Abstract
MicroRNAs (miRNAs) are non-coding RNA molecules ~22 nucleotides in length that post-transcriptionally regulate gene expression by complementary binding to target mRNAs. MiRNAs have been identified in a diverse range of both metazoan and plant species. Functionally, miRNAs modulate multiple cellular processes including development, hematopoiesis, immunity, and oncogenesis. More recently, DNA viruses were found to encode and express miRNAs during host infection. While the function of most viral miRNAs are not well understood, early analysis of target genes pointed to immune modulation suggesting that viral miRNAs are a component of the immune evasion repertoire which facilitates viral persistence. In addition to directly targeting immune functions, viral encoded miRNAs contribute to immune evasion by targeting pro-apoptotic genes, and in the case of herpesviruses, by controlling viral latency. Here we summarize the recently discovered targets of viral miRNAs and discuss the complex nature of this novel emerging regulatory mechanism.
MiRNAs are encoded by DNA viruses
Viral miRNA biogenesis, like cellular, initiates in the nucleus, where the RNase III endonuclease Drosha cleaves pri-miRNA hairpins into pre-miRNAs. These pre-miRNAs are exported into the cytoplasm by the Exportin 5/Ran GTPase pathway where they are further cleaved by another RNase III endonuclease, Dicer, into a short dsRNA duplex. Finally, one strand of the duplex is incorporated into the RNA-induced silencing complex (RISC) which targets 3′UTR’s of mRNAs containing complementary sequences, leading to translational silencing and/or transcript cleavage [1,2].
In 2004 Tuschl and colleagues discovered the first viral encoded miRNAs in Epstein Barr Virus (EBV) infected Burkitt’s lymphoma cells [3]. To date, the miRNA registry miRBase (http://www.mirbase.org/) [4,5] contains 176 viral miRNAs, of these, 173 are encoded by DNA viruses which replicate in the nucleus (e.g., herpesvirus and polyomavirus). While polyomaviruses encode 2 miRNAs, members of all three herpesvirus subfamilies (Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae) encode between 7 (HSV-1) and 35 (EBV) miRNAs (recently reviewed in [6,7]).
Despite high-throughput sequencing attempts, RNA viruses (e.g. Influenza, HIV, and HCV) and cytoplasmic replicating DNA viruses (Poxviruruses), have not been found to encode miRNAs. The absence of viral miRNAs from these viruses may reflect their inability to access nuclear Drosha and the requirement for RNA viruses to protect their genome from Drosha/Dicer processing. Interestingly, the majority of identified viral miRNAs are encoded by herpesviruses, suggesting that they play an essential role in the herpesvirus lifecycle. This review will focus on the known targets of viral miRNAs, with a strong emphasis on the host-immune pathways that these miRNAs help to regulate in order to promote immune evasion.
Viral miRNAs inhibit cell-mediated immunity
The ability of viruses to repress host immune responses is essential for persistent infection. Herpesviruses and polyomaviruses have co-evolved miRNAs as a means to suppress cell-mediated immunity, an important component of the host response to intracellular pathogens, either by miRNA inhibition of effector cell recognition (T-cell and NK cell) or by miRNA induced expression of cytokines such as IL-6 and IL-10 [8–13].
Elegant genetic studies on the betaherpesvirus, human cytomegalovirus (HCMV) identified that miR-UL112-1, represses expression of the major histocompatibility complex class I-related chain B (MICB), a ligand that promotes NK cell killing [11]. The extent of MICB regulation is significant since cells infected with a recombinant HCMV containing a miR-UL112-1 deletion are more efficiently recognized and killed by NK cells. Unlike the majority of miRNA targets, which contain a full seed sequence match (nucleotides 2 to 8 of a miRNA), the MICB 3′ UTR target site only contains a partial miR-UL112-1 seed match which mediates repression. HCMV miR-UL112-1 was the first example for miRNA-dependent immune evasion through targeting of a host immune effector molecule. Recently, KSHV (miR-K12-7) and EBV (miR-BART2-5p) miRNAs have also been shown to downregulate MICB [8]; furthermore interrupting MICB targeting by KSHV and EBV miRNAs using miRNA sponges, increased NK cell killing of latently infected lymphoma cells. It appears that inhibiting NK cell killing through MICB repression is important for herpesvirus infection because both HCMV and KSHV also express proteins that inhibit MICB surface expression [14,15]. These findings suggest that herpesviruses have co-evolved miRNAs as host immune regulators to escape NK cell recognition.
Like NK cells, cytotoxic T lymphocytes (CTLs) are important effectors of cell-mediated immunity, whose response to viral infection is also inhibited by viral miRNAs. Two polyomaviruses, the human JC virus (JCV) and simian virus 40 (SV40), express miRNAs that downregulate the viral large T-antigen, an early viral product that elicits strong CTL responses [10,12]. To examine the effect of this downregulation on virus replication, Sullivan and colleagues generated mutant SV40 viruses with non-targeting miRNA and showed that these viruses had an increased susceptibility to CTL lysis. Interestingly, a murine polyomavirus was also found to encode miRNAs that target early viral transcripts, but the role of these miRNAs during infection remains unclear [16]. In addition to polyomaviruses, the EBV miR-BHRF1-3 modulates expression of the T-cell attractant chemokine CXCL11 [13]. However, the importance of CXCL11 downregulation in EBV infected B-cells needs to be validated with CTL assays.
Cell-mediated immunity in response to viral infection is also orchestrated by regulated expression of cytokines. KSHV miR-K12-3 and miR-K12-7 targeting of LIP, an isoform of the transcription factor C/EBPβ, was shown to induce expression of IL-6 and IL-10 in human myelomonocytic cell lines [9]. Both cytokines inhibit dendritic cell maturation, and as a result antigen presentation, thereby contributing to KSHV immune evasion [17]. In addition, IL-10 potently suppresses cytokine production in a number of effector cells including T-cells, NK cells, and macrophages [18,19]. Viral miRNA-dependent modulation of cytokines, which inhibit cell-mediated immunity, suggests a novel immune evasion mechanism that needs to be further studied in appropriate animal models.
Viral miRNAs target apoptotic inducers and cell cycle regulators
Apoptosis, triggered by viral infection, is an alternate mechanism of the innate immune response to eliminate viral spread. To date, pro-apoptotic targets of viral miRNAs have only been reported for the oncogenic gammaherpesviruses EBV and KSHV. Herpesviruses encode numerous proteins with anti-apoptotic activity; therefore it is not surprising that they also express miRNAs to further regulate cell-death pathways.
The first reported pro-apoptotic target of EBV miRNAs was the EBV latent membrane protein 1 (LMP1), a transforming factor that promotes cell proliferation and survival by activating nuclear factor-kappa B (NFκ-B) [20]. While LMP1 is required for EBV immortalization, LMP1 over-expression strongly induces apoptosis and inhibits NFκ-B [21]. Lo and colleagues showed that three EBV miRNAs (miR-BART16, miR-BART17-5p, and miR-BART1-5p) target and downregulate LMP1 expression, thereby attenuating the pro-apoptotic affect of LMP1 and its inhibitory effect on NFκ-B. Thus, it appears that EBV miRNAs fine tune LMP1 expression in order to ensure cell survival.
EBV miRNAs also target a host pro-apoptotic factor, p53 up-regulated modulator of apoptosis (PUMA), a member of the ‘BH3-only’ subclass of bcl2 proteins [22]. Bioinformatic analysis predicted PUMA as a potential miR-BART5 target. Inhibition of miR-BART5 expression using antagomirs induced apoptosis of EBV infected cells, suggesting that EBV miRNAs play an important role in blocking apoptosis. This study is the first to demonstrate that a viral miRNA can directly inhibit apoptosis by targeting a host pro-apoptotic protein.
Like EBV, KSHV miRNAs have been shown to modulate apoptotic pathways. The first reported apoptotic target was Bcl-2-associated factor (BCLAF1), a transcriptional repressor that induces apoptosis when over-expressed [23]. Ziegelbauer and colleagues demonstrated that KSHV miR-5, miR-K12-9, and miR-K12-10b target and repress BCLAF1 expression in human umbilical vein endothelial cells (HUVEC). Interestingly, examination of BCLAF1 miRNA targeting under differing experimental conditions revealed that BCLAF1 has both pro-and anti-apoptotic function.
In addition, miRNA-dependent BCLAF1 regulation was shown to impact KSHV latency [23], a herpesvirus intrinsic immune evasion mechanism which is discussed in the following section. BCLAF1 and three additional regulators of apoptosis LDOC1, BCL2L11, and BCL6B were also found to be inhibited in miR-K12-11 expressing cells [24] further supporting apoptosis as a major regulatory target for viral miRNAs.
To promote cell viability and proliferation during infection, herpesviruses not only inhibit apoptosis but also modulate cell cycle regulation. KSHV miRNAs have been found to target two proteins involved in cell cycle progression, Thrombospondin 1 (THBS1) and p21. THBS1, a tumor suppressor with strong anti-proliferative and anti-angiogenic activity, was shown to be targeted by KSHV miR-K12-1, miR-K12-3-3p, miR-K12-6-3p and miR-K12-11 [25]. THBS1 activates latent TGFβ, and Samols and colleagues demonstrated that KSHV miRNA-mediated repression of THBS1 inhibits TGFβ activity. The second target p21, a p53-inducible gene that functions as a cell cycle inhibitor and tumor suppressor, was found to be targeted by KSHV miR-K1 [26]. Knockdown of endogenous miR-K1, with miRNA sponges in KSHV infected cells, resulted in a modest increase of p53 mediated cell cycle arrest, implicating miR-K1 in cell cycle regulation. While KSHV miRNA regulation of the cell cycle is not a direct tool of immune evasion, it greatly affects the host response to infection and is a contributing mechanism to viral pathogenesis, especially in KSHV associated tumorigenesis.
Herpesvirus miRNAs and latency an “intrinsic” immune evasion mechanism
Herpesvirus infection persists for the life of the host, therefore avoiding host immune surveillance is essential for viral persistence. One immune evasion mechanism employed by all herpesviruses is the establishment of latency, during which only a minimal number of genes, including miRNAs are expressed. Maintenance of latency requires the suppression of viral gene products that promote the switch from latency to lytic replication. Conceptually, since herpesvirus miRNAs are non-immunogenic they are ideal tools for negatively regulating viral gene expression during latency, an idea that was proposed during the discovery of the first herpesvirus miRNAs [3,27,28]. The first experimental evidence supporting this idea came from two independent studies on HCMV, which demonstarted that miR-UL112-1 mediates repression of IE72 expression, a major trans-activating gene that promotes lytic replication [28,29]. Furthermore, it was shown that HCMV DNA replication was reduced in response to overexpression of miR-UL112-1, indicating a direct correlation between miRNA regulation and latency control [29].
More recently, Stern-Ginossar and colleagues [30] reported that miR-UL112-1 represses another viral protein UL114, a uracil DNA glycosylase that is involved in viral DNA replication [31]. However, mutant viruses lacking UL114 still showed a reduction of viral DNA synthesis in cells that ectopically expressed UL112-1. Interestingly, HCMV UL112-1 also targets MICB, as described above, linking this miRNA to multiple pathways of immune evasion.
Several KSHV miRNAs have been identified that modulate the latent-lytic switch. As mentioned in the section above, three KSHV miRNAs were found to regulate the expression of BCLAF1 [23]. In this study, it was demonstrated that inhibiting KSHV miRNA targeting of BCLAF1 decreased the ability of KSHV-infected endothelial cells (SLK) to undergo lytic reactivation. This was the first described role for viral miRNAs in promoting or sensitizing latently infected cells to lytic reactivation [23].
In contrast to a lytic role, KSHV miRNAs have also been reported to promote latency. Using KSHV recombinant viruses that lack 10 of the 12 miRNA genes, two independent studies found elevated levels of lytic genes, including transcription activator (RTA), during de novo infection of HEK293 cells and dermal microvascular endothelial cells (DMVECs) [32,33]. Because RTA is a master regulator of lytic reactivation, Lu and colleagues examined its direct regulation by individual KSHV miRNAs and found that miR-K5 moderately downregulates RTA mRNA levels. In addition, genome wide analysis of the recombinant viruses revealed reduced repressive marks on histones as well as a global reduction of DNA methylation, suggesting that KSHV miRNAs contribute to latency by regulating epigenetic modification of the viral episome. Further insight into this mechanism was provided by the finding that KSHV miR-K12-4-5p targets retinoblastoma (Rb)-like protein 2 (Rbl2), a negative regulator of DNA methyltransferases, thereby inducing DNA methyltransferase activity.
Lei and colleagues, using a similar mutant virus, showed that KSHV miR-K1 can activate the NF-κB pathway, which inhibits lytic reactivation and is required for PEL cell survival, by downregulating expression of its inhibitor IκBα [32].
In a separate study KSHV miR-K9*, but not miR-K5, was also found to modulate RTA expression, suggesting multiple KSHV miRNAs target RTA expression [34]. However, careful analysis of the extent by which miR-K9* regulates RTA suggest that KSHV miRNAs, while contributing to the maintenance of viral latency do not function as the major switch from latent to lytic replication [34]. The numerous miRNA targets affecting the latent-lytic switch of KSHV suggest that viral miRNAs may function as a priming mechanism which allows rapid reactivation from latency in response to environment stimuli.
A promising system to study latency control in animal models is HSV-1, which expresses only non-coding RNAs including multiple miRNAs during latency [35,36]. The majority of these miRNAs are processed from the latency-associated transcript (LAT), a non-coding RNA that is antisense to two lytic genes: ICP0, a transcriptional regulator, and ICP34.5, a neurovirulence factor (for review of LAT see [37]). Overexpression experiments confirmed that ICP0 is downregulated by HSV-1 miR-H2 [35]. In addition, miR-H6, expressed from a transcript separate from LAT was found to target ICP4, a major viral transactivator required for lytic activation. These findings together with phenotypes of LAT deletion mutants affecting reactivation and apoptosis, prior to the identification of HSV-encoded miRNAs, suggest that HSV miRNAs play a major role in negatively regulating lytic gene expression in order to maintain latency.
Summary and Outlook
Viral miRNA regulation is an emerging component of the complex relationship that governs viral-host interactions. From the targets identified to date (Table 1, Figure 1) it is apparent that viral miRNAs play an important role in immune evasion by inhibiting immune surveillance and extending the life of the infected host cell. However, determining the targets of these miRNAs is only one step in understanding their function. Because viral miRNA regulation is likely dependent on the context of infection (i.e. cell-type and viral genome expression), future studies using recombinant viruses, appropriate cell lines, and where available animal models are needed to further understand their impact on viral pathogenesis in vivo. Studying this novel class of viral post-transcriptional regulators may point to novel therapeutic strategies, especially for oncogenic herpesviruses. Additionally, a detailed understanding, of how miRNAs function in immune evasion will be crucial for any herpesvirus vaccine development.
Table 1.
Immunomodulatory viral miRNAs
| DNA Virus Family | Virus | miRNA | Target | Function | References |
|---|---|---|---|---|---|
| Latency | |||||
| Alphaherpesvirus | HSV-1 | miR-H2-3p | Viral ICP0 | Immediate-early transactivator | [35] |
| miR-H6 | Viral ICP4 | Immediate-early transactivator | [35] | ||
| Cell-mediated Immunity | |||||
| Betaherpesvirus | HCMV | miR-UL112-1 | Host MICB | NK cell ligand | [11] |
| Latency | |||||
| miR-UL112-1 | Viral IE72 | Immediate-early transactivator | [28,29] | ||
| Cell-mediated Immunity | |||||
| Gammaherpesvirus | EBV | miR-BART2-5p | Host MICB | NK cell ligand | [8] |
| miR-BHRF1-3 | Host CXCL11 | Chemokine, T-cell attractant | [13] | ||
| Apoptotic inducer | |||||
| miR-BART1-5p | Viral LMP1 | Transforming factor | [20] | ||
| miR-BART16 | |||||
| miR-BART17-5p | |||||
| miR-BART5 | Host PUMA | Pro-apoptotic factor | [22] | ||
| Cell-mediated Immunity | |||||
| KSHV | miR-K12-7 | Host MICB | NK cell ligand | [8] | |
| miR-K12-3 | Host C/EBPβ (LIP) | Inhibits IL6 and IL10 expression | [9] | ||
| miR-K12-7 | |||||
| Apoptotic inducer and Latency | |||||
| miR-K12-5 | Host BCLAF1 | Pro-apoptotic factor | [23] | ||
| miR-K12-9 | Promotes Lytic reactivation | ||||
| miR-K12-10b | |||||
| Cell cycle regulator | |||||
| miR-K12-1 | Host THBS1 | Tumor Suppressor | [25] | ||
| miR-K12-3-3p | |||||
| miR-K12-6-3p | |||||
| miR-K12-11 | |||||
| miR-K1 | Host p21 | Cell cycle inhibitor | [26] | ||
| Latency | |||||
| miR-K5 | Viral RTA | Master lytic switch | [33] | ||
| miR-K9* | |||||
| miR-K1 | Host IκBα | Inhibits NF-κβ | [32] | ||
| Cell-mediated Immunity | |||||
| Polyomavirus | SV40 | miRNA 5p and 3p | Viral Large TAg | Transforming factor | [12] |
| JCV | miRNA 5p and 3p | Viral Large Tag | [10] | ||
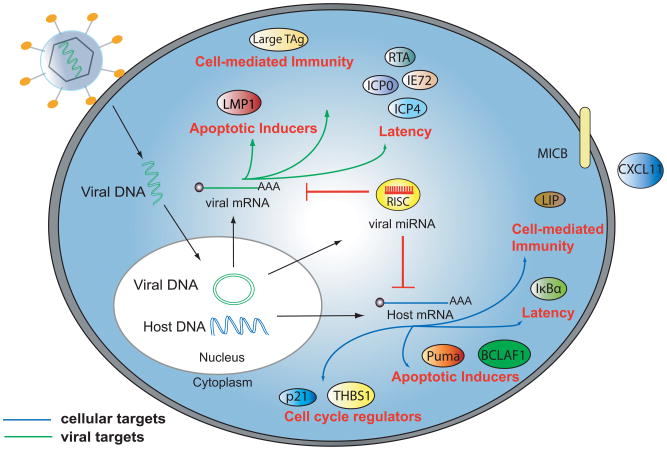
Figure 1. DNA virus miRNAs regulate the host immune response.
Host cells are infected by DNA viruses that replicate in the nucleus. After nuclear processing, viral miRNAs are exported into the cytoplasm where they are incorporated into RISC. Viral miRNAs then target multiple arms of the immune system by regulating expression of both host and viral mRNAs involved in cell-mediated immunity, apoptosis, cell cycle regulation, and viral latency.
Acknowledgments
We like to thank Karlie Plaisance for critical reading and thoughtful suggestions. Research was supported by R01 CA88763, RO1 CA 119917, and RC2CA148407 to R.R. I.B. was supported by the BMID and BEID NIH T32 training programs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1•.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. A recent in-depth review covering the known parameters of miRNA target recognition, as well as the biological functions of miRNA mediated transcript repression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3••.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. This study was the first to identify viral miRNAs. The authors cloned and sequenced small RNAs from an EBV infected cell line and found two clusters of miRNAs expressed from the EBV genome. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Boss IW, Plaisance KB, Renne R. Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol. 2009;17:544–553. doi: 10.1016/j.tim.2009.09.002. A comprehensive review of herpesvirus miRNAs, their targets, and the potential roles they play during host-cell infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. A recent study which found that multiple herpesviruses could repress MICB through miRNA mediated targeting. Providing further evidence that MICB regulation by herpesviruses is an important mechanism of immune evasion. [DOI] [PubMed] [Google Scholar]
- 9•.Qin Z, Kearney P, Plaisance K, Parsons CH. Pivotal Advance: Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol. 2009 doi: 10.1189/jlb.0409251. The first report of cytokine secretion induced by viral miRNA targeting of a host factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. This study reported that an HCMV miRNA could target the host protein MICB, whose expression is important in immune surveillance. This is the first reported example of host immune regulation by a viral miRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. One of the few studies not related to herpesvirus miRNA regulation, which identified that polyomavirus miRNA regulation of a viral product is important for escaping a CTL response. [DOI] [PubMed] [Google Scholar]
- 13.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ. Down-regulation of NKG2D and NKp80 ligands by Kaposi’s sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105:1656–1661. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387:157–167. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirone M, Lucania G, Aleandri S, Borgia G, Trivedi P, Cuomo L, Frati L, Faggioni A. Suppression of dendritic cell differentiation through cytokines released by Primary Effusion Lymphoma cells. Immunol Lett. 2008;120:37–41. doi: 10.1016/j.imlet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 19.Mosmann TR. Properties and functions of interleukin-10. Adv Immunol. 1994;56:1–26. [PubMed] [Google Scholar]
- 20•.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. This study found that EBV miRNA regulation of the viral protein LMP1 is important for reducing its pro-apoptotic affects, thereby promoting host-cell survival. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Wang X, Lo AK, Wong YC, Cheung AL, Tsao SW. Latent membrane protein-1 of Epstein-Barr virus inhibits cell growth and induces sensitivity to cisplatin in nasopharyngeal carcinoma cells. J Med Virol. 2002;66:63–69. doi: 10.1002/jmv.2112. [DOI] [PubMed] [Google Scholar]
- 22.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–134. doi: 10.1038/ng.266. This paper found that KSHV miRNA regulation of host BCLAF1 sensitized infected cells to lytic reactivation, indicating a novel function for viral miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Samols MA, Hu J, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007 doi: 10.1371/journal.ppat.0030065. The authors of this study found that KSHV miRNAs target the tumor suppressor THBS1, suggesting a mechanism for increased proliferation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Gottwein E, Cullen BR. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol. doi: 10.1128/JVI.00202-10. Along with ref. 25, this study finds increasing evidence that KSHV miRNAs can regulate cell cycle control by targeting host cell cycle inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. This study provides the first evidence that herpesvirus miRNAs can help to maintain latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol. 2009;83:10684–10693. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Courcelle CT, Courcelle J, Prichard MN, Mocarski ES. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J Virol. 2001;75:7592–7601. doi: 10.1128/JVI.75.16.7592-7601.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 12:193–199. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. 84:2697–2706. doi: 10.1128/JVI.01997-09. This study provides the first example of viral miRNA mediated epigenetic modification of the viral genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. The first identification of HSV-1 miRNAs encoded and expressed by the non-coding LAT transcript during latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bloom DC. HSV LAT and neuronal survival. Int Rev Immunol. 2004;23:187–198. doi: 10.1080/08830180490265592. [DOI] [PubMed] [Google Scholar]