Abstract
DNA methyltransferase I (DNMT1) is the major methyltransferase responsible for methylating DNA and is overexpressed in many cancers. DNMT1 is also a therapeutic target for chemotherapy and chemoprevention. We hypothesized that loss of DNMT1 copy number could result in reduced DNMT1 levels and greater sensitivity to DNMT1 inhibitors. We examined DNMT1 expression in pancreatic cancers by immunohistochemistry and western blotting. We also examined DNMT1 copy number in 20 pancreatic cancer cell lines using Affymetrix SNP arrays and correlated copy number with DNMT1 expression. We tested eight pancreatic cancer cell lines with DNMT1 inhibitors and measured growth inhibition. We identified overexpression of DNMT1 relative to normal pancreatic duct in 78.7% of pancreatic cancers (37/47) by immunohistochemistry and in 16/20 pancreatic cancer cell lines by western blot. Pancreatic cancer cell lines with loss of DNMT1 alleles tended to have lower DNMT1 expression (three of nine cell lines) compared to those without DNMT1 copy number loss (one of eleven). 5-aza-deoxycytidine (5-Aza-dC) treatment (1–10 uM) depleted DNMT1 in seven of eight pancreatic cancer cell lines. Three of four pancreatic cancers cell lines with low/normal DNMT1 expression were sensitive to growth inhibition by low dose 5-Aza-dC (1 uM), whereas only one of four cell lines with high DNMT1 expression had growth inhibition, and this occurred without evidence of DNMT1 depletion suggesting a different mechanism for growth inhibition in this cell line. Loss of DNMT1 alleles may reduce DNMT1 levels in some pancreatic cancers. Pancreatic cancers with low DNMT1 expression tend to be more sensitive to low-dose 5-Aza-dc.
Keywords: pancreatic cancer, DNMT1, 5-aza-deoxycytidine, loss of heterozygosity, EGCG
Introduction
DNA methylation regulates gene expression, genomic imprinting and chromatin structure.1 During DNA replication, DNMT1 binds to and methylates the cytosines of palindromic CpG dinucleotides of the daughter strand of newly replicated DNA thereby preserving the parental methylation pattern.2 DNMT1 also interacts with histone proteins to influence chromatin structure.3,4 DNMT1 levels are regulated by transcriptional and posttranscriptional mechanisms5,6 and levels vary by the cell cycle.7 Aberrant DNA methylation contributes to the development of pancreatic8–16 and other cancers,17–21 and a likely contributor to the increased DNA methylation in cancer cells is DNMT1 overexpression.22–25 Both transcriptional activation26 and inhibition of protein degradation are thought to be important in the overexpression of DNMT1 in cancer cells.27,28
The importance of DNMT1 in cancer development is further supported by mouse models, as adenomatous polyposis coli (APC)-min mice hypomorphic for DNMT1 are protected from developing intestinal neoplasia.29 However, the resulting DNA hypomethylation increases the risk of liver tumors,29 highlighting the role of both DNA hypermethylation and DNA hypomethylation in cancer development. Global DNA hypomethylation of repetitive DNA is thought to contribute to the development of some cancers by generation of genomic instability and loss of imprinting.29,30 Methyl group deficiency such as from low vitamin B12 or folate may increase the risk of developing certain cancers including pancreatic cancer.31,32 In addition, pancreatic cancers that acquire defective methylene tetrahydrofolate reductase (MTHFR) function by losing MTHFR alleles are more hypomethylated and have more chromosomal losses than those without.30 Thus, DNMT1 alterations have differing consequences for tumor development depending on the tumor type. DNMT1 expression contributes to cell viability, though other DNMTs compensate when DNMT1 deficient cells.2,33,34 Since DNA hypermethylation often inactivates tumor suppressor genes, DNMT1 inhibitors have been tested as cancer therapeutics. Cytosine analogues such as 5-Aza-dC are incorporated into DNA during replication and covalently link DNMT1 to DNA, effectively preventing DNMT1 from methylating DNA.35 5-Aza-dC has activity against certain cancers and is approved for the treatment of myelodysplastic syndrome (MDS). Drugs that inhibit DNMT1 without the toxicity of 5-Aza-dC would potentially have more therapeutic applications.36 Tea polyphenols such as epigallocatechin-3-O-gallate (EGCG) inhibit DNA methyltransferase-mediated DNA methylation, and act by directly inhibiting DNMTs.37,38 EGCG is one of the most abundant components in green tea extract, and has been shown to inhibit cell growth, malignant invasion and metastasis of pancreatic and other cancers and contribute to apoptosis and growth arrest in experimental systems.39
Although DNMT1 overexpression has been described in several cancers,28,40,41 the mechanism for this overexpression is not known. Aberrant signaling induced by mutant KRAS has been implicated as a cause of DNMT1 overexpression, and KRAS mutations occur in approximately 90% of pancreatic ductal adenocarcinomas.42,43 In addition, both KRAS mutations and aberrant promoter methylation are first observed in precursor lesions of the pancreas.14,15,44–47 However, DNMT1 overexpression also occurs in cancers without KRAS mutations.28
Another cause of altered protein levels in cancer cells is alterations in chromosome copy number. Pancreatic and other adenocarcinomas undergo widespread chromosomal losses during tumor development.48–52 Alterations in gene copy number can influence responses to therapeutic agents. For example, loss of thymidylate synthetase alleles may influence responses to 5-fluorouracil53 and loss of MTHFR copies could have adverse effects on folate metabolism.30 Pancreatic and other cancers frequently lose copies of chromosome 19p, the locus of DNMT1.54,55 We hypothesized that pancreatic cancers with loss of a DNMT1 allele may have lower DNMT1 levels than pancreatic cancers without DNMT1 allelic loss.
In this study we examined the expression of DNMT1 protein in a panel of pancreatic cancers and correlated levels with chromosome 19p copy number and KRAS mutation status. We also examined the response of pancreatic cancer lines with low and high DNMT1 expression to the DNMT1 inhibitors 5-Aza-dC and EGCG.
Results
Expression of DNMT1 in pancreatic cancer cell lines
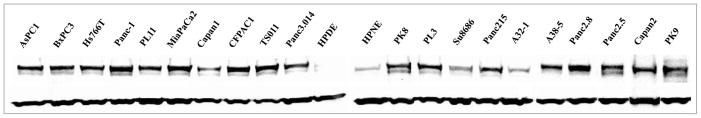
We examined the expression of DNMT1 protein in 20 pancreatic cancer cell lines and two non-neoplastic pancreatic lines (HPDE and HPNE) by western blot (Fig. 1). Sixteen of 20 (80%) of the pancreatic cancer lines had higher DNMT1 levels than HPDE and HPNE after normalizing protein abundance by GAPDH. By western blot most of the pancreatic cancers had approximately 2–5 fold higher levels than the reference non-neoplastic cell lines. Four of the pancreatic cancer lines A32-1, Capan1, Su8686 and Panc3.014 had DNMT1 levels approximately the same as that of the non-neoplastic lines.
Figure 1.
Expression of DNMT1 in cell lines. Western blot analyses for DNMT1 in pancreatic cancer (20) and non-neoplastic cell lines (2). DNMT1 (~190 kDa) was detected with GAPDH (~40 kDa) served as a loading control.
DNMT1 immunohistochemistry in pancreatic cancers
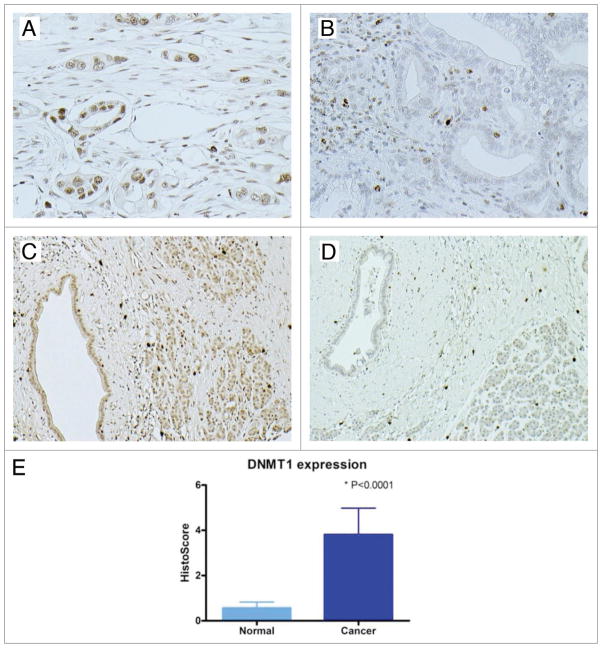
Next we analyzed DNMT1 expression in pancreatic cancer tissues by immunohistochemistry. DNMT1 labeling was detected in only a minority of normal pancreatic duct and acinar cells. Lymphocytes express sufficient DNMT1 to be observed by immunohistochemistry providing a useful positive control. DNMT1 labeling indicative of overexpression compared to normal pancreas was observed in 37 of 47 pancreatic cancers, a similar proportion to that observed in the pancreatic cancer cell lines. Although cancer samples showed significant higher DNMT1 score than normal control (p < 0.001) (Fig. 2), there were no differences in the clinicopathological parameters between DNMT1 expressing and non-expressing cases (data not shown).
Figure 2.
Immunohistochemical staining of DNMT1 protein in pancreatic cancer. (A) Nuclear labeling of DNMT1 and (B) no DNMT1 labeling in the malignant tumor cells; (C) Expression of DNMT1 and (D) Lack of DNMT1 expression in normal ductal epithelial cells; (E) A “Histo-score” was generated as the product of intensity times area.
Chromosome 19p copy number, KRAS mutation and DNMT1 expression
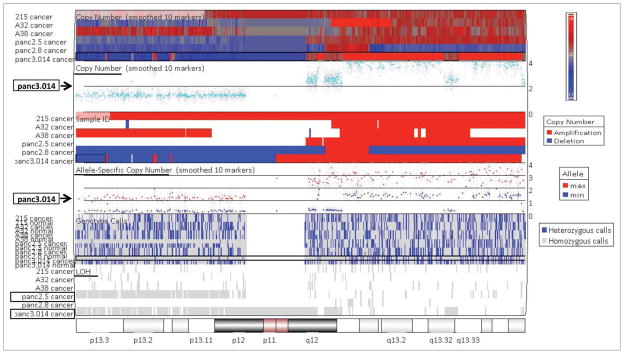
We hypothesized that one mechanism for differences in the expression of DNMT1 in different pancreatic cancers would be loss of a DNMT1 allele through loss of the portion of chromosome 19p containing DNMT1. Loss of chromosome 19p is common in pancreatic cancers.51,52 We examined the copy number profiles of pancreatic cancer cell lines using Affymetrix SNP arrays looking in particular for evidence copy number loss and allelic loss (loss of heterozygosity or LOH) at the DNMT1 locus on chromosome 19p13.2. Copy number profiles of eight of these lines was determined using the Affymetrix 50K SNP arrays,53 with Affymetrix 250K SNP arrays used for the remaining lines (Fig. 3). LOH at chromosome 19p was observed in 75.0% (15/20) of these pancreatic cancer lines, but seven of the cancers with 19p LOH did not have 19p copy number loss because of duplication of the retained chromosome. One cell line A32-1 had partial copy number loss with two 19p alleles indicating that the cell line contained a mixed population of cells some with and some without 19p allelic loss. There was no difference in the levels of DNMT1 protein in cell lines with and without 19p LOH. We next compared pancreatic cancers with and without loss of 19p. Nine of 20 (45.0%) of the pancreatic cancer lines had 19p loss and three of them (A32-1, Capan1 and Panc3.014) had low DNMT1 levels. In contrast, only 1 of the eleven lines without copy number loss at 19p had low DNMT1 protein levels (33.3 vs. 9.1%; p > 0.05) (Tables 1 and 2). Overall, although the association was not statistically significant, our data is consistent with the hypothesis that 19p copy number loss could cause low DNMT1 levels in some pancreatic cancers, but in other cancers additional mechanisms could still cause DNMT1 overexpression. Interestingly, we also identified an isolated loss of 19p in the HPDE cell line raising the possibility that copy number loss in this line could contribute to low DNMT1 expression.
Figure 3.
Copy number analysis and LOH detection at the DNMT1 locus (19p13.2) by Affymetrix 250K SNP arrays in pancreatic cancer cell lines (representative shown). Copy number loss was observed in Panc3.014 compared to the matched lymphocytes. LOH appeared on this locus in Panc3.014 and Panc2.5 cell lines.
Table 1.
E xpression of DNMT1, loss of copy number and LINE1 methylation in pancreatic cancer cell lines
| Cell line | Relative low expression of DNMT1 | Allelic loss | Loss of copy number | KRAS status | % LINE1 methylation |
|---|---|---|---|---|---|
| Cancer | |||||
| A32-1 | + | −* | +* | M | 71.48 |
| Capan1 | + | + | + | M | 54.98 |
| Capan2 | − | + | + | M | ND |
| CFPAC1 | − | + | + | M | 61.28 |
| Panc3.014 | + | + | + | M | 65.76 |
| Panc-1 | − | + | + | M | 60.74 |
| PK9 | − | + | + | W | 77.81 |
| PL3 | − | + | + | W | 55.00 |
| TS011 | − | + | + | M | ND |
| Panc215 | − | − | − | M | 57.60 |
| A38-5 | − | − | − | M | ND |
| AsPC1 | − | + | − | W | 70.72 |
| BxPC3 | − | − | − | M | 31.94 |
| Hs766 | − | + | − | M | 53.54 |
| MiaPaCa2 | − | + | − | M | 71.53 |
| Panc2.5 | − | + | − | M | ND |
| Panc2.8 | − | − | − | M | ND |
| PK8 | − | + | − | W | 75.29 |
| PL11 | − | + | − | W | 66.55 |
| Su8686 | + | + | − | M | 47.34 |
| Non-neoplastic | |||||
| HPDE | + | + | + | W | 68.69 |
| HPNE | + | − | − | W | 64.79 |
M, mutation type; W, wild-type; ND, not determined;
, this cell line has partial loss of copy number.
Table 2.
Correlation of copy number loss, KRAS mutation and LINE1 methylation with low expression of DNMT1 in pancreatic cancer cell lines
| Relative low expression of DNMT1 (+) | Relative low expression of DNMT1 (−) | p value | |
|---|---|---|---|
| Loss of copy number (+) | 33.3% (3/9) | 66.7% (6/9) | |
| Loss of copy number (−) | 9.1% (1/11) | 90.9% (10/11) | Chi test, p = 0.178 |
| KRAS mutant type | 26.7% (4/15) | 73.3% (11/15) | |
| KRAS wild-type | 0.0% (0/5) | 100% (5/5) | Chi test, p = 0.197 |
| % LINE1 methylation | 59.9 ± 10.8% | 62 ± 12.9% | t-test, p = 0.776 |
Since mutational activation of the KRAS pathway has been implicated as a cause of DNMT1 overexpression, we also examined the relationship between KRAS status and DNMT1 expression level, but found no significant relationship between mutant KRAS and DNMT1 expression level. Of the 20 cell lines examined, five were wild-type for KRAS and none of these five had low DNMT1 levels (Tables 1 and 2).
Effects of DNMT1 on global DNA methylation in pancreatic cancer
Since reduced expression of DNMT1 can lead to a loss of DNA methylation, we next examined the relationship between DNMT1 levels and a global measure of DNA methylation using the methylation of the repetitive element LINE1 (a CpG-rich repeat sequence abundant throughout the genome). The percentage methylation of LINE1 elements of each cancer line was determined using COBRA. Interestingly, there was no difference in the percentage of LINE1 methylation among cell lines with elevated DNMT1 (62.0 +/− 12.9%) vs. those with normal or reduced DNMT1 (59.9 +/− 10.8%, Student’s t-test, p > 0.05) (Tables 1 and 2).
Effects of 5-Aza-dC and EGCG on viability of cells with deficient and proficient DNMT1 in pancreatic cancer
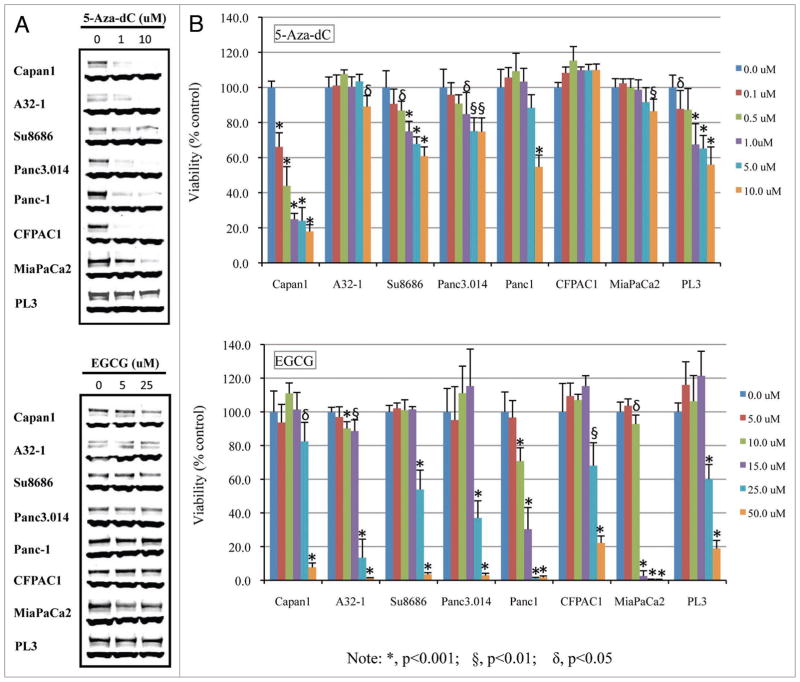
We next determined if the sensitivity of pancreatic cancer cell lines to DNMT1 inhibition correlated with levels of DNMT1. We first tested the DNA methyltransferase inhibitor 5-Aza-dC in eight pancreatic cancer lines, four with low/normal DNMT1 levels and four with high DNMT1 levels. Cells were treated with increasing concentrations of 5-Aza-dC (0, 1, 10 uM) for 48 h. Treatment led to a dose-dependent depletion of DNMT1 in seven of the eight cell lines, with PL3 showing minimal depletion. All seven of these cell lines had partial depletion of DNMT1 at 1 uM concentrations of 5-Aza-dC, and almost complete depletion of DNMT1 was observed in Capan1, A32-1 and CFPAC1 at the highest 5-Aza-dC dose tested (10 uM) (Fig. 4A).
Figure 4.
Response of pancreatic cancer cell lines with deficient and proficient DNMT1 to the DNA methylation inhibitor 5-Aza-dC and tea polyphenol (EGCG). (A) Effect of increasing concentration of 5-Aza-dC (0, 1, and 10 uM) and EGCG (0, 5, and 25 uM) on expression of DNMT1 protein. GAPDH used as a loading control; (B) Effect of DNMT1 on cell viability. Cells were treated with indicted doses of 5-Aza-dC for 96 h (fresh drug added every 48 h) and then cell viability was determined using the MTS assay.
We next examined the effects of 5-Aza-dC on the viability of the same eight cell lines. All the cells were treated with 0.1 to 10 uM 5-Aza-dC for 48 and 96 h (with fresh drugs added after 48 h) and then the cell proliferation was assessed using the MTS assay. The cell lines with high DNMT1 cells showed little growth inhibition to 48 h of 5-Aza-dC treatment, whereas the four cell lines with low/normal DNMT1 levels had a modest inhibition of growth in response to 5-Aza-dC. One of these one cell lines (Capan1) had ~20% decrease in growth with low-dose 5-Aza-dC (0.5 uM) (data not shown). After 96 h incubation, Capan1 cell number was reduced by doses as low as of 0.1 uM (p < 0.001) (Fig. 4B). Similarly, the cell lines Su8686 and Panc3.014 which also had low/normal DNMT1 levels were somewhat sensitive to low dose 5-Aza-dC showing an approximately 20% reduction of cell viability at 1.0 uM (p < 0.001, p < 0.05, respectively). Thus, at 1.0 uM 5-Aza-dC concentrations three of the four cell lines with low/normal DNMT1 levels (Capan1, Su8686 and panc3.014) were sensitive to 5-Aza-dC. In contrast, only one of the cell lines with high DNMT1 (PL3) had a similar response to low-dose 5-Aza-dC (p < 0.05). Interestingly, PL3 did not show much depletion of DNMT1 with 5-Aza-dC treatment. It is possible that the effective concentration of 5-Aza-dC was lower in this cell line because of drug efflux, or that the cell line retained the ability to produce enough DNMT1 to overcome the effect of the drug. It also suggests that its sensitivity to the drug could be for reasons other than DNMT1 inhibition, although the lack of response of PL3 raises the possibility that 5-Aza-dC sensitivity does not depend on DNMT1 levels. None of the remaining cell lines showed inhibition to 5-Aza-dC even at 96 h. Overall these data support the hypothesis that cancers with low/normal DNMT1 expression are usually more sensitive to 5-Aza-dC than those with high DNMT1 expression.
Next, we examined the responses of these same pancreatic cancer lines to another DNMT1 inhibitor, EGCG. DNMT1 protein levels and cell viability were measured in response to a range of concentration of EGCG (0, 5 and 25 uM for western blot) and (0, 5, 10, 15, 25 and 50 uM for MTS assay, respectively). In contrast to the response to 5-Aza-dC, there was minimal depletion of DNMT1 protein at the concentrations tested for most of the cell lines (Fig. 4A). The MTS assay indicated a greater sensitivity to EGCG than to 5-Aza-dC (Fig. 4B). There was no clear relationship between the DNMT1 level of cell lines and their EGCG response. This is consistent with the mechanism of inhibition of DNMT1 by direct inhibition rather than by protein depletion. Panc-1 and MiaPaCa2 were the most sensitive to EGCG with MiaPaCa2 showing almost complete inhibition at a concentration of 15 uM after 96 h in culture.
Discussion
In this study we find that many pancreatic cancers overexpress DNMT1 relative to normal or non-neoplastic pancreas cells. Indeed, a similar fraction of pancreatic cancers were found to overexpress DNMT1 by western blot analysis of pancreatic cancer cell lines (16 of 20, 80.0%) or by immunohistochemistry (37 of 47, 78.7%). We attempted to identify molecular differences between DNMT1 overexpressing and non-overexpressing cell lines. First, we hypothesized that pancreatic cancers with chromosomal loss of a copy of DNMT1 would be more likely to have low DNMT1 levels. We also correlated DNMT1 expression with KRAS mutational status. We found that even KRAS wild-type cancers had DNMT1 overexpression and there was no evidence that KRAS mutation was associated with DNMT1 levels. For the chromosome loss analysis, we found that of 20 pancreatic cancers examined for chromosome 19p loss using Affymetrix SNP arrays, three of nine with 19p copy number loss had low DNMT1 levels, but only one of the eleven cell lines without loss of a DNMT1 locus had low DNMT1 levels. Although this difference did not reach statistical significance, these data are consistent with the hypothesis that copy number loss could result in lower DNMT1 levels if other mechanisms that cause DNMT1 overexpression are not present to compensate for loss of a DNMT1 allele.
We next determined if low DNMT1 levels influenced pancreatic cancer responses to the DNMT1 inhibitors, 5-Aza-dC and EGCG. We found that 5-Aza-dC depleted DNMT1 protein in all cell lines except the line PL3 and found that cell lines with high DNMT1 levels showed less growth inhibition than cell lines with low/normal DNMT1. In contrast, although pancreatic cancer growth could be inhibited by EGCG, responses to EGCG did not correlate with DNMT1 levels which is consistent with the mechanism of EGCG inhibition of DNMT1.38 Recent studies have demonstrated that pancreatic cancer cell growth can be inhibited by EGCG.39 It should be noted that both 5-Aza-dC and EGCG are likely to have off target effects besides DNMT1 that could contribute to their responses in vitro. These data are consistent with the hypothesis that there is a dose-dependent depletion of DNMT1 with 5-Aza-dC and pancreatic cancers with low DNMT1 are more likely to deplete their DNMT1 levels at lower doses of 5-Aza-dC than cancers with high DNMT1 levels. Indeed, the level of inhibition of DNMT1 may be critical to the cellular response to 5-Aza-dC, as recent studies suggest that complete depletion of DNMT1 may be required to kill cancer cells.34,60,61 Our results suggest that by reducing DNMT1 levels chromosome 19p copy number loss could render cancer cells more sensitive to DNMT1 depleting agents such as 5-Aza-dC.
Numerous studies have evaluated the potential therapeutic benefit of using DNMT1 inhibitors to treat cancers. While DNMT1 inhibitors such as 5-Aza-dC have modest activity against myeloid neoplasms, they have not proven to be effective as single agents against solid neoplasms in clinical trials despite the fact that they inhibit cancer cell proliferation and cause cell death in vitro. Further clinical trials are necessary to determine if DNMT1 inhibitors may have more potent effects when used in combination with other chemotherapeutics.62 Our results provide preliminary evidence that pancreatic cancer growth inhibition from DNMT1 inhibitors may depend on levels of DNMT1 protein. It would be useful to determine if solid tumor responses to DNMT1 inhibitors were more likely in cancers with low DNMT1 levels.
In conclusion, we find that while many pancreatic cancers overexpress DNMT1, some pancreatic cancers have low/normal DNMT1 levels. Pancreatic cancers with low/normal DNMT1 levels appear to be more sensitive to DNMT1 inhibition by 5-Aza-dC.
Methods
Cell lines and chemicals
Twenty human pancreatic cancer cell lines A32-1, A38-5, AsPC1, BxPC3, Capan1, Capan2, CFPAC1, Hs766T, MiaPaCa2, Panc-1, Panc2.5, Panc2.8, Panc3.014, Panc215, PK8, PK9, PL3, PL11, Su8686 and TS011 were grown in standard culture conditions. The majority of these cell lines were generated from primary pancreatic cancers at this institution either by Dr. Elizabeth Jaffee (Panc1, Panc2.5, Panc2.8, Panc3.014, PL3, PL11 and TS011) or Dr. James Eshleman (A32-1, A38-5 and Panc215). PK8 and PK9 were obtained from Dr. Akira Hori, University of Sendai Japan, who created these cell lines and the commercial cell lines (AsPC1, BxPC3, Capan1, Capan2, CFPAC1, Hs766T, MiaPaCa2 and Panc1) were obtained from ATCC. The human non-neoplastic pancreatic ductal epithelium (HPDE) cell line and the Nestin-expressing human pancreatic cell line (HPNE) were generously provided by Dr. Ming-Sound Tsao (University of Toronto, Ontario Canada) and Dr. Michel M. Ouellette (University of Nebraska Medical Center, Omaha, NE), respectively. HPNE has a pancreatic fibroblast phenotype. To our knowledge, HPDE is the only non-neoplastic pancreatic duct line available. 5-Aza-dC and EGCG were purchased from Sigma and were dissolved in 1× phosphate-buffered saline (PBS; pH 7.0).
Drug treatment
Eight pancreatic cancer cell lines (A32-1, Capan1, Su8686, Panc3.014, CFPAC1, MiaPaCa2, Panc-1 and PL3) were treated with 5-Aza-dC and EGCG, respectively. Cells in log phase growth were seeded in 10 cm cell culture dishes. After overnight incubation, the cells were cultured with 0.1–10 uM 5-Aza-dC and 5–50 uM EGCG, with drug and media replaced every 48 h.
Western blot
Cells were collected and washed with 1× PBS. Cells were lyzed using ultrasound in RIPA buffer (Sigma), and then centrifuged at 13,000 rpm for 30 min. The supernatant was collected, and the protein concentration was determined using the Bio-Rad protein assay reagent. An equal amount of protein (40 ug) from each sample was separated by 4–12% gradient SDS gel electrophoresis (Invitrogen), transferred to nitrocellulose membranes, and incubated in blocking solution (5% milk in TBS with 0.1% Tween 20) for 60 min. Membranes were incubated overnight with a polyclonal antibody against DNMT1 (rabbit-anti DNMT1)41,56 (generously provided by Dr. Bill Nelson, Johns Hopkins University) at 1:200 dilution or a monoclonal antibody against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) at 1:2,000 dilution. Secondary HRP-linked antibodies (Santa Cruz Biotechnology, Amersham Biosciences) were applied at 1:2,000 dilution and proteins were detected using an ECL kit (Amersham Biosciences).
Immunohistochemistry
The expression of DNMT1 protein was examined by immunohistochemical labeling of tissue microarrays (TMAs) (kindly performed by Dr. Angelo DeMarzo and Ms. Jessica L. Hicks, Johns Hopkins University). Antigen retrieval was performed in high temperature target retrieval buffer, heated at 95°C in a steamer for 40 min. After blocking endogenous peroxidase activity with a H2O2 solution, the sections were incubated with an anti-DNMT1 polyclonal antibody at 1:1,000 for 45 min. Labelling was detected with the Envision Plus Detection Kit (DAKO, Carpinteria, CA, USA). All sections were counterstained with hematoxylin. The results of immunohistochemical staining were scored based on intensity of staining as 0 (negative), 1 (weak) or 2 (strong), and based on the percentage of positive epithelial cells as 0 (<5%), 1 (6–25%), 2 (26–50%), 3 (51–75%) or 4 (>76%), respectively. A “histo-score” was generated as the product of intensity multiplied by area.
Total genomic copy number and LOH analysis
Genomic DNA extracted from pancreatic cancer cell lines, matching normal samples (peripheral blood lymphocytes or immortalized lymphocyte) and non-neoplastic cell lines HPDE and HPNE was digested and hybridized to the Affymetrix GeneChip Human Mapping 250K Sty I array and/or Nsp I array according to the manufacture’s instruction. Genotype calls were determined by BRLMM algorithm using the Affymetrix Chromosome Copy Number Analysis Tool 4.0 (CNAT 4.0). Genotyping call and probe intensities were imported into Partek® Genomics Suite ver. 6.4 (Partek GS). To create copy number- and LOH-profiles, primary pancreatic cancer line SNP calls were compared to their paired normal sample where available and the AsPC1, BxPC3, Capan2, CFPAC1, Panc-1, Su8686, HPDE and HPNE were compared to the SNP array data of 270 individuals from the HapMap collection. In order to detect regions of copy number gain and loss we applied the Partek Genomic Suite (v6.3) Segmentation tool with segmentation parameters set at: minimum genomic markers: 10, p-value threshold: 0.001, and signal to noise: 0.3. LOH region was detected by Hidden Markov Model with parameters set at maximum probability: 0.99, genomic decay 10, genotyping error rate: 0.1.
Cell viability assay
Cell growth was assessed utilizing a CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS, Promega), which was done following the manufacturer’s instructions. 3,000 cells in 100 ul medium were plated in each well of 96-well plates and treated as described above. Parallel cell-free reactions were set up and used for background correction. Measurements were performed in six replicates.
DNA isolation and bisulfite treatment
DNA was isolated from cell line samples using the DNeasy Tissue Kit (Qiagen) and bisulfite modification was done as previously described.57
KRAS mutation detection
PCR amplification of the KRAS gene first exon was performed using an upstream KRAS primer (5′-ACT GAA TAT AAA CTT GTG GTA GTT GGA CCT-3′) that encoded a G to C substitution at the first position of codon 11 and a downstream primer 5′-TCA AAG AAT GGT CCT GCA CC-3′.58 15 ul PCR reactions were done with Platinum Taq polymerase. The PCR generated a DNA fragment of 157 nucleotides and the upstream KRAS primer generated a BstN1 restriction enzyme site (CCTGG) overlapping the first two nucleotides of codon 12. Upon incubation with BstN1, the fragments containing the wild-type codon 12 sequences were cleaved, resulting in two bands of 128 and 29 nucleotides. Fragments containing mutations at either the first or second positions of codon 12 were not cleaved.
Assay of global DNA methylation
The level of methylation of the long interspersed nucleotide element-1 (LINE1) repetitive elements was measured using combined bisulfite restriction analysis (COBRA) and previously reported primers and PCR conditions. Two separate assays were done and results were averaged.59
Statistics
Chi-square tests were used to determine differences in the proportion of copy number loss and KRAS mutation between normal and reduced DNMT1 group. The average percentage of LINE1 methylation was compared using Student’s t-test. A two-tailed p value of less than 0.05 was used to assess statistical significance. Statistical analysis was performed using the Excel statistics software (Microsoft, Redmond, WA), STATA version 8.2 software.
Acknowledgments
This work was supported by the National Cancer Institute grant (CA120432, CA62924), the V foundation, and the Michael Rolfe Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/10750
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–5. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 3.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 4.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 5.Suetake I, Kano Y, Tajima S. Effect of aphidicolin on DNA methyltransferase in the nucleus. Cell Struct Funct. 1998;23:137–42. doi: 10.1247/csf.23.137. [DOI] [PubMed] [Google Scholar]
- 6.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–13. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Sun L, Jost JP. In differentiating mouse myo-blasts DNA methyltransferase is posttranscriptionally and posttranslationally regulated. Nucleic Acids Res. 1996;24:2718–22. doi: 10.1093/nar/24.14.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–65. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–66. [PubMed] [Google Scholar]
- 10.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–42. [PubMed] [Google Scholar]
- 11.Sato N, Goggins M. Epigenetic alterations in intraductal papillary mucinous neoplasms of the pancreas. J Hepatobiliary Pancreat Surg. 2006;13:280–5. doi: 10.1007/s00534-005-1056-2. [DOI] [PubMed] [Google Scholar]
- 12.Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2006;13:286–95. doi: 10.1007/s00534-005-1057-1. [DOI] [PubMed] [Google Scholar]
- 13.Sato N, Matsubayashi H, Abe T, Fukushima N, Goggins M. Epigenetic downregulation of CDKN1C/p57KIP2 in pancreatic ductal neoplasms identified by gene expression profiling. Clin Cancer Res. 2005;11:4681–8. doi: 10.1158/1078-0432.CCR-04-2471. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Yeo CJ, Hruban RHMG. Preproenkephalin and p16 gene CpG island hypermethylation in pancreatic intraepithelial neoplasia (PanIN) and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–81. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato N, Ueki T, Fukushima N, Iacobuzio-Donahue CA, Yeo CJ, Cameron JL, Hruban RHMG. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas increases with histological grade. Gastroenterology. 2002;123:1365–72. doi: 10.1053/gast.2002.34160. [DOI] [PubMed] [Google Scholar]
- 16.Sato NFN, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2007;21:238–44. doi: 10.1038/modpathol.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–40. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 18.Matsubayashi H, Skinner H, Iacobuzio-Donahue C, Abe T, Sato N, Riall TS, et al. Pancreaticobiliary cancers with deficient methylenetetrahydrofolate reductase genotypes. Clin Gastro Hepatol. 2005;3:752–60. doi: 10.1016/s1542-3565(05)00359-9. [DOI] [PubMed] [Google Scholar]
- 19.Matsubayashi H, Sato N, Brune K, Blackford AL, Hruban RH, Canto M, et al. Age- and disease-related methylation of multiple genes in non-neoplastic duodenal tissues. Clin Cancer Res. 2005;11:573–83. [PubMed] [Google Scholar]
- 20.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 21.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–6. [PubMed] [Google Scholar]
- 23.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–71. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 24.Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, Hirohashi S. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105:527–32. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 25.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–22. [PubMed] [Google Scholar]
- 26.Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science. 1999;283:387–90. doi: 10.1126/science.283.5400.387. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res. 2008;6:873–83. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–10. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 29.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580–5. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsubayashi H, Skinner HG, Iacobuzio-Donahue C, Abe T, Sato N, Riall TS, et al. Pancreaticobiliary cancers with deficient methylenetetrahydrofolate reductase genotypes. Clin Gastroenterol Hepatol. 2005;3:752–60. doi: 10.1016/s1542-3565(05)00359-9. [DOI] [PubMed] [Google Scholar]
- 31.Stolzenberg-Solomon RZ, Albanes D, Nieto FJ, Hartman TJ, Tangrea JA, Rautalahti M, et al. Pancreatic cancer risk and nutrition-related methyl-group availability indicators in male smokers. J Natl Cancer Inst. 1999;91:535–41. doi: 10.1093/jnci/91.6.535. [DOI] [PubMed] [Google Scholar]
- 32.Stolzenberg-Solomon RZ, Pietinen P, Barrett MJ, Taylor PR, Virtamo J, Albanes D. Dietary and other methyl-group availability factors and pancreatic cancer risk in a cohort of male smokers. Am J Epidemiol. 2001;153:680–7. doi: 10.1093/aje/153.7.680. [DOI] [PubMed] [Google Scholar]
- 33.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–7. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 34.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–6. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 35.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 37.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–70. [PubMed] [Google Scholar]
- 38.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–30. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 39.Qanungo S, Das M, Haldar S, Basu A. Epigallocatechin-3-gallate induces mitochondrial membrane depolarization and caspase-dependent apoptosis in pancreatic cancer cells. Carcinogenesis. 2005;26:958–67. doi: 10.1093/carcin/bgi040. [DOI] [PubMed] [Google Scholar]
- 40.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–60. [PubMed] [Google Scholar]
- 42.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–7. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hruban RH, van Mansfeld ADM, Offerhaus GJA, van Weering DHJ, Allison DC, Goodman SN, et al. K-ras oncogene activation in adenocarcinomas of the human pancreas: a study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–54. [PMC free article] [PubMed] [Google Scholar]
- 44.Brune K, Goggins M, O’Mailey L, Abe T, Canto M, Klein A, et al. Detailed pathologic evaluation of non-invasive precursor lesions of the pancreas in patients with a strong family history of pancreatic cancer. Am J Surg Pathol. 2006;30:1067–76. [PMC free article] [PubMed] [Google Scholar]
- 45.DiGiuseppe JA, Hruban RH, Offerhaus GJ, Clement MJ, van den Berg FM, Cameron JL, van Mansfeld AD. Detection of K-ras mutations in mucinous pancreatic duct hyperplasia from a patient with a family history of pancreatic carcinoma. Am J of Pathol. 1994;144:889–95. [PMC free article] [PubMed] [Google Scholar]
- 46.Hong SM, Kelly D, Griffith M, Omura N, Li A, Li CP, et al. Multiple genes are hypermethylated in intraductal papillary mucinous neoplasms of the pancreas. Modern Path. 2008;21:1499–507. doi: 10.1038/modpathol.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2008;21:238–44. doi: 10.1038/modpathol.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harada T, Chelala C, Bhakta V, Chaplin T, Caulee K, Baril P, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–60. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calhoun ES, Hucl T, Gallmeier E, West KM, Arking DE, Maitra A, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–8. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 50.Pole JC, Courtay-Cahen C, Garcia MJ, Blood KA, Cooke SL, Alsop AE, et al. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene. 2006;25:5693–706. doi: 10.1038/sj.onc.1209570. [DOI] [PubMed] [Google Scholar]
- 51.Abe T, Fukushima N, Brune K, Boehm C, Sato N, Matsubayashi H, et al. Genome-wide allelotypes of familial pancreatic adenocarcinomas and familial and sporadic intraductal papillary mucinous neoplasms. Clin Cancer Res. 2007;13:6019–25. doi: 10.1158/1078-0432.CCR-07-0471. [DOI] [PubMed] [Google Scholar]
- 52.Kowalski J, Morsberger LA, Blackford A, Hawkins A, Yeo CJ, Hruban RH, Griffin CA. Chromosomal abnormalities of adenocarcinoma of the pancreas: identifying early and late changes. Cancer Genet Cytogenet. 2007;178:26–35. doi: 10.1016/j.cancergencyto.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Brody JR, Hucl T, Gallmeier E, Winter JM, Kern SE, Murphy KM. Genomic copy number changes affecting the thymidylate synthase (TYMS) gene in cancer: a model for patient classification to aid fluoropyrimidine therapy. Cancer Res. 2006;66:9369–73. doi: 10.1158/0008-5472.CAN-06-2165. [DOI] [PubMed] [Google Scholar]
- 54.Iacobuzio-Donahue CA, van der Heijden MS, Baumgartner MR, Troup WJ, Romm JM, Doheny K, et al. Large-scale allelotype of pancreaticobiliary carcinoma provides quantitative estimates of genome-wide allelic loss. Cancer Res. 2004;64:871–5. doi: 10.1158/0008-5472.can-03-2756. [DOI] [PubMed] [Google Scholar]
- 55.Calhoun ES, Hucl T, Gallmeier E, West KM, Arking DE, Maitra A, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–8. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 56.Agoston AT, Argani P, De Marzo AM, Hicks JL, Nelson WG. Retinoblastoma pathway dysregulation causes DNA methyltransferase 1 overexpression in cancer via MAD2-mediated inhibition of the anaphase-promoting complex. Am J Pathol. 2007;170:1585–93. doi: 10.2353/ajpath.2007.060779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parsi MA, Li A, Li CP, Goggins M. DNA methylation alterations in endoscopic retrograde cholangiopancreatography brush samples of patients with suspected pancreaticobiliary disease. Clin Gastroenterol Hepatol. 2008;6:1270–8. doi: 10.1016/j.cgh.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brune K, Hong SM, Li A, Yachida S, Abe T, Griffith M, et al. Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiol Biomarkers Prev. 2008;17:3536–42. doi: 10.1158/1055-9965.EPI-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsubayashi H, Sato N, Brune K, Blackford AL, Hruban RH, Canto M, et al. Age- and disease-related methylation of multiple genes in nonneoplastic duodenum and in duodenal juice. Clin Cancer Res. 2005;11:573–83. [PubMed] [Google Scholar]
- 60.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, et al. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004;24:1270–8. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, et al. Overcoming resistance to inter-feron-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–9. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]