Abstract
We adapted the method of epitope mapping by site-directed masking, which was described for purified soluble antigens [Paus,D. and Winter,G. (2006) Proc. Natl Acad. Sci. USA, 103, 9172–9177.], to map the binding site of an inhibitory monoclonal antibody on the cell surface protein ecto-nucleotidase NTPDase3. Using homology modeling, we built a 3D structure of NTPDase3 and designed 21 single cysteine mutations distributed over the surface of the enzyme. The mutant proteins were expressed in cells, biotinylated with a cysteine-specific reagent, and then extracted with detergent and immobilized on streptavidin-coated plates. Tethering NTPDase3 via cysteine residues located in a surface patch near the active site cleft masked the epitope and blocked antibody binding, as evaluated by enzyme inhibition assay and by ELISA. We then constructed 18 single alanine substitution mutations within the defined patch and found that W403A, D414A, E415A and R419A decreased the inhibitory effect of the antibody, whereas the double mutation W403A/R419A abolished both antibody binding and enzyme inhibition, suggesting the critical role of these residues for interaction with the antibody. Lack of competition between the antibody and a non-hydrolyzable substrate analog AMPPCP, as well as location of the epitope adjacent to the active site, suggest a noncompetitive mechanism of inhibition by steric hindrance. The described technique should be useful for systematic epitope mapping in cell membrane proteins for which either a 3D structure is available, or a sufficiently accurate 3D model can be obtained by homology modeling.
Keywords: alanine scanning mutagenesis, ecto-nucleotidase, epitope mapping, NTPDase, site-directed masking
Introduction
Identification of discontinuous B-cell epitopes remains a challenging task with no common solution despite a variety of available experimental techniques and epitope prediction methods (Gershoni et al., 2007; Ponomarenko et al., 2008; Yang and Yu, 2009). The recently invented technique of site-directed masking offers a systematic means of epitope mapping, if a three-dimensional structure of a protein antigen is available (Paus and Winter, 2006). In this technique, a panel of single cysteine mutations distributed evenly on a protein antigen surface are constructed, and then used to chemically tether the antigen via a cysteine residue and a linker to a solid phase, hence masking an area around the cysteine and preventing access of antibody to this area. The mutations which block antibody binding define the location of the antibody epitope, which can be further refined by alanine scanning mutagenesis. We adapted this technique, which was originally described for purified soluble antigens, to map epitopes in unpurified proteins expressed on the cell surface. We used this method to define the binding site of an inhibitory monoclonal antibody to human ecto-nucleotidase NTPDase3, and to gain insight into the mechanism of inhibition.
Plasma membrane nucleoside triphosphate diphosphohydrolases (NTPDase1, 2, 3 and 8) are integral membrane proteins with two membrane-spanning domains and a catalytic site facing the extracellular space (Robson et al., 2006). These enzymes hydrolyze the γ and β phosphates of extracellular nucleotides, and function as the major regulators and terminators of purinergic signaling (Kukulski et al., 2005). They are viewed as potential therapeutic targets, and efforts were made to develop subtype-selective NTPDase inhibitors (Gendron et al., 2002). Modulation of nucleotide signaling by specific NTPDase inhibitors is thought to have a profound impact on different physiological systems under normal and pathological conditions, and thus could find applications as both experimental and pharmacological tools. Many synthetic compounds have been reported to inhibit NTPDases, including nucleotide analogs and mimetics (Brunschweiger et al., 2008), sulfonated dyes (Iqbal et al., 2005; Munkonda et al., 2007) and polyoxometalates (Muller et al., 2006), but all of them have relatively low affinity to the enzymes, and none can clearly discriminate between different isoenzymes [discussed in (Munkonda et al., 2009)].
We have recently described a monoclonal antibody hN3-B3S as a novel and specific inhibitor of human NTPDase3 that decreased enzymatic activity by 60–90% depending on the assay conditions (Munkonda et al., 2009). Considering the location of NTPDases on the plasma membrane, which makes them readily accessible, as well as high affinity and selectivity characteristic of antibodies, the antibodies could function as potent and highly specific inhibitors of NTPDases both in vitro and in vivo. In order to better understand the mechanism of inhibition and to define the means for improvement of inhibitory activity of hN3-B3S, we report here the mapping of the antibody epitope in human NTPDase3 using a modification of the site-directed masking technique and a 3D structure of NTPDase3 generated by homology modeling. Our data suggest that hN3-B3S does not compete with substrate, but rather binds NTPDase3 in close proximity to the active site and inhibits the enzymatic activity by steric hindrance. The described method is likely to be useful for systematic epitope mapping in unpurified cell surface proteins for which a 3D structure is available, or can be obtained with adequate accuracy by homology modeling.
Materials and methods
Monoclonal antibody
Production and characterization of monoclonal antibodies hN3-B3S and hN3-H10S has been described (Munkonda et al., 2009). These antibodies are specific to human NTPDase3 and are unique in their ability to inhibit enzymatic activity of NTPDase3 by 60–90% depending on conditions. hN3-B3S and hN3-H10S antibodies are produced by two hybridoma clones, but they performed similarly in all assays, and therefore are likely to be identical in structure. In the majority of experiments, we used the antibody produced by hN3-B3S clone. The antibody was affinity purified from hybridoma medium by Protein A chromatography, combined with glycerol (10% final concentration), aliquoted and stored at −80°C.
Site-directed mutagenesis, transfection and preparation of COS cell membranes
The cDNA clone encoding wild-type human NTPDase3 ligated into the pcDNA3 mammalian expression vector was described (Smith and Kirley, 1998), GenBank AF034840.2. For mutagenesis, C10S mutation of the wild-type NTPDase3 was used as a background. This mutation removes Cys10 from the N-terminal cytoplasmic tail and eliminates artificial dimerization of the protein via intermolecular disulfides formed by oxidation of Cys10, which occurs to varying degree during preparation of cell membranes containing wild-type NTPDase3. The C10S mutant is a fully active, wild-type-like enzyme (Murphy et al., 2002). Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions. The presence of correct mutations was confirmed by sequencing.
COS-1 cells were transfected with 4 µg plasmid DNA per 100 mm plate using Lipofectamine and Plus reagents (Invitrogen). Transfection with an empty pcDNA3 vector was also performed as a control. Approximately 48 h post-transfection, the COS-1 cells were harvested by scraping in the buffer (30 mM MOPS-NaOH, pH 7.4, 2 mM EDTA, 250 mM sucrose), and the crude cell membrane preparations were obtained by centrifugation as described (Smith and Kirley, 1998).
Protein concentrations were determined using the Bio-Rad protein assay reagent with the modifications of Stoscheck (Stoscheck, 1990), using bovine serum albumin as the standard.
ATPase assay
ATPase activity in biotinylated membrane preparations was determined by measuring the amount of inorganic phosphate (Pi) released from ATP (2.5 mM ATP, 5 mM MgCl2 and 20 mM MOPS-NaOH, pH 7.4) at 37°C using the Fiske and Subbarow technique (Fiske and Subbarow, 1925) as described (Smith and Kirley, 1999a,b). In all other experiments, 0.3 mM ATP was used, and the released Pi was assayed with a more sensitive malachite green procedure (Baykov et al., 1988).
Biotinylation of cell surface proteins
Cell surface proteins were biotinylated with Nα-(3-maleimidylpropionyl)biocytin (biotin maleimide, Molecular Probes) that selectively reacts with sulfhydryl groups, and which is impermeable to the plasma membrane under conditions used, thus limiting biotinylation to the extracellular parts of cell surface proteins. One hundred millimeter plates containing transfected COS-1 cells were placed on ice, washed twice with ice-cold PBS, pH 7.0 and incubated with 250 µM biotin maleimide in PBS, pH 7.0, for 30 min at 4°C. After incubation, the cells were washed with ice-cold PBS and incubated with a quenching buffer (5 mM cysteine in PBS) for 10 min at 4°C. Cells were scraped in buffer (30 mM MOPS-NaOH, pH 7.4, 2 mM EDTA, 250 mM sucrose) and processed for cell membrane preparation.
Capture of biotinylated proteins with streptavidin agarose beads
Biotinylated membrane preparations (60 µg) were solubilized with 1% Triton X-100 containing 1 mM EDTA and 1% of Protease Inhibitor Cocktail Set III (Calbiochem) in a total volume of approximately 100 µl for 15 min at 21°C, and then centrifuged in an Airfuge for 20 min at 100 000×g. The supernatants were diluted with 1.2 ml TBS buffer (150 mM NaCl, 50 mM Tris–HCl, pH 7.5) containing 1% Triton X-100, and incubated with 15 µl ImmunoPure Immobilized Streptavidin agarose beads (Pierce) for 20 h with end-over-end rotation at 4°C. Beads were then washed three times with TBS containing 1% Triton X-100, and biotinylated proteins were eluted by re-suspending the beads in SDS loading buffer containing 200 mM DTT and boiling for 5 min. Beads were removed by centrifugation, and the supernatant was analyzed by western blotting for detection of biotinylated NTPDase3.
ELISA
Biotinylated NTPDase3 was immobilized on either transparent plastic High Binding EIA/RIA plates (Corning) for the ATPase assay, or black plastic High Binding EIA/RIA plates (Corning) for the assay of monoclonal antibody binding to the immobilized NTPDase3 using chemiluminescence detection. Plates were coated with 100 µl of 20 µg/ml streptavidin (Prozyme) in 50 mM sodium carbonate buffer, pH 9.6, and incubated for 20 h at 4°C. Subsequently, plates were washed twice with TBS, incubated with the blocking solution (4% BSA (Sigma) in TBS containing 0.02% NaN3 and 1 µg/ml streptavidin) for 2 h at 21°C, and either used for NTPDase3 immobilization, or stored at 4°C. To extract membrane-bound NTPDase3 for immobilization, 60 µg of biotinylated cell membrane preparation was combined with buffer (0.4% Triton X-100, 2 mM EDTA, 2% Protease Inhibitor Cocktail Set III (Calbiochem)) to a total volume of 120 µl, incubated for 15 min at 21°C, and then centrifuged in an Airfuge for 20 min at 100 000×g. The supernatant was diluted with 1.2 ml TBS containing 0.1% Triton X-100, and 100 µl were applied to plates, which have been pre-washed four times with TBS and once with TBS containing 0.1% Triton X-100. Binding of biotinylated NTPDase3 to immobilized streptavidin was accomplished by incubating plates for 18 h at 4°C. Membrane preparations from COS cells transfected with either the C10S mutant of NTPDase3 (which does not possess a free cysteine residue in the extracellular part and thus could not be biotinylated), or the empty pcDNA3 vector, were used as controls for non-specific binding to streptavidin-coated plates.
To assay the effect of the monoclonal antibody on ATPase activity of tethered NTPDase3, the plates were washed five times with TBS containing 0.1% Triton X-100 and three times with TBS. Subsequently, 100 µl of affinity-purified antibody (1 µg/ml hN3-B3S in TBS containing BSA at 1 mg/ml) was applied to odd-numbered wells. Affinity-purified polyclonal antibody KLH1 that recognizes the C-terminal peptide of human NTPDase3 (Smith and Kirley, 1999a,b), or a solution with no antibody (TBS containing BSA at 1 mg/ml) were applied to the even numbered wells as controls. After 3 h incubation at 21°C, plates were washed three times with TBS, and 100 µl of substrate solution (0.3 mM ATP, 5 mM MgCl2 and 20 mM MOPS-NaOH, pH 7.4) was added and incubated at 37°C in a water bath for 10–40 min, depending on the level of ATPase activity. While the subsequent determination of released Pi could be performed in the plates, we obtained more accurate results by transferring the reaction solution from the plate into 0.2 ml tubes and adding 25 µl of malachite green reagent (Baykov et al., 1988) into 16 tubes in 1 min intervals. After 16 min incubation, absorption of the samples was recorded in 1 min intervals at 630 nm using a Beckman Coulter DU800 spectrophotometer and a microcell, and the amount of released Pi was calculated.
For the assay of monoclonal antibody binding to tethered NTPDase3, black plastic plates were used to prevent light pollution from neighboring wells during chemiluminescence detection. After immobilization of biotinylated NTPDase3, the plates were washed five times with TBS containing 0.1% Triton X-100 and three times with TBS, blocked with 5% non-fat milk in TBS for 30 min at 21°C, and incubated with the antibody (1 µg/ml hN3-B3S in TBS containing 5% milk) for 18 h at 4°C. Plates were washed five times with TBS containing 0.1% Tween-20, and incubated with the secondary Stabilized Goat Anti-Mouse HRP-Conjugated (Pierce) antibody (1:2000 dilution in TBS containing 5% milk) for 2 h at 21°C. Plates were washed five times with TBS containing 0.1% Tween-20, and 100 µl of SuperSignal West Dura Extended Duration Substrate (Pierce) was added. Chemiluminescence intensity was recorded using FluorChem IS-8800 system (Alpha Innotech), and quantified using AlphaEaseFC software (Alpha Innotech).
SDS–PAGE and western blotting
Pre-cast 10-well or 15-well 4–15% gradient mini-gels (Bio-Rad) were used for SDS–PAGE. After electrophoresis, the proteins were electrotransferred to Immun-Blot™ PVDF membrane (Bio-Rad) and processed as described (Smith and Kirley, 1999a,b). NTPDase3 was detected using KLH11 rabbit polyclonal antiserum (Ivanenkov et al., 2005) at 1:5000 dilution and incubation overnight at 21°C, and a secondary Stabilized Goat Anti-Rabbit HRP-Conjugated antibody (Pierce) at 1:2000 dilution for 2 h at 21°C. To analyze the binding of the monoclonal hN3-B3S antibody to NTPDase3 mutants by western blotting, the electrophoresis was performed under non-reducing conditions, since reduction of disulfides abolishes the epitope recognition (Munkonda et al., 2009). hN3-B3S was used at 1 µg/ml and incubated overnight at 21°C, and a secondary Stabilized Goat Anti-Mouse HRP-Conjugated antibody (Pierce) was used at 1:2000 dilution and incubated for 2 h at 21°C. Immunoreactivity was detected by chemiluminescence using SuperSignal West Dura Extended Duration Substrate (Pierce), and was recorded using FluorChem IS-8800 system (Alpha Innotech), or Blue Autorad Film (BioExpress).
Slot blotting
Binding of monoclonal antibodies to non-denatured NTPDase3 proteins was analyzed using COS cell membranes immobilized on PVDF membrane by vacuum filtration. Aliquots of COS cell membranes diluted with TBS (0.1 µg total protein in 200 μl) were filtered in individual slots of a BioRad Bio Dot SF slot blot apparatus. After washing the slots with TBS buffer three times, the membranes were blocked with 5% milk in TBS and processed as described above for western blots.
Computer analyses
We used the FFAS fold recognition server (http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl) (Rychlewski et al., 2000; Jaroszewski et al., 2005) to build a 3D model of human NTPDase3 by threading the primary sequence of the extracellular portion of NTPDase3 through the recently described crystal structure of the extracellular portion of rat NTPDase2 (Zebisch and Strater, 2008) (PDB ID code 3CJ1, www.pdb.org). Several epitope prediction methods, DiscoTope (Haste Andersen et al., 2006), BEPro (PEPITO) (Sweredoski and Baldi, 2008) and ElliPro (Ponomarenko et al., 2008), were used in the design of alanine mutations for fine epitope mapping. PyMoL (www.pymol.org) was used to view 3D protein structures and prepare figures.
Results
3D model of NTPDase3 and design of cysteine mutations
We adapted the method of epitope mapping by site-directed masking, which was originally described for purified proteins (Paus and Winter, 2006), to proteins expressed on the cell surface (Fig. 1). We applied this technique to map the binding site of the inhibitory monoclonal antibody hN3-B3S, specific for human NTPDase3. Because this method requires a 3D structure of the protein of interest, but no such structure is available for human NTPDase3, we used the FFAS fold recognition server (Rychlewski et al., 2000; Jaroszewski et al., 2005) to make a 3D model of human NTPDase3 by threading the primary sequence of the extracellular portion of NTPDase3 through the crystal structure of the extracellular portion of rat NTPDase2 (Zebisch and Strater, 2008). Using this model, we designed 21 cysteine mutations scattered over the surface of NTPDase3 (Fig. 4). Most of these residues are well-exposed to solvent, and many of them represent homologous substitutions of cysteine for serine or threonine. Since our recent study suggested the involvement of the residue S297 and the 5th disulfide bond loop (C399-C422) in antibody binding (Munkonda et al., 2009), we targeted this area with a cluster of mutations (S297C, T299C, W403C, D414C, E415C, V416C and R419C).
Fig. 1.
Epitope mapping technique. (A) Cell surface protein (black circles) anchored to the plasma membrane (‘cell membrane’, double line) via a transmembrane domain (zigzag). A series of single cysteine mutations (‘cys’) distributed evenly on the protein surface is made. While the majority of cysteine mutations are likely to be located outside the antibody epitope (e.g. mutation ‘2’, right panel), some mutations are likely to be found within the epitope (e.g. mutation ‘1’ in left panel; the epitope is shown in grey). (B) Cell surface biotinylation results in covalent attachment of a biotin linker (‘biotin’) to cysteine residues. (C) The protein is extracted from the plasma membrane with detergent and tethered via the biotin linker to a streptavidin-coated plate (‘streptavidin plate’), thus masking the area around the tethered cysteine and blocking the access of antibody to this area. (D) Mutations that block antibody (‘mAb’) binding indicate the location of the epitope (mutation ‘1’ in the left panel, but not mutation ‘2’ in the right panel).
Epitope mapping by site-directed masking using ATPase inhibition assay
COS-1 cells were transfected with the plasmids containing cDNA encoding the desired mutations in NTPDase3, and after expression of the enzyme, the cells were biotinylated with a sulfhydryl-specific and plasma membrane-impermeable reagent (biotin maleimide), thus limiting biotinylation to the sulfhydryls in the extracellular parts of cell surface proteins. After biotinylation, specific ATPase activity of most NTPDase3 mutants (except A77C) was more than 25% of that for C10S, which was used as a background for cysteine mutations and is similar to the wild type (Fig. 2A). NTPDase3 mutants were then extracted from membranes with Triton X-100 and immobilized on streptavidin-coated plates via biotin–streptavidin interaction. For most mutants (except S87C, A267C and S394C), ATPase activity was sufficiently high for assay (Fig. 2B), and also remarkably stable, allowing several consecutive assays in the same plate with only a moderate decrease in activity. ATPase activity of immobilized enzymes (Fig. 2B) varied greatly between mutants, and did not correlate with moderate variations in activity in biotinylated membranes (Fig. 2A), suggesting differences in either reactivity or accessibility for biotinylation of the cysteine residues in different mutants.
Fig. 2.
Site-directed tethering of biotinylated NTPDase3 to streptavidin-coated plates, and the effect of hN3-B3s inhibitory antibody on ATPase activity of tethered enzymes. (A) ATPase activity of biotinylated cysteine mutants. Crude membrane preparations were obtained from COS transfected with mutant NTPDase3 plasmid cDNA, as indicated below the bars, and biotinylated before harvesting. C10S NTPDase3 mutant was used as the background for all cysteine mutations. This mutant does not contain a free cysteine residue in the extracellular part of the protein and is shown for comparison (see ‘Materials and Methods’ for details). ATPase activities were assayed using the same amount of membrane protein, and the activities are expressed in µmole of inorganic phosphate (Pi) released per h and per mg of crude membrane protein. (B) ATPase activity of biotinylated NTPDase3 mutants immobilized on streptavidin-coated plates. Equal amount of biotinylated membrane preparation of each mutant was treated with Triton X-100, and the extracted proteins were immobilized on streptavidin-coated plates. ATPase activity is expressed in nmole of inorganic phosphate (Pi) released per h and per plate well. (C) Effect of hN3-B3s inhibitory antibody on ATPase activity of immobilized NTPDase3 mutants. Biotinylated NTPDase3 mutants immobilized on streptavidin-coated plates were incubated with either hN3-B3S, or a BSA solution with no antibody (control), followed by ATPase assay. Values represent means ± SD for three or four plate wells. LA, enzymatic activity is too low to measure accurately. NA, no ATPase activity above the C10S background was detected.
We showed recently that hN3-B3S monoclonal antibody inhibited ATPase activity of human NTPDase3 by 60–90%, depending on the assay conditions (Munkonda et al., 2009). Tethering of NTPDase3 mutants via cysteine residues to plates masks an area centered on each cysteine residue. Consequently, location of a cysteine tether in the antibody binding site would prevent antibody binding and ATPase activity inhibition, thus designating the epitope. Therefore, we analyzed the inhibitory effect of hN3-B3s antibody on ATPase activity of immobilized NTPDase3 mutants (Fig. 2C). The mutants can be divided into two groups: in the first group (A77C, G122C, S162C, S178C, S246C, S279C, S340C, S387C and T439C), hN3-B3S decreases ATPase activity more than 30%, whereas in the second group (S101C, A112C, S297C, T299C, W403C, D414C, E415C, V416C and R419C), ATPase activity is no longer affected by the antibody. These data suggest that cysteine residues of the second group are located either in the epitope, or in close proximity to it.
Epitope mapping by site-directed masking using antibody binding assay
While ATPase inhibition assay proved to be a convenient approach to delineate the antibody binding site, its application is limited to enzymes, such as NTPDase3, and to inhibitory antibodies, such as hN3-B3S. We investigated whether the site-directed masking approach could be applied to other cell surface proteins which either do not possess enzymatic activity, or their activity is not affected by antibody binding. Thus, we attempted to map the epitope by analyzing the binding of hN3-B3S to tethered NTPDase3 mutants (Fig. 3). NTPDase3 mutants tethered to plates were incubated with hN3-B3S antibody, and the bound antibody was detected using the secondary goat anti-mouse HRP-conjugated antibody and a chemiluminescence reaction (Fig. 3A). Black plates were used in this assay to prevent light pollution from adjacent wells. Chemiluminescence intensity in plates was expressed in arbitrary units, where one unit corresponds to the intensity of the S87C mutant (Fig. 3B). Large variations in ATPase activity of immobilized mutants (Fig. 2B) suggest large variations in the amount of immobilized NTPDase3 protein. For that reason, antibody binding to tethered mutants cannot be compared by merely detecting the amount of antibody bound to plates, but rather has to be expressed as a ratio of the amount of bound antibody to the amount of immobilized enzyme. Therefore, we attempted to estimate the amount of immobilized NTPDase3 using KLH1 polyclonal antibody against the C-terminal peptide (amino acid residues 515–529) of NTPDase3 (Smith and Kirley, 1999a,b). However, this antibody produced substantial background even with the A267C mutant, which neither bound to the streptavidin-coated plate, nor displayed measurable ATPase activity on plates, likely due to lack of cysteine biotinylation. The reason for this background signal is not clear, but it precluded the use of KLH1 for quantitatively evaluating the amount of immobilized NTPDase3. Therefore, a simple qualitative estimate of immobilized enzyme was made by analyzing the amount of biotinylated NTPDase3 in the Triton X-100 membrane extracts (Fig. 3C). We assumed that the presence of biotinylated NTPDase3 in the extracts used for immobilization implies that some amount of the enzyme has to be bound to streptavidin-coated plates. Accordingly, the mutants that are biotinylated (revealing a protein band in lanes ‘B’, Fig. 3C) are assumed to be bound to streptavidin-coated plates (marked with ‘+’ in Fig. 3D), whereas the rest of mutants are assumed to be non-bound to plates (marked with ‘0’ in Fig. 3D). This method is likely to underestimate the amount of immobilized enzyme, since it categorizes G122C, S394C and D414C as ‘non-bound’, although G122C and S394C bind hN3-B3s (Fig. 3A), and D414C exhibits substantial ATPase activity (Fig. 2B). Although this limitation may exclude some mutants from further analysis (e.g. D414C, see below), it does not result in any false conclusions (see below). Qualitative estimation of the ratio of the amount of bound hN3-B3S antibody (chemiluminescence intensity, Fig. 3B) to the amount of immobilized enzyme (‘+’ or ‘0’ immobilized enzyme, Fig. 3D) results in values above zero, zero, and undefined (marked with ‘+’, ‘0’ and ‘0/0’, respectively, in Fig. 3E). Antibody binding to immobilized NTPDase3 (marked with ‘+’, Fig. 3E) suggests epitope accessibility (marked with ‘a’ in Fig. 3F). The zero ratio of bound antibody/immobilized protein suggests that the epitope is masked (marked with ‘M’ in Fig. 3F). The 0/0 ratio for mutants A267C and D414C (Fig. 3E) makes the evaluation of epitope accessibility uncertain, and therefore these mutants have been excluded from further analysis.
Fig. 3.
Binding of hN3-B3S antibody to tethered NTPDase3 mutants. (A) Plates with immobilized NTPDase3 mutants were incubated with hN3-B3S monoclonal antibody. The bound antibody was detected using the secondary goat anti-mouse HRP-conjugated antibody and a chemiluminescence reaction. The signal in three replicate wells is shown for each mutant. (B) Quantitative estimate of the bound hN3-B3S shown in panel (A). Chemiluminescence intensity was determined using AlphaEaseFC software (Alpha Innotech) and expressed in arbitrary units, where one unit corresponds to the intensity of the S87C mutant. The background intensity of C10S is approximately the same as in wells where the membranes of COS cells transfected with an empty vector (pcDNA3) were used, and considered to be zero. The numbers represent means of three wells. (C) Susceptibility of cysteine mutants for biotinylation. Proteins were extracted from biotinylated cell membranes with Triton X-100 and incubated with streptavidin agarose beads. Proteins bound to beads (shown in lanes B) were eluted by SDS and analyzed together with non-bound proteins (shown in lanes N) by western blotting using the polyclonal KLH11 antibody for NTPDase3 detection. The amount of NTPDase3 in lane ‘B’ indicates the efficiency of biotinylation. (D) Qualitative estimate of the amount of NTPDase3 immobilized on streptavidin plates. The presence of biotinylated NTPDase3 (lanes ‘B’ in Panel C) implies that a certain amount of this protein is immobilized on streptavidin plates (denoted by ‘+’ in Panel D). The lack of biotinylation (no NTPDase3 is detected in lanes ‘B’, Panel C) implies that little or no NTPDase3 immobilized on plates (marked with ‘0’ in Panel D). (E) Qualitative estimation of the ratio of the amount of hN3-B3S antibody bound to plates (chemiluminescence intensity, Panel B) to the amount of NTPDase3 immobilized on plates (Panel D). Values above zero, zero and undefined are denoted by ‘+’, ‘0’ and ‘0/0’, respectively. (F) Antibody binding to immobilized NTPDase3 (denoted by ‘+’, Panel E) suggests epitope accessibility (denoted by ‘a’ in Panel F). The lack of antibody binding to immobilized enzyme (marked with ‘0’ in Panel E) suggests that the epitope is masked (marked with ‘M’ in Panel F). The 0/0 ratio for mutants A267C and D414C (Panel E) makes the evaluation of epitope accessibility uncertain.
The results of epitope mapping by site-directed masking using both ATPase inhibition and antibody binding assays are displayed on the 3D model of the extracellular portion of human NTPDase3, as shown in Fig. 4. The cysteine tethers which abolished both hN3-B3S binding and ATPase inhibition by the antibody are clustered on the same surface area of the extracellular portion of NTPDase3 molecule. This area delineates the approximate location of the antibody epitope, and it is likely facing away from the plasma membrane, since the two transmembrane domains are connected to the opposite side of the molecule (as indicated by arrowheads labeled ‘NT’ and ‘CT’ in Fig. 4A). Remarkably, this epitope area covers the active site cleft, indicated by the arrows in Fig. 4A and E.
Fig. 4.
Mapping the antibody binding site on NTPDase3 by site-directed masking. The Figure shows a 3D model of NTPDase3 produced by the FFAS fold recognition server (Rychlewski et al., 2000; Jaroszewski et al., 2005) by threading the primary sequence of the extracellular portion of NTPDase3 through the crystal structure of the extracellular portion of rat NTPDase2 (PDB ID code 3CJ1, www.pdb.org). Approximate locations of the connection points with N-terminal and C-terminal transmembrane helices are shown by arrowheads labeled NT and CT, respectively. Panels (A)–(D) show the surface of NTPDase3 model produced by rotation around the ‘Y’ axis in Panel A, by approximately 90° for each successive view. (E) View from the top of the extracellular portion of NTPDase3. Arrows in (A) and (E) indicate the active site cleft. Residues which have been singly mutated to cysteine are indicated. Locations of cysteine tethers which abolished both ATPase inhibition and hN3-B3S binding are colored red, whereas cysteine tethers which abolished ATPase inhibition, but allowed detectable antibody binding, are colored magenta. Cysteine tethers which had no significant effect on either hN3-B3S binding, or ATPase inhibition are shown in blue. Mutants that could not be analyzed due to low or no activity after immobilization on plates (S87C, A267C and S394C) are not shown.
Fine epitope mapping by alanine scanning mutagenesis
Site-directed masking relies on blocking antibody access to a relatively large surface area centered on each cysteine tether, and thus limits the resolution of the resultant epitope map. In order to obtain a detailed map of the hN3-B3S antibody binding site, we constructed 18 single alanine mutations within the epitope area delineated by site-directed masking and investigated their effect on antibody binding and inhibition of enzymatic activity (Fig. 5A). These residues were chosen for mutagenesis based on masking results and computational analyses using several epitope prediction methods (DiscoTope, ElliPro and BEPro). The residue F413 is not represented in the NTPDase3 3D model, and therefore the position of F413A mutation is not shown in Fig. 5A. The mutant NTPDase3 proteins were expressed in COS-1 cells, and all mutants exhibited specific activity in cell membrane preparations similar to that of C10S NTPDase3, which was used as a background for alanine mutagenesis (data not shown). Inhibition of ATPase activity by hN3-B3S antibody was less efficient for several mutants (W403A, D414A, E415A and R419A) than for control C10S NTPDase3 (Fig. 5B and D), suggesting that these residues participate in interaction with the antibody. Three other mutations (I296A, V416A and F413A) do not affect the level of ATPase inhibition with hN3-B3S, but abolish antibody binding to NTPDase3 denatured with SDS under non-reducing conditions and subjected to western blotting (Fig. 5C). This suggests that these residues also participate in interaction with the antibody. Double alanine mutations within the epitope further decreased the inhibitory effect of hN3-B3S on ATPase activity (Fig. 5E). Remarkably, ATPase activity of the double alanine mutant (C10S)/W403A/R419A was not inhibited by hN3-B3S antibody, suggesting that W403 and R419 represent an antigenic ‘hot spot’, which is critical for antibody binding. This was further confirmed by slot blot assay (Fig. 5F), demonstrating that the (C10S)/W403A/R419A mutant lost the ability to bind hN3-B3S antibody. In this experiment, a monoclonal antibody produced by the hybridoma clone hN3-H10S was also tested. It demonstrated the same reactivity with NTPDase3 mutants as hN3-B3S antibody, thus further suggesting that both clones and antibodies are likely to be identical.
Fig. 5.
Fine mapping the antibody binding site on NTPDase3 by alanine mutagenesis. (A) The surface of the NTPDase3 model as viewed from above the active site cleft. Location of the substrate analog AMPPNP bound to the active site is modeled using the crystal structure of the AMPPNP complex with rat NTPDase2 (Zebisch and Strater, 2008) (PDB ID code 3CJA, www.pdb.org) and shown in magenta and blue. Residues which were singly mutated to alanine are indicated. Colored green are alanine mutations which do not affect the level of inhibition of ATPase activity by hN3-B3S antibody, as shown in panel (B). Marked in orange are four alanine mutations which decreased the level of inhibition by hN3-B3S, and therefore increased the residual ATPase activity in the presence of the antibody, as revealed in panel (B). Two mutations shown in yellow and the F413A mutation, which is not represented in the model, do not significantly affect the inhibition of ATPase activity by hN3-B3S, but they abolished the interaction of denatured non-reduced protein with the antibody in western blotting (C), suggesting their location in the epitope. (B) Inhibition of ATPase activity of alanine mutants by the hN3-B3S antibody. Membrane preparations from transfected cells were incubated with hN3-B3s (0.3 µg/ml) for 20 min at 37°C followed by ATPase activity assay. C10S mutant was used as a background for all alanine mutations and shown for comparison (see ‘Materials and Methods’ for the rationale of using C10S). Values represent means ± SD of three assays. (C) Western blotting of alanine mutants. Membrane proteins of transfected cells were denatured with SDS under non-reducing conditions, subjected to electrophoresis and electroblotting, and probed with hN3-B3S antibody. (D) Concentration dependence of the inhibition of ATPase activity of single alanine mutants with hN3-B3S antibody. Membrane preparations from transfected cells were incubated with hN3-B3s at the indicated concentrations for 20 min at 37°C and assayed for ATPase activity. (E) Titration of the inhibition of ATPase activity of double alanine mutants with hN3-B3S antibody. Assay was performed as in panel (D). (F) Binding of hN3-B3S to alanine mutants in slot blot assay. Membrane preparations from transfected cells (0.1 µg total protein in 200 µl per slot) were applied to PVDF membrane by vacuum filtration, and probed with antibodies produced by either hN3-B3S, or hN3-H10S hybridoma clones, followed by incubation with a secondary goat anti-mouse HRP-conjugated antibody and chemiluminescence detection. C10S mutant is shown for comparison. pcDNA3 indicates membranes from cells transfected with an empty expression vector.
Mutagenesis of putative active site residues in the 5th disulfide loop
In order to better understand the mechanism of NTPDase3 inhibition by the monoclonal antibody, we tested a hypothesis that antibody interaction with the residues (E415, V416 and R419) in the 5th disulfide loop (delimited by C399 and C422) may deform the loop and transmit this deformation to the adjacent residues (Y417 and Y421), which are supposed to interact with the substrate (ATP) or the product (AMP) in the active site (Zebisch and Strater, 2008). Changes in orientation of Y417 and Y421 may impair the binding of ATP and/or AMP and thus result in inhibition of enzymatic activity. This hypothesis is based on the study of crystal structures of rat NTPDase2 in complexes with a non-hydrolyzable ATP analog AMPPNP, or the product AMP (Zebisch and Strater, 2008). It was inferred from the 3D structures of NTPDase2 that residue R394 (homologous to Y417 in NTPDase3) is involved in interaction with the adenine base and the ribose of ATP, and that residue Y398 (homologous to Y421 in NTPDase3) interacts with the adenine base of the product AMP (Zebisch and Strater, 2008). We hypothesized that if the changes in orientation of residues Y417 and Y421 in NTPDase3 are responsible for the inhibitory effect of hN3-B3S antibody, then substitution of these residues by the smaller alanine residues should lessen the inhibitory effect because changes in their orientation would have less influence on the position of the adenine base. In addition, we expected to see a detrimental effect of Y417A and Y421A mutations on enzyme activity due to impairment of interaction with the adenine base. Surprisingly, specific ATPase activities of Y417A and Y421A mutants were higher than those of C10S NTPDase3 by approximately 100 and 50%, respectively. In addition, antibody hN3-B3S inhibited ATPase activity of Y417A and Y421A mutants by 60–70%, similar to that observed for C10S NTPDase3. Based on these experiments, it seems unlikely that the inhibition mechanism involves residues Y417 and Y421 of the 5th disulfide loop.
hN3-B3S does not compete with substrate analog AMPPCP in binding NTPDase3
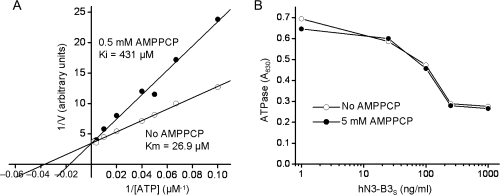
Location of the epitope in close proximity to the active site cleft (Fig. 5A) suggests that the antibody inhibits enzymatic activity by obstructing the active site for substrate access and/or product release. Therefore, we attempted to characterize whether the inhibition is competitive or noncompetitive. Because of the low dissociation rate of the antibody–antigen complex, an experimental setting in which the enzyme is pre-incubated with the antibody followed by adding substrate and recording the enzymatic reaction is difficult to implement for competition assay. Therefore, we first determined the half-saturating concentration (Ki) for a non-hydrolyzable ATP analog, AMPPCP, and then analyzed competition of AMPPCP and hN3-B3S antibody for binding NTPDase3 immobilized on plates. In a representative experiment shown in Fig. 6A, A Km value of 26.9 µM was obtained for ATP. AMPPCP competitively inhibited ATPase activity of C10S NTPDase3 with Ki value of 431 µM. We then immobilized the biotinylated A77C mutant of NTPDase3 on streptavidin plates for competition assays (Fig. 6B). In these experiments, the plates were incubated with various concentrations of hN3-B3S antibody in either the absence, or the presence of AMPPCP at a concentration of 5 mM, which is more than 10 times higher than the Ki for AMPPCP (431 µM). The incubation continued for 18 h, which should be sufficient to reach equilibrium. Subsequently, plates were washed several times to remove all AMPPCP, as well as non-bound hN3-B3S antibody, and processed for ATPase assay. Competition of AMPPCP with hN3-B3S for binding NTPDase3 should result in less hN3-B3S bound to NTPDase3 in plates which contained AMPPCP, when compared with plates which did not contain AMPPCP. Since hN3-B3S inhibits ATPase activity, the differences in the amount of bound hN3-B3S can be inferred from the differences in ATPase activity between plates. As shown in Fig. 6B, hN3-B3S inhibited ATPase to approximately the same degree regardless of the absence or the presence of AMPPCP, suggesting a lack of competition between AMPPCP and hN3-B3S antibody. Therefore, since AMPPCP closely resembles ATP, it is very likely that hN3-B3S antibody inhibits ATPase activity of NTPDase3 in a noncompetitive manner.
Fig. 6.
Antibody hN3-B3S does not compete with substrate analog AMPPCP for binding to NTPDase3. (A) Competitive inhibition of ATPase activity of NTPDase3 with the non-hydrolyzable substrate analog, AMPPCP. Reactions were carried out in TBS (150 mM NaCl, 50 mM Tris–HCl, pH 7.5) containing 0.1 mg/ml BSA. A representative Lineweaver-Burk plot of ATP hydrolysis by C10S NTPDase3 in the absence (open circles) and in the presence (filled circles) of 0.5 mM AMPPCP is shown. Calculated Km for NTPDase3 and Ki for AMPPCP are indicated. (B) Lack of competition between AMPPCP and hN3-B3s for binding NTPDase3 immobilized on plates. Biotinylated A77C mutant of NTPDase3 was immobilized on streptavidin-coated plates and incubated for 18 h at 4°C with hN3-B3S at the indicated concentrations in the absence (open circles), or in the presence (filled circles) of 5 mM AMPPCP. Subsequently, plates were washed three times with TBS to remove both bound and free AMPPCP, as well as free hN3-B3S, and ATPase activity was assayed. The graph shows that the bound hN3-B3S inhibits ATPase activity to approximately the same degree regardless of the absence, or the presence of AMPPCP at a concentration of 5 mM, which exceeds the half-saturation concentration (Ki of approximately 0.4 mM, panel (A)) by more than 10-fold. This suggests that AMPPCP and hN3-B3s do not compete for the same binding site on NTPDase3.
Discussion
Rational design of preventive vaccines, immunodiagnostics and targeted therapeutics requires identification and understanding of the molecular basis of antibody–antigen recognition. A common task in relevant studies is epitope mapping, characterization of the region of an antigen recognized by an antibody. Despite many available experimental techniques (Gershoni et al., 2007) and computer prediction methods (Yang and Yu, 2009), epitope mapping remains challenging and has no general solution. The recently described method of site-directed masking (Paus and Winter, 2006) provides a systematic approach for epitope mapping in soluble purified proteins, for which 3D structure is available. We adapted this method for epitope mapping in unpurified cell surface proteins and showed that such analysis can be based on a 3D homology model if the crystal structure of the antigen is not available. Using this technique, we identified the binding site of an inhibitory monoclonal antibody hN3-B3S to human NTPDase3.
In order to map the epitope of the hN3-B3S antibody, we coupled site-directed masking with either ATPase assay, which could be applied to this inhibitory antibody and human NTPDase3, or antibody binding assay, which is suitable to most cell surface proteins. Both assays mapped the epitope to the same surface area near the active site cleft (Fig. 4E). However, the epitope area mapped by antibody binding assay seems to be slightly smaller than the epitope mapped by ATPase inhibition assay. Thus, tethering of NTPDase3 via any cysteine residue shown in red or magenta in Fig. 4E abolished ATPase inhibition by hN3-B3s antibody. However, the cysteine tethering sites shown in magenta in Fig. 4E (T299C, S101C and A112C) did not abolish antibody binding (Fig. 3A), while others shown in red (S297C, W403C, D414C, E415C, V416C and R419C) did abolish antibody binding (Fig. 3A). This suggests that the antibody binding site on the NTPDase3 surface is located further away from residues T299C, S101C and A112C than the site which is important for the inhibition of enzymatic activity. We hypothesize therefore that hN3-B3S antibody binds NTPDase3 via residues in the 5th disulfide loop (W403, D414, E415, V416 and R419) and I296 (Figs 4E and 5A), but it causes inhibition by a part of the antibody molecule which extends toward residues T299, S101 and A112, covering, but not directly interacting with, residues in the active site cleft. This hypothesis suggests that the inhibitory mechanism is mediated by steric hindrance.
Two additional observations are consistent with this model. First, we tested a hypothesis that the inhibitory mechanism operates within the 5th disulfide loop located between C399 and C422. According to this hypothesis, antibody binding to the residues of the 5th disulfide loop (E415, V416 and R419) may deform the loop and change the position of the adjacent residues Y417 and Y421, which according to homology with the crystal structure of rat NTPDase2 (Zebisch and Strater, 2008) form a part of the active site, and thus cause inhibition. However, mutation of Y417 or Y421 to alanine neither had a detrimental effect on NTPDase3 specific activity, nor changed the extent of ATPase inhibition by the antibody, indicating that these properties are not sensitive to the nature and orientation of residues in positions 417 and 421. Therefore, it is unlikely that the inhibition of enzymatic activity is caused by deformation of the 5th disulfide loop. Second, we showed in competitive ELISA experiments that hN3-B3S antibody does not compete with a non-hydrolyzable substrate analog AMPPCP for binding to NTPDase3 (Fig. 6B), suggesting that the antibody does not bind the residues within the active site. These observations are consistent with a noncompetitive mechanism of inhibition by steric hindrance. Similar to the effect of hN3-B3S on NTPDase3 activity, partial inhibition by antibodies was reported for other enzymes (Devaux et al., 1987; McNally et al., 1995). Although the actual mechanism of inhibition was not determined in these studies, the authors proposed that antibodies bound close to, but distinct from, the active site, and inhibited the enzymatic activity by hindering the access to the active site.
Among the identified seven epitope residues, four residues W403, D414, E415 and R419 are likely to be critical for antibody binding, whereas other three residues I296, F413 and V416 contribute to lesser degree (Fig. 5). It is therefore surprising that although all four critical residues are identical in human NTPDase3, mouse NTPDase3 and rat NTPDase3, only human NTPDase3 is recognized by hN3-B3S antibody (Munkonda et al., 2009). However, a similar phenomenon was described in a comprehensive study on mapping the epitopes for several monoclonal antibodies in a human G protein-coupled receptor CCR5 (Paes et al., 2009). Those authors found that all critical residues for three monoclonal antibodies were identical between human and murine CCR5, although none of the antibodies reacted with murine CCR5. They suggested that identical amino acid sequences can present immunogenically different structures, possibly due to differences in conformation or post-translational processing (Paes et al., 2009).
In conclusion, we adapted the site-directed method described by Paus and Winter (Paus and Winter, 2006) for soluble and purified antigens to epitope mapping in unpurified proteins expressed on the cell surface. We also demonstrated that the epitope mapping can be based on a 3D structure which is modeled on a homologue if the crystal structure of the antigen is not available. This method could be especially useful for systematic identification of those conformational epitopes which have no significant linear motifs, and therefore are not suitable for the analysis using peptide libraries on phage (Rowley et al., 2004; Gershoni et al., 2007), or peptide arrays on a solid phase (Carter and Loomis-Price, 2004). Using this method, we located the epitope of the inhibitory hN3-B3S monoclonal antibody in close proximity to the active site of NTPDase3, suggesting a noncompetitive mechanism of inhibition by steric hindrance. This knowledge will be useful for engineering more efficient inhibitory antibodies specific to human NTPDase3, as well as for developing inhibitory antibodies to other NTPDase isoenzymes. Such inhibitors would be valuable instruments for basic research analyzing the complex network of extracellular receptors and enzymes involved in purinergic signaling, as well as potential tools for clinical applications.
Funding
This work was supported by National Institutes of Health R01 grant HL72382 to T.L.K.; by a grant from the Canadian Institutes of Health Research (CIHR) to J.S. who was also the recipient of a New Investigator award from the CIHR and of a Junior 2 scholarship from the Fonds de Recherche en Santé du Québec (FRSQ); and a Dalton-Zannoni Fellowship supported by the University of Cincinnati-American Society of Pharmacology and Experimental Therapeutics (UC-ASPET) summer undergraduate research training program directed by Prof. Ronald W. Millard, Pharmacology & Cell Biophysics, to Aimi Toyama.
Footnotes
Edited By Greg Winter
References
- Baykov A.A., Evtushenko O.A., Avaeva S.M. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. doi:10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- Brunschweiger A., Iqbal J., Umbach F., Scheiff A.B., Munkonda M.N., Sévigny J., Knowles A.F., Muller C.E. J. Med. Chem. 2008;51:4518–4528. doi: 10.1021/jm800175e. doi:10.1021/jm800175e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J.M., Loomis-Price L. Curr. Protoc. Immunol. 2004 doi: 10.1002/0471142735.im0904s60. Chapter 9, Unit 9 4. [DOI] [PubMed] [Google Scholar]
- Devaux C.A., Covell D.G., Barbet J., el Gamil M., Sachs D.H. Mol. Cell. Biochem. 1987;74:117–128. doi: 10.1007/BF00224949. [DOI] [PubMed] [Google Scholar]
- Fiske C.H., Subbarow Y. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- Gendron F.P., Benrezzak O., Krugh B.W., Kong Q., Weisman G.A., Beaudoin A.R. Curr. Drug Targets. 2002;3:229–245. doi: 10.2174/1389450023347713. doi:10.2174/1389450023347713. [DOI] [PubMed] [Google Scholar]
- Gershoni J.M., Roitburd-Berman A., Siman-Tov D.D., Tarnovitski Freund N., Weiss Y. BioDrugs. 2007;21:145–156. doi: 10.2165/00063030-200721030-00002. doi:10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haste Andersen P., Nielsen M., Lund O. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. doi:10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J., Vollmayer P., Braun N., Zimmermann H., Muller C.E. Purinergic Signal. 2005;1:349–358. doi: 10.1007/s11302-005-8076-x. doi:10.1007/s11302-005-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanenkov V.V., Meller J., Kirley T.L. Biochemistry. 2005;44:8998–9012. doi: 10.1021/bi047487z. doi:10.1021/bi047487z. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. doi:10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski F., Lévesque S.A., Lavoie E.G., Lecka J., Bigonnesse F., Knowles A.F., Robson S.C., Kirley T.L., Sévigny J. Purinergic Signal. 2005;1:193–204. doi: 10.1007/s11302-005-6217-x. doi:10.1007/s11302-005-6217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally A.J., Motter K., Jordan F. J. Biol. Chem. 1995;270:19736–19743. doi: 10.1074/jbc.270.34.19736. [DOI] [PubMed] [Google Scholar]
- Muller C.E., Iqbal J., Baqi Y., Zimmermann H., Rollich A., Stephan H. Bioorg. Med. Chem. Lett. 2006;16:5943–5947. doi: 10.1016/j.bmcl.2006.09.003. doi:10.1016/j.bmcl.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Munkonda M.N., Kauffenstein G., Kukulski F., Lévesque S.A., Legendre C., Pelletier J., Lavoie E.G., Lecka J., Sévigny J. Biochem. Pharmacol. 2007;74:1524–1534. doi: 10.1016/j.bcp.2007.07.033. doi:10.1016/j.bcp.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Munkonda M.N., Pelletier J., Ivanenkov V.V., Fausther M., Tremblay A., Kunzli B., Kirley T.L., Sévigny J. FEBS J. 2009;276:479–496. doi: 10.1111/j.1742-4658.2008.06797.x. doi:10.1111/j.1742-4658.2008.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.M., Ivanenkov V.V., Kirley T.L. J. Biol. Chem. 2002;277:6162–6169. doi: 10.1074/jbc.M110105200. doi:10.1074/jbc.M110105200. [DOI] [PubMed] [Google Scholar]
- Paes C., Ingalls J., Kampani K., Sulli C., Kakkar E., Murray M., Kotelnikov V., Greene T.A., Rucker J.B., Doranz B.J. J. Am. Chem. Soc. 2009;131:6952–6954. doi: 10.1021/ja900186n. doi:10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus D., Winter G. Proc. Natl Acad. Sci. USA. 2006;103:9172–9177. doi: 10.1073/pnas.0600263103. doi:10.1073/pnas.0600263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. BMC Bioinformat. 2008;9:514. doi: 10.1186/1471-2105-9-514. doi:10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson S.C., Sévigny J., Zimmermann H. Purinergic Signal. 2006;2:409–430. doi: 10.1007/s11302-006-9003-5. doi:10.1007/s11302-006-9003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M.J., O'Connor K., Wijeyewickrema L. Biotechnol. Annu. Rev. 2004;10:151–188. doi: 10.1016/S1387-2656(04)10006-9. doi:10.1016/S1387-2656(04)10006-9. [DOI] [PubMed] [Google Scholar]
- Rychlewski L., Jaroszewski L., Li W., Godzik A. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.M., Kirley T.L. Biochim. Biophys. Acta. 1998;1386:65–78. doi: 10.1016/s0167-4838(98)00063-6. [DOI] [PubMed] [Google Scholar]
- Smith T.M., Kirley T.L. Biochemistry. 1999a;38:1509–1516. doi: 10.1021/bi9821768. doi:10.1021/bi9821768. [DOI] [PubMed] [Google Scholar]
- Smith T.M., Kirley T.L. Biochemistry. 1999b;38:321–328. doi: 10.1021/bi9820457. doi:10.1021/bi9820457. [DOI] [PubMed] [Google Scholar]
- Stoscheck C.M. Anal. Biochem. 1990;184:111–116. doi: 10.1016/0003-2697(90)90021-z. doi:10.1016/0003-2697(90)90021-Z. [DOI] [PubMed] [Google Scholar]
- Sweredoski M.J., Baldi P. Bioinformatics. 2008;24:1459–1460. doi: 10.1093/bioinformatics/btn199. doi:10.1093/bioinformatics/btn199. [DOI] [PubMed] [Google Scholar]
- Yang X., Yu X. Rev. Med. Virol. 2009;19:77–96. doi: 10.1002/rmv.602. doi:10.1002/rmv.602. [DOI] [PubMed] [Google Scholar]
- Zebisch M., Strater N. Proc. Natl Acad. Sci. USA. 2008;105:6882–6887. doi: 10.1073/pnas.0802535105. doi:10.1073/pnas.0802535105. [DOI] [PMC free article] [PubMed] [Google Scholar]