Abstract
Type I interferons (IFNs) play a crucial role in the innate immune avant-garde against viral infections. Virtually all viruses have developed means to counteract the induction, signaling, or antiviral actions of the IFN circuit. Over 170 different virus-encoded IFN antagonists from 93 distinct viruses have been described up to now, indicating that most viruses interfere with multiple stages of the IFN response. Although every viral IFN antagonist is unique in its own right, four main mechanisms are employed to circumvent innate immune responses: (i) general inhibition of cellular gene expression, (ii) sequestration of molecules in the IFN circuit, (iii) proteolytic cleavage, and (iv) proteasomal degradation of key components of the IFN system. The increasing understanding of how different viral IFN antagonists function has been translated to the generation of viruses with mutant IFN antagonists as potential live vaccine candidates. Moreover, IFN antagonists are attractive targets for inhibition by small-molecule compounds.
Innate immunity during infection
The innate immune system forms the first line of defense against invading micro-organisms such as viruses. It dampens initial virus replication and ensures survival of the host until specialized adaptive responses are developed. Type I interferons (IFNs) are secreted key cytokines on the innate immune axis that protect uninfected cells and stimulate leukocytes residing at the interface of innate and adaptive immunity, such as macrophages and dendritic cells (DC) [1]. These cells prod the adaptive immune system to mount a full, specialized response against the invading microbe.
The ability to outrun innate immunity before adaptive immune responses are mounted is crucial for the survival of virtually all the mammalian viruses, regardless of their genome type and complexity. Relatively simple viruses such as RNA viruses from the Picornaviridae family, as well as DNA viruses with large genomes, such as members from the Poxviridae family, have been shown to inhibit the IFN system. This review covers the latest insights into how virus-encoded antagonists sidetrack the IFN machinery and how this knowledge is currently used to generate second generation live vaccines and antiviral compounds.
BOX 1: The IFN circuit.
The IFN circuit consists of three distinct steps. The first step consists of recognition of pathogen-associated molecular patterns (PAMP), resulting in the synthesis and secretion of IFN-β (Figure 2). Subsequently, secreted IFN binds to the IFN-α receptor (IFNAR) on the same or surrounding cells, resulting in the transcription of hundreds of IFN-stimulated effector molecules (Figure 3).
Viral nucleic acid or proteins are recognized by Toll-like receptors on the plasma membrane or in endosomes of predominantly antigen presenting cells (APC). Moreover, most cells express cytoplasmic sensors retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5) that recognize viral RNA [2]. Cytoplasmic microbial B-form DNA can be recognized by the DNA-sensors DAI and AIM2 [3, 4, 5] or cellular RNA polymerase III, which converts it into 5′-triphosphate containing RNAs that are recognized by RIG-I [6, 7].
Upon activation, RIG-I and MDA-5 engage mitochondrial antiviral signaling adapter (MAVS) [8]. In turn, MAVS activates two kinase complexes that ultimately phosphorylate and activate the two key transcription factors for IFN-β induction: nuclear factor κB (NFκB) and IFN-regulatory factor 3 (IRF-3) [2]. The first kinase complex consists of TNF receptor associated factor 3 (TRAF-3), TRAF family member associated NF-κB activator (TANK), TANK-binding kinase 1 (TBK-1), and inhibitor of κB kinase (IκB) ɛ (IKKɛ) [8]. The second complex phosphorylates IκB and thereby activates NF-κB. It consists of TRAF-6, receptor interacting protein 1 (RIP-1), NF-κB essential modulator (NEMO), TGF-β activated kinase 1 (TAK-1), IKKα, and IKKβ [8]. Upon activation, NF-κB and IRF-3 translocate to the nucleus and drive IFN-β transcription (Figure 2).
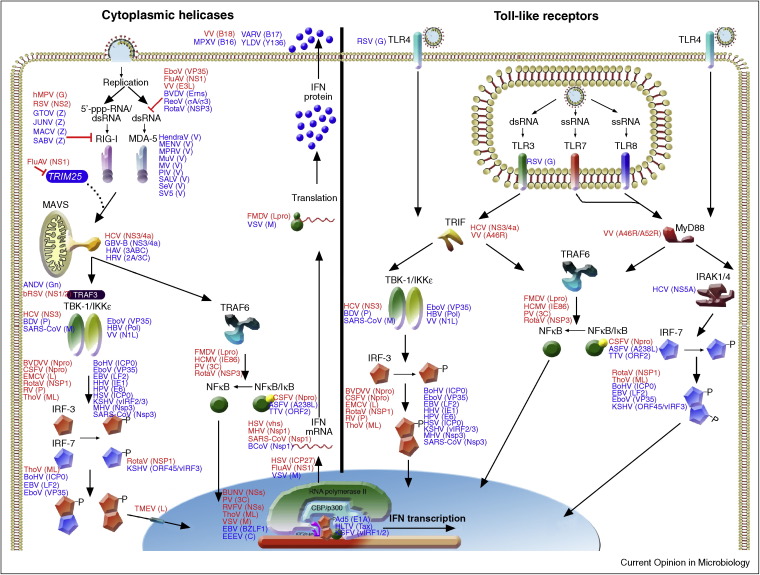
Figure 2.
Schematic representation of type I IFN induction through RLRs and TLRs. Viruses and their antagonistic proteins are indicated at the steps of the IFN pathway they affect. Antagonistic proteins are shown adjacent to their targets in alphabetical order. Antagonists in red indicate proof for IFN antagonist by recombinant viruses lacking the IFN antagonist. Antagonists in blue indicate proof by over expression and/or wild-type virus infection.
Upon binding of extracellular IFN-β, the IFNAR recruits janus kinase 1 (JAK-1) and tyrosine kinase 2 (TYK-2) to its cytoplasmic domain. These kinases phosphorylate the key transcription factors signal transducers and activators of transcription (STAT) 1 and 2, which together with IRF-9 form the IFN-stimulated gene factor 3 (ISGF3) complex [8]. This active transcription complex translocates to the nucleus, where it drives the expression of ISGs [9].
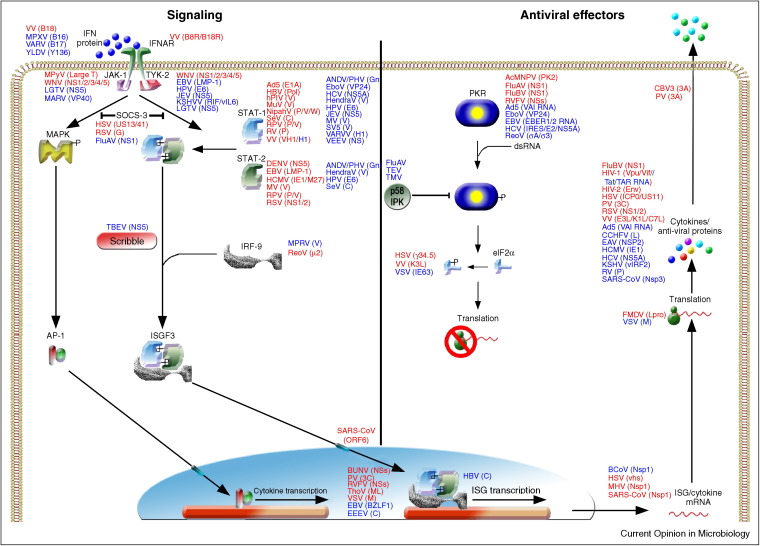
To date, over 500 publications have reported the identification and/or characterization of more than 170 different virus-encoded IFN antagonists by 93 distinct viruses (suppl. Table S1). Over the past four years, the number of reports describing new viral IFN antagonists has almost doubled, underpinning the speed with which the field is developing. Table S1 was compiled from all current literature and includes viral proteins that act as IFN antagonists and whether experimental proof comes from expression studies or infections with mutant viruses lacking the putative IFN antagonists. This latter issue is of great importance since single gene over expression may result in aberrant expression, localization and interactions of viral proteins compared to virus infected cells. Hence, proteins identified by overexpression may not retain the same phenotype in the context of infection. Figure 2, Figure 3 summarize the steps of the IFN system that different viruses interfere with. Although it lays beyond the scope of this review to discuss all antagonists in detail, the ones discussed exemplify the different strategies used by viruses to subvert the IFN response.
Figure 3.
Schematic representation of type I IFN signaling. Viruses and their antagonistic proteins are indicated at the steps of the IFN pathway they affect. Antagonistic proteins are shown adjacent to their targets in alphabetical order. Antagonists in red indicate proof for IFN antagonist by recombinant viruses lacking the IFN antagonist. Antagonists in blue indicate proof by over expression and/or wild-type virus infection.
Main molecular mechanisms of viral antagonism
The majority of IFN antagonists exert their action by one of four different strategies: (i) general inhibition of cellular gene expression, (ii) sequestration of molecules in the IFN circuit, (iii) proteolytic cleavage of innate immune components, or (iv) targeting these components for proteasomal degradation (Table S1).
General block of gene expression is a feature seen in various RNA viruses (FluAV, VSV, PV, RVFV, and BunV) and seems to provide an efficient means to prevent synthesis of not only IFN, but also additional antiviral molecules detrimental to virus replication [10]. Whereas host shut-off in some large dsDNA viruses (such as herpes simplex virus) is partly related to its necessity to temporally regulate expression of its own genes [11], RNA viruses seem to mainly block viral gene expression to circumvent the expression of antiviral proteins [10].
Viruses interfere with all three steps of the IFN system
Evidently, inhibition of early steps in the cascade implicitly inhibits the induction of down-stream molecules and amplification of the IFN signal. Most viral IFN antagonists are exclusively produced in the infected cells, implying that surrounding uninfected cells can still be stimulated to establish an antiviral state by systemic IFN. For that reason some large dsDNA viruses express soluble IFN-binding proteins that compete with the cellular IFN receptor for its ligand (Table S1). These ‘viroceptors’ neutralize secreted IFN, thereby preventing autocrine amplification. Possibly, these strategies enable some of these viruses to persist in their host for longer times or disseminate to hard-to-infect cells.
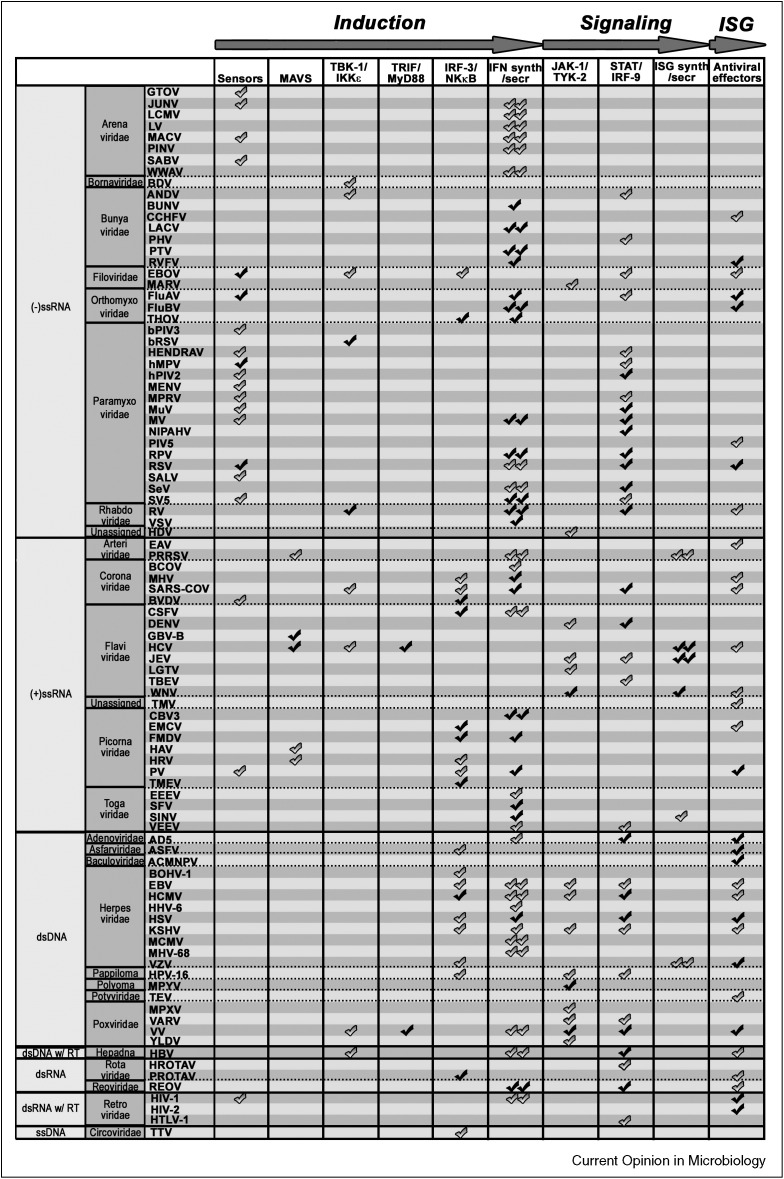
The fact that nearly 50% of the viruses for which IFN antagonists have been identified interfere with multiple steps of the IFN response (Figure 1 ), exemplifies the necessity for viruses to successfully circumvent the IFN response. Moreover, approximately one third of the viruses encode an antagonist that interferes with more than one step of the IFN response system (Figure 2, Figure 3 ), underpinning the pleiotropic nature of viral proteins.
Figure 1.
Viral antagonism with the IFN circuit. A check symbol is displayed at various steps of the IFN cascade (X-axis) that are impaired by a particular virus (Y-axis). The plot is organized by viral genome type. Double symbols indicate inhibition of IFN induction or signaling at yet unknown step of the pathway. Black symbols indicate proof for viral antagonism of the indicated step in the pathway by recombinant viruses lacking the IFN antagonist. Grey symbols indicate proof by over expression and/or wild-type virus infection.
Conservation of IFN antagonists
Several viral IFN antagonists are conserved within distinct RNA virus families, whereas dsDNA viruses do not use common proteins to antagonize the IFN response (Table S1). This may partly be explained by bias in data available for different virus families. On the contrary, it may be explained by stricter genome restrictions for RNA viruses. Evolutionary pressure to encode maximum functionality by relatively limited genome capacity drives a high degree of multi-functionality in RNA virus proteins. Genome size restrictions make it such that they cannot derive new large genes encoding IFN antagonists from their hosts as is the case for large dsDNA viruses (e.g. soluble IFN receptor decoys). This considerably restricts the variability of accessory proteins found in RNA viruses and may explain why some related RNA viruses use the same viral protein to antagonize the IFN system, whereas this conservation is limited in large dsDNA viruses (Table S1). Nine common RNA virus antagonists stand out (Table S1).
The NP proteins of Arenaviridae are conserved inhibitors of IFN induction, although their precise molecular mechanism of antagonism remains unclear [12, 13, 14]. Moreover, New World Arenaviruses express Z proteins that bind RIG-I and interfere with its association with MAVS [15]. Phleboviruses and orthobunyaviruses share an NSs protein to antagonize IFN transcription [16, 17•, 18, 19], and influenza A and B viruses express an NS1 protein to inhibit the IFN-inducible antiviral effector protein PKR, as well as the viral sensor RIG-I [20, 21, 22•, 23, 24]. By far the largest virus family with conserved IFN antagonists is that of the Paramyxoviridae. Differences in mechanism exist between different family members, yet their V, P, and C proteins predominantly prevent MDA-5 dimerization and target STAT molecules for proteasomal degradation [25, 26, 27, 28].
Several (+)RNA viruses share common antagonists within their virus family as well. Coronaviruses express the largest known viral RNA genome (>30 000 nt). Their Nsp1 protein is the first protein expressed in infected cells and is an important virulence factor in vivo [29, 30••]. It degrades cellular mRNA, thereby limiting the expression of antiviral genes [30••, 31]. Interestingly, SARS-CoV recently transmitted from bats to humans and expresses several accessory proteins that are not expressed by related coronaviruses such as MHV. Two of these accessory gene products (ORF3b and ORF6) have reported IFN antagonistic activity [32, 33]. An ORF6 deletion virus convincingly demonstrated that the ORF6 protein is involved in preventing nuclear translocation of STAT1 by binding to the nuclear transport protein karyopherin α2 and contributes to pathogenicity. In addition to Coronaviridae, three genera within the Flaviviridae family also share common antagonists within their genus. Pestiviruses BVDV and CSFV Npro proteins target IRF-3 for proteasomal degradation [34, 35]. Moreover, their Erns protein was reported to sequester dsRNA [36, 37, 38]. By contrast, all flaviviruses modulate JAK–STAT signaling, albeit by slightly different mechanisms [39, 40, 41, 42, 43]. The hepaciviruses HCV and GBV-B both cleave MAVS with their main protease NS3/4a, thereby rendering it unable to confer the signal from RIG-I to TBK-1/IKKɛ [44, 45]. Using a similar strategy, enteroviruses commonly cleave MAVS and other innate immune molecules with their main protease 3C [46, 47, 48]. By contrast, non-enteroviruses within the Picornaviridae family express an L protein to target IRF-3 and NFκB [49, 50, 51].
Virus-host coevolution
Despite all the sophisticated mechanisms used by viruses to inhibit the IFN response, a hallmark of virus infection is the induction of IFN in infected hosts. Thus, viral IFN antagonism is not complete, which probably underscores the need for viruses to economize resources to assure optimal replication and transmission. Moreover, hosts have also been under selection pressure to overcome viral antagonism of IFN, and this is exemplified by examples of positive selection in some of the key components of the IFN system targeted by viruses, such as PKR, which appears to have been rapidly evolving in mammalian hosts to escape from antagonism by K3L, a poxvirus PKR inhibitor [52••].
Differential prevalence of antagonistic strategies among different virus classes
Some inhibitory strategies seem to have a higher prevalence in certain virus classes. For example, nearly all viruses with (−)ssRNA genomes interfere with sensory molecules, whereas this is much less so for viruses with other genomes (Figure 1). By contrast, (+)ssRNA and dsDNA viruses preferably target IRFs as means to interfere with IFN induction. Finally, (+)ssRNA viruses target the IFN signaling step to a lesser extent than other viruses (Figure 1). Notable exceptions are members of the Flaviviridae. Although these patterns may be explained by selection bias for studied viruses, they may also indicate that different virus families may utilize certain dominant strategies to antagonize the IFN response. These strategies not necessarily provide the broadest possible antagonism (e.g. block of general gene expression), but may be strongly influenced by the type of infection (e.g. acute, chronic, and persistent), vector/host species, and infected cell types.
Upregulation of cellular factors that downregulate the IFN response
Recently, several viruses have been reported to partly circumvent the IFN circuit by activation or upregulation of cellular inhibitors of the JAK–STAT pathway and PKR. The cellular PKR inhibitor p58(IPK) is activated during FluAV, TMV, and TEV infection [53, 54] and contributes to negative regulation of PKR by direct protein–protein interaction [55, 56]. By contrast, FluAV, RSV, HCV, and HSV induce SOCS-1/3 expression, which negatively regulates the JAK–STAT pathway [57, 58, 59, 60, 61]. It remains unclear if inhibition by cellular components is a general cellular stress-induced negative feedback loop that is activated during virus infection to resolve or fine-tune the IFN response, or if these viruses specifically modulate cells to use their own proteins against them.
IFN antagonists are promising vaccines and antiviral targets
Better understanding of molecular mechanisms by which IFN antagonists influence viral replication and pathogenesis has indicated that IFN antagonist knockout viruses are promising candidates for live virus vaccines. Several groups have shown that viruses lacking fully functional IFN antagonists are rapidly cleared in vivo as the result of a potent IFN response, while their replication competent nature ensures the establishment of long-lasting immune memory. Moreover, these viruses can be grown in large quantities in IFN-deficient hosts for potential vaccine stocks. So far, Influenza A/B virus NS1 deletion mutants have been tested in mice and showed that they could protect against challenge with wild-type virus [62, 63, 64, 65•]. Also, recent evidence from studies with SARS-CoV nsp-1, JEV E, and RSV NS2 mutant viruses suggests that these viruses likewise represent good vaccine candidates [29, 65•, 66•]. The generation of attenuated viruses is aimed at achieving an optimal balance between minimal clinical symptoms and maximum replication in the nasal mucosa. Mucosal immunity is especially effective against respiratory viruses such as influenza viruses, RSV, and coronaviruses [67].
Vaccination is the most cost-effective method for preventing virus infection and disease. However, under certain circumstances, such as in immune compromised patients, antiviral drugs are crucial to treat acute life-threatening cases. Viruses lacking IFN antagonists are severely attenuated. This makes viral IFN antagonist proteins prime drug targets. Screens with large libraries of small-molecule inhibitors have already identified potential new lead compounds against various clinically relevant viruses. Several studies in tissue culture indicate that this is a viable strategy for the development of antiviral drugs [68, 69, 70, 71, 72], although the potential of such compounds to inhibit virus replication in vivo remains unclear.
Concluding remarks
The recent surge in publications on viral IFN antagonists underpins the necessity of every virus to interfere with the innate immune system and provides potential targets for antiviral drugs and rational vaccine design. However, only a limited number of these described antagonists have been extensively validated in animal models by infection studies with recombinant viruses lacking their IFN antagonists. Nevertheless, recent studies with recombinant Ebola virus and SARS-CoV mutants lacking IFN antagonists, initially identified by over expression studies, prove the potential of these approaches [30••, 31, 73, 74, 75, 76]. In depth characterization of IFN antagonists in vivo will be crucial to the development of new vaccine candidates and identification of lead compounds that diminish IFN antagonist function during viral infection.
Since various IFN antagonists target multiple proteins, drugs against these viral factors may prove to work as a double edged sword. The benefit of such a drug would not only be that it inhibits two distinct modes of antagonism, but also that resistant virus mutants are less likely to arise since they would have to facilitate both functions. In particular viral antagonists that also have a direct role in virus replication would be good drug targets. Obtaining crystal structures of viral antagonists in complex with their different viral and cellular ligands will be crucial for the rational design of such drugs.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Research in the A.G.-S. laboratory is partly supported by NIH funding: R01 AI46954, U54 AI57158 (North East Biodefense Center), U19AI83025, U01AI070469, U01AI074539, P01AI058113, P01AI082325 and by CRIP (Center for Research on Influenza Pathogenesis, NIAID contract HHSN266200700010C).
Disclosure: Mount Sinai School of Medicine owns intellectual property in influenza virus reverse genetics and vaccines. A.G.-S. owns equity and receives financial compensation for providing consulting services and for serving on the advisory board of Vivaldi BioScience, a biotechnology company that develops influenza vaccines based on NS-1-modification technology.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in, or financial conflict with, the subjects discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mib.2010.05.009.
Appendix A. Supplementary data
References
- 1.Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 2.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stetson D.B., Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes-Alnemri T., Yu J.W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K.A., Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Sadler A.J., Williams B.R.G. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyles D.S. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol Mol Biol Rev. 2000;64:709–724. doi: 10.1128/mmbr.64.4.709-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith R.W.P., Malik P., Clements J.B. The herpes simplex virus ICP27 protein: a multifunctional post-transcriptional regulator of gene expression. Biochem Soc Trans. 2005;33:499–501. doi: 10.1042/BST0330499. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Sobrido L., Zuniga E.I., Rosario D., Garcia-Sastre A., de la Torre J.C. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2006;80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Sobrido L., Giannakas P., Cubitt B., Garcia-Sastre A., de la Torre J.C. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Sobrido L., Emonet S., Giannakas P., Cubitt B., Garcia-Sastre A., de la Torre J.C. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2009;83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan L., Briese T., Lipkin W.I. Z proteins of New World Arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol. 2010;84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blakqori G., Delhaye S., Habjan M., Blair C.D., Sanchez-Vargas I., Olson K.E., ttarzadeh-Yazdi G., Fragkoudis R., Kohl A., Kalinke U. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol. 2007;81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Ikegami T., Narayanan K., Won S., Kamitani W., Peters C.J., Makino S. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 2009;5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that the NSs protein of RVFV not only suppresses general transcription, but also downregulates PKR at a post-transcriptional level. Control of PKR expression was shown to be essential for efficient virus replication.
- 18.Habjan M., Pichlmair A., Elliott R.M., Overby A.K., Glatter T., Gstaiger M., Superti-Furga G., Unger H., Weber F. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol. 2009;83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikegami T., Narayanan K., Won S., Kamitani W., Peters C.J., Makino S. Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and post-transcriptional downregulation of protein kinase PKR. Ann N Y Acad Sci. 2009;1171(Suppl. 1):E75–E85. doi: 10.1111/j.1749-6632.2009.05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min J.Y., Li S., Sen G.C., Krug R.M. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363:236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Li S., Min J.Y., Krug R.M., Sen G.C. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 22•.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E., Farzan M., Inoue S., Jung J.U., Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; The FluAV NS1 protein interacts with the coil–coil domain of the E3 ligase TRIM25, thereby preventing the activation of RIG-I and subsequent IFN induction.
- 23.Mibayashi M., Martinez-Sobrido L., Loo Y.M., Cardenas W.B., Gale M., Jr, Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan S.L., Katze M.G. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J Interferon Cytokine Res. 1998;18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 25.Andrejeva J., Childs K.S., Young D.F., Carlos T.S., Stock N., Goodbourn S., Randall R.E. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakatsu Y., Takeda M., Ohno S., Shirogane Y., Iwasaki M., Yanagi Y. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J Virol. 2008;82:8296–8306. doi: 10.1128/JVI.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caignard G., Guerbois M., Labernardiere J.L., Jacob Y., Jones L.M., Wild F., Tangy F., Vidalain P.O. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-α/β signaling. Virology. 2007;368:351–362. doi: 10.1016/j.virol.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Kubota T., Yokosawa N., Yokota Si., Fujii N., Tashiro M., Kato A. Mumps virus V protein antagonizes interferon without the complete degradation of STAT1. J Virol. 2005;79:4451–4459. doi: 10.1128/JVI.79.7.4451-4459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using mutant SARS-CoVs, the authors show that the nsp1 virulence factor modifies mRNAs, thereby targeting them for rapid turnover by the host degradation machinery.
- 31.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. SARS-CoV ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rER/Golgi membrane. J Virol. 2007;18:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z., Rijnbrand R., Jangra R.K., Devaraj S.G., Qu L., Ma Y., Lemon S.M., Li K. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology. 2007;366(2):277–292. doi: 10.1016/j.virol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szymanski M.R., Fiebach A.R., Tratschin J.D., Gut M., Ramanujam V.M.S., Gottipati K., Patel P., Ye M., Ruggli N., Choi K.H. Zinc binding in pestivirus Npro is required for interferon regulatory factor 3 interaction and degradation. J Mol Biol. 2009;391:438–449. doi: 10.1016/j.jmb.2009.06.040. [DOI] [PubMed] [Google Scholar]
- 36.Luo X., Ling D., Li T., Wan C., Zhang C., Pan Z. Classical swine fever virus Erns glycoprotein antagonizes induction of interferon-beta by double-stranded RNA. Can J Microbiol. 2009;55:698–704. doi: 10.1139/w09-013. [DOI] [PubMed] [Google Scholar]
- 37.Luo X., Pan R., Wan C., Liu X., Wu J., Pan Z. Glycosylation of classical swine fever virus Erns is essential for binding double-stranded RNA and preventing interferon-beta induction. Virus Res. 2009;146:135–139. doi: 10.1016/j.virusres.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Magkouras I., Matzener P., Rumenapf T., Peterhans E., Schweizer M. RNase-dependent inhibition of extracellular, but not intracellular, dsRNA-induced interferon synthesis by Erns of pestiviruses. J Gen Virol. 2008;89:2501–2506. doi: 10.1099/vir.0.2008/003749-0. [DOI] [PubMed] [Google Scholar]
- 39.Lin R.J, Chang B.L., Yu H.P., Liao C.L., Lin Y.L. Blocking of interferon-induced Jak–Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Best S.M., Morris K.L., Shannon J.G., Robertson S.J., Mitzel D.N., Park G.S., Boer E., Wolfinbarger J.B., Bloom M.E. Inhibition of interferon-stimulated JAK–STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werme K., Wigerius M., Johansson M. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK–STAT signalling. Cell Microbiol. 2008;10:696–712. doi: 10.1111/j.1462-5822.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- 42.Mazzon M., Jones M., Davidson A., Chain B., Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 43.Laurent-Rolle M., Boer E.F., Lubick K.J., Wolfinbarger J.B., Carmody A.B., Rockx B., Liu W., Ashour J., Shupert W.L., Holbrook M.R. The NS5 protein of the virulent West Nile Virus NY99 strain is a potent antagonist of type I interferon-mediated JAK–STAT signaling. J Virol. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z., Benureau Y., Rijnbrand R., Yi J., Wang T., Warter L., Lanford R.E., Weinman S.A., Lemon S.M., Martin A., Li K. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol. 2007;81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drahos J., Racaniello V.R. Cleavage of IPS-1 in cells infected with human rhinovirus. J Virol. 2009;83:11581–11587. doi: 10.1128/JVI.01490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulmann D., Magulski T., Schwarz R., Heitmann L., Flehmig B., Vallbracht A., Dotzauer A. Hepatitis A virus protein 2B suppresses beta interferon (IFN) gene transcription by interfering with IFN regulatory factor 3 activation. J Gen Virol. 2008;89:1593–1604. doi: 10.1099/vir.0.83521-0. [DOI] [PubMed] [Google Scholar]
- 48.Neznanov N., Chumakov K.M., Neznanova L., Almasan A., Banerjee A.K., Gudkov A.V. Proteolytic cleavage of the p65-RelA subunit of NF-κB during poliovirus infection. J Biol Chem. 2005;280:24153–24158. doi: 10.1074/jbc.M502303200. [DOI] [PubMed] [Google Scholar]
- 49.Hato S.V., Ricour C., Schulte B.M., Lanke K.H., de B.M., Zoll J., Melchers W.J., Michiels T., van Kuppeveld F.J. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol. 2007;9:2921–2930. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- 50.Ricour C., Borghese F., Sorgeloos F., Hato S.V., van Kuppeveld F.J.M., Michiels T. Random mutagenesis defines a domain of Theiler's virus leader protein that is essential for antagonism of nucleocytoplasmic trafficking and cytokine gene expression. J Virol. 2009;83:11223–11232. doi: 10.1128/JVI.00829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de los Santos T., Diaz-San Segundo F., Grubman M.J. Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J Virol. 2007;81:12803–12815. doi: 10.1128/JVI.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Elde N.C., Child S.J., Geballe A.P., Malik H.S. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]; PKR has rapidly coevolved to parry poxvirus eIF2α mimicry by K3L by selection across combinations of multiple dynamic interaction surfaces in the protein. It indicates that mammalian hosts are also adapting to viral evasion strategies.
- 53.Goodman A.G., Smith J.A., Balachandran S., Perwitasari O., Proll S.C., Thomas M.J., Korth M.J., Barber G.N., Schiff L.A., Katze M.G. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J Virol. 2007;81:2221–2230. doi: 10.1128/JVI.02151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bilgin D.D., Liu Y., Schiff M., nesh-Kumar S.P. P58IPK, a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev Cell. 2003;4:651–661. doi: 10.1016/s1534-5807(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 55.Melville M.W., Tan S.L., Wambach M., Song J., Morimoto R.I., Katze M.G. The cellular inhibitor of the PKR protein kinase, P58IPK, is an influenza virus-activated co-chaperone that modulates heat shock protein 70 activity. J Biol Chem. 1999;274:3797–3803. doi: 10.1074/jbc.274.6.3797. [DOI] [PubMed] [Google Scholar]
- 56.Melville M., Hansen W., Freeman B., Welch W., Katze M. The molecular chaperone hsp40 regulates the activity of P58IPK, the cellular inhibitor of PKR. Proc Natl Acad Sci U S A. 1997;94:97–102. doi: 10.1073/pnas.94.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bode J.G., Ludwig S., Ehrhardt C., Albrecht U., Erhardt A., Schaper F., Heinrich P.C., Haussinger D. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 58.Yokota S., Yokosawa N., Okabayashi T., Suzutani T., Miura S., Jimbow K., Fujii N. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J Virol. 2004;78:6282–6286. doi: 10.1128/JVI.78.12.6282-6286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oshansky C.M., Krunkosky T.M., Barber J., Jones L.P., Tripp R.A. Respiratory syncytial virus proteins modulate suppressors of cytokine signaling 1 and 3 and the type I interferon response to infection by a toll-like receptor pathway. Viral Immunol. 2009;22:147–161. doi: 10.1089/vim.2008.0098. [DOI] [PubMed] [Google Scholar]
- 60.Pauli E.K., Schmolke M., Wolff T., Viemann D., Roth J., Bode J.G., Ludwig S. Influenza A virus inhibits type I IFN signaling via NF-kappaB-dependent induction of SOCS-3 expression. PLoS Pathog. 2008;4:e1000196. doi: 10.1371/journal.ppat.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pothlichet J., Chignard M., Si-Tahar M. Cutting edge: innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-dependent pathway. J Immunol. 2008;180:2034–2038. doi: 10.4049/jimmunol.180.4.2034. [DOI] [PubMed] [Google Scholar]
- 62.Donelan N.R., Basler C.F., Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hai R., Martinez-Sobrido L., Fraser K.A., Ayllon J., Garcia-Sastre A., Palese P. Influenza B virus NS1-truncated mutants: live-attenuated vaccine approach. J Virol. 2008;82:10580–10590. doi: 10.1128/JVI.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wressnigg N., Voss D., Wolff T., Romanova J., Ruthsatz T., Mayerhofer I., Reiter M., Nakowitsch S., Humer J., Morokutti A. Development of a live-attenuated influenza B ΔNS1 intranasal vaccine candidate. Vaccine. 2009;27:2851–2857. doi: 10.1016/j.vaccine.2009.02.087. [DOI] [PubMed] [Google Scholar]
- 65•.Liang J.J., Liao C.L., Liao J.T., Lee Y.L., Lin Y.L. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine. 2009;27:2746–2754. doi: 10.1016/j.vaccine.2009.03.007. [DOI] [PubMed] [Google Scholar]; These papers show that, like for FluAV, FluBV, and SARS-CoV, recombinant JEV and RSV lacking one of their IFN antagonists form promising live vaccine candidates.
- 66•.Ling Z., Tran K.C., Teng M.N. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol. 2009;83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to [65•].
- 67.Yuki Y., Kiyono H. Mucosal vaccines: novel advances in technology and delivery. Expert Rev Vaccines. 2009;8:1083–1097. doi: 10.1586/erv.09.61. [DOI] [PubMed] [Google Scholar]
- 68.Basu D., Walkiewicz M.P., Frieman M., Baric R.S., Auble D.T., Engel D.A. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J Virol. 2009;83:1881–1891. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemm J.A., O’Boyle D., II, Liu M., Nower P.T., Colonno R., Deshpande M.S., Snyder L.B., Martin S.W., St. Laurent D.R., Serrano-Wu M.H. Identification of Hepatitis C Virus NS5A inhibitors. J Virol. 2010;84:482–491. doi: 10.1128/JVI.01360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emert-Sedlak L., Kodama T., Lerner E.C., Dai W., Foster C., Day B.W., Lazo J.S., Smithgall T.E. Chemical library screens targeting an HIV-1 accessory factor/host cell kinase complex identify novel antiretroviral compounds. ACS Chem Biol. 2009;4:939–947. doi: 10.1021/cb900195c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller N.H., Pattabiraman N., Ansarah-Sobrinho C., Viswanathan P., Pierson T.C., Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of West Nile Virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52:3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bolken T.C., Laquerre S., Zhang Y., Bailey T.R., Pevear D.C., Kickner S.S., Sperzel L.E., Jones K.F., Warren T.K., Amanda Lund S. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World Arenaviruses. Antiviral Res. 2006;69:86–97. doi: 10.1016/j.antiviral.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prins K.C., Delpeut S., Leung D.W., Reynard O., Volchkova V.A., Reid S., Ramanan P., Cardenas W.B., Amarasinghe G.K., Volchkov V.E., Basler C.F. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hartman A.L., Bird B.H., Towner J.S., Antoniadou Z.A., Zaki S.R., Nichol S.T. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartman A.L., Dover J.E., Towner J.S., Nichol S.T. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hartman A.L., Towner J.S., Nichol S.T. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.