
- Bassam BJ, Caetano-anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Length distribution of uineges in the flower and fruit cDNA library of fruiting mei. This graph shows the description of the length distribution of uineges in the flower and fruit cDNA library of fruiting mei.
Composition of contigs. This table shows the composition of contigs in this study.
Types and frequency of SSRs in this study. Tables show the details of classified repeat types (considering sequence complementary).
Classification of P. mume unigenes with known or putative functions. This graph shows the detail classification of P. mume unigenes with known or putative functions.