Abstract
Objectives
To compare genotypes of Mycobacterium bovis strains from humans in Southern California with genotypes of M. bovis strains in cattle in Mexico and the USA to explore the possible origins of human infections.
Methods
We conducted a descriptive analysis of M. bovis genotypes from a binational population of humans and cattle using spacer oligonucleotide typing (spoligotyping).
Results
One hundred six human M. bovis spoligotypes were compared to spoligotypes from 496 Mexican cattle and 219 US cattle. Twelve spoligotype patterns were identified among human cases and 126 spoligotype patterns were detected in cattle. Over 91% (97/106) of the human M. bovis isolates had spoligotypes that were identical to those found in Mexican cattle. Four human cases had spoligotypes that matched both cattle born in Mexico and in the USA. Nine human cases had spoligotypes that did not match cattle born in Mexico or the USA.
Conclusions
Our data indicate that the population of M. bovis strains causing human TB disease in Southern California is closely related to the M. bovis strain population found in Mexican cattle and supports existing epidemiological evidence that human M. bovis disease in San Diego likely originated from Mexican cattle.
Keywords: Molecular epidemiology, Mycobacterium bovis, Tuberculosis, Spoligotyping
1. Introduction
Tuberculosis (TB) is currently one of the leading causes of death due to infectious disease globally, with 9.3 million incident cases and over 1.7 million deaths reported in 2007.1 TB in humans is caused mostly by Mycobacterium tuberculosis, but this was not always the case. In the early 1900 s an estimated 30% of TB cases in Europe were caused by the cattle TB pathogen, Mycobacterium bovis,2 a closely related Mycobacterium species largely transmitted to humans via inhalation of infectious droplets from infected cattle and consumption of contaminated, unpasteurized dairy products.3
The introduction of milk pasteurization and ‘test and slaughter’ cattle control programs in the early 1900 s all but eradicated M. bovis from cattle and humans in most of the USA and other developed nations.2,4 A recent study of all human TB cases in the USA from 1995 through 2005 estimated that only 1.4% of cases were still being caused by M. bovis, most of which were among individuals born outside of the USA.5
However, in certain regions of the USA along the border with Mexico, and in Mexican-born individuals in New York City, M. bovis TB prevalence has been shown to be significantly higher than the national prevalence.6–12 In San Diego, California in particular, over 45% of all culture-confirmed TB cases in children and 8% of all TB cases were recently found to be due to M. bovis.13
Epidemiological evidence suggests that M. bovis TB in Southern California is largely the result of the consumption of unpasteurized dairy products, such as unpasteurized cheese commonly referred to as queso fresco, from infected cattle in Mexico,10,13–15 but evidence linking the M. bovis pathogen populations in cattle and humans is lacking. The aim of this study was to utilize molecular genotyping to examine the relationships between the population of M. bovis strains collected from cattle in Mexico and the USA, and the population of M. bovis strains isolated from human TB cases in San Diego.
2. Methods
2.1. Study design
We conducted a retrospective analysis of all known M. bovis genotypes in humans from San Diego County, and a convenience sample of genotypes from cattle in Mexico and the USA, represented by spacer oligonucleotide types (spoligotypes). The study protocol was approved by an Institutional Review Board at the University of California, San Diego.
2.2. Data source
We obtained human M. bovis isolates from the San Diego County Tuberculosis Surveillance Program for the years 2004 through 2007, which included all known M. bovis TB cases. M. bovis TB cases represented approximately 10% of all TB cases during that period. All TB isolates from patient specimens were initially identified as M. tuberculosis complex based on the AccuProbe hybridization protection assay (GenProbe, San Diego, CA, USA). We identified specimens as either M. bovis or M. tuberculosis based on culture morphology, the results of the niacin strip test, the nitrate reduction test and their susceptibility to pyrazinamide,16 and confirmed species designations with genotyping17 conducted by the California Department of Public Health, Microbial Diseases Laboratory.
M. bovis genotypes from cattle born in Mexico were collected as convenience samples as part of cattle TB surveillance efforts in Mexico from 1997 through 2007. Mexican isolates were genotyped by the Programa Nacional de Epidemiología (CENIDFA-INIFAP) in Mexico. We also obtained Mexican cattle M. bovis spoligotypes from peer-reviewed, published studies14,18–24 listed in PubMed from 1997 through 2008. To our knowledge these genotypes represent all known reported M. bovis genotypes by spoligotyping from cattle in Mexico.
For this study, we also examined all available spoligotypes in the US national surveillance database of M. bovis isolates sent to the National Veterinary Services Laboratories (NVSL), Ames, Iowa (US Department of Agriculture, Animal and Plant Health Inspection Services) from 1997 through 2008. The NVSL surveillance database represents a convenience sample of all cattle suspected of having TB in the USA from 1997 to 2008.
2.3. Spoligotyping
Spoligotyping was performed independently in Mexico and in the USA. Spoligotyping was performed by all laboratories using the Spoligo Kit (Isogen Bioscience BV, Maarsen, the Netherlands) using standard methods described in detail in Kamerbeek et al.25 Briefly, the direct repeat (DR) region of the M. bovis genome was amplified using a polymerase chain reaction (PCR), then probed for the presence or absence of 43 different spacer sequences using a dot blot method. Primers were labeled with biotin, and amplified products were detected using a streptavidin–POD conjugate (Boehringer Mannheim, Mannheim, Germany) or a chemiluminescent detection system (Amersham ECL, Rockford, IL, USA) with X-ray film.
2.4. Mycobacterial interspersed repeat units (MIRU)
Spoligotypes of M. bovis isolates from humans in San Diego County were further subtyped by MIRU typing as described elsewhere.26–28 Briefly, loci consisting of multiple copies of tandem repeats distributed around the M. bovis chromosome were amplified by PCR before reactions were multiplexed and fragment lengths were evaluated using a capillary sequencer. The standard 20-locus set was tested. Cattle spoligotypes were not further subdivided by MIRU genotyping as MIRU data were not available for the cattle isolates and isolates are no longer available for testing.
2.5. Data management and analysis
We converted all spoligotype data to a 15-digit number using an octal coding system and compiled the data into a database using MS Excel software (Microsoft, Seattle, WA, USA). Each spoligotype was cataloged with a unique identifier along with information on its source (human or cattle), cattle breeding purpose (dairy, beef, unknown), and state within Mexico or USA from which the case originated. For each cohort, cattle and human, we classified M. bovis isolates with identical spoligotype patterns as clusters of related strains. Isolates with spoligotypes that did not match any other human or cattle spoligotypes were considered unrelated or ‘orphan strains’.24,29–32 All spoligotype patterns were submitted to the public M. bovis spoligotype database (www.mbovis.org), and given a unique identifier number (SB number) to identify them in this publication.
3. Results
We obtained a total of 106 M. bovis genotypes from 109 human M. bovis TB cases in San Diego County from 2004 through 2007 (spoligotypes for three cases could not be resolved). These human M. bovis spoligotypes were compared to spoligotypes from 496 M. bovis isolates from Mexican cattle and 219 M. bovis isolates from US cattle.
3.1. Mycobacterium bovis isolates from humans
Twelve different spoligotype patterns were identified among the 106 human cases in San Diego County from 2004 through 2007 (Table 1). Overall 98% (104/106) of cases were Hispanic; two cases occurred in non-Hispanic whites. Sixty-two percent of cases (66/106) reported being born in Mexico while the remainder reported being born in the USA.
Table 1.
Frequencies of spoligotype patterns for Mycobacterium bovis cases reported in San Diego County from 2004 to 2007 showing matches to cattle originating in Mexico and the USA
| Spoligotype name | Spoligotype (octal) | No. of MIRU subtypesa | Number of cases, n (% of total) | Match to Mexican cattle | Match to US cattle |
|---|---|---|---|---|---|
| SB0145 | 640013777777600 | 6 | 51 (48.1) | Yes | No |
| SB1040 | 640013377777600 | 4 | 39 (36.8) | Yes | No |
| SB1758 | 676573777776600 | 1 | 3 (2.8) | No | No |
| SB0673 | 264073777777600 | 2 | 2 (1.9) | Yes | Yes |
| SB0140 | 664073777777600 | 2 | 2 (1.9) | Yes | Yes |
| SB0152 | 400000077777600 | 1 | 2 (1.9) | Yes | No |
| SB1759 | 640013776377600 | 1 | 2 (1.9) | No | No |
| SB0971 | 664073777700600 | 1 | 1 (0.9) | Yes | No |
| SB1760 | 640012377777600 | 1 | 1 (0.9) | No | No |
| SB1761 | 640013357777600 | 1 | 1 (0.9) | No | No |
| SB1762 | 672573777777600 | 1 | 1 (0.9) | No | No |
| SB1345 | 676713677777600 | 1 | 1 (0.9) | No | No |
| Total | 106 |
Isolates were subtyped by mycobacterial interspersed repetitive units (MIRU) analysis. Six out of the 106 cases could not be resolved by MIRU analysis.
Eighty-five percent (90/106) of the human M. bovis cases were clustered in two spoligotypes, SB0145 and SB1040. Eleven of the remaining 16 human M. bovis cases were in clusters of two or three isolates, and four cases had orphan spoligotypes that did not match any other human or cattle cases. Isolates were further subdivided by MIRU subtype into 22 different strain types. The MIRU pattern for six out of the 106 human isolates could not be resolved.
3.2. Mycobacterium bovis isolates from cattle
M. bovis genotypes were obtained from Mexican cattle populations in 13 of 31 Mexican states (Table 2). The majority were from Chihuahua (n = 92) and Jalisco (n = 61), and the northern and eastern Mexican states. Baja California contributed 10 isolates. We could not determine the state of origin for 117 Mexican cattle genotypes.
Table 2.
Total number of Mycobacterium bovis strains and spoligotypes reported from cattle originating in Mexico. Table ordered by number of cattle spoligotypes from each Mexican state that matched human cases, 1997–2008.
| State | Number of isolates by cattle type |
Number of unique spoligotypes (% per state) | Number of unique cattle spoligotypes matching human spoligotypes in San Diego (% matching per state) | |||
|---|---|---|---|---|---|---|
| Dairy | Beef | Unknown | Total (%) | |||
| Jalisco | 39 | 5 | 17 | 61 (12.3) | 48 (78.7) | 5 (10.4) |
| Tamaulipas | 1 | 23 | 5 | 29 (5.8) | 11 (35.5) | 4 (30.1) |
| Veracruz | 0 | 5 | 18 | 23 (4.6) | 12 (54.5) | 4 (26.7) |
| Durango | 0 | 25 | 5 | 30 (6.0) | 13 (44.8) | 4 (30.1) |
| Aguascalientes | 13 | 15 | 2 | 30 (6.0) | 18 (60.0) | 4 (21.1) |
| Nueva Leon | 0 | 22 | 7 | 29 (5.8) | 17 (58.6) | 4 (21.1) |
| Sonora | 7 | 11 | 13 | 31 (6.3) | 10 (32.3) | 3 (25.0) |
| Chihuahua | 65 | 17 | 10 | 92 (18.5) | 17 (18.5) | 3 (18.8) |
| Coahuila | 0 | 9 | 3 | 12 (2.4) | 6 (50.0) | 3 (50.0) |
| Baja Californiaa | 2 | 4 | 4 | 10 (2.0) | 8 (80.0) | 3 (37.5) |
| Campeche | 0 | 3 | 0 | 3 (0.6) | 2 (66.7) | 2 (100) |
| Querétaro | 8 | 0 | 0 | 8 (1.6) | 7 (88.9) | 0 (0) |
| Mexico State | 11 | 0 | 10 | 21 (4.2) | 18 (85.7) | 0 (0) |
| Unknownb | 0 | 28 | 89 | 117 (23.6) | 24 (21.2) | 5 (26.3) |
| Total | 146 | 167 | 183 | 496 | ||
Baja California includes both Baja California and Baja California Sur.
Cattle isolates obtained from animals for which state of origin was not available or untraceable.
Of 496 M. bovis cases in Mexican cattle, we identified 126 different spoligotype patterns. Ninety-three Mexican cattle spoligotypes were orphan strains (Table 3). Approximately one third (146/496) of Mexican cattle tested were dairy cattle and one third were beef cattle (167/496). The breeding purpose for 37% (183/496) of cattle tested was unknown.
Table 3.
Mycobacterium bovis spoligotypes reported among isolates from cattle originating from Mexico, 1997–2008
| Spoligotype ID | Spoligotype (octal) | No. of isolates, n |
Match to human cases | |||
|---|---|---|---|---|---|---|
| Dairy | Beef | Unknown | Total (% of all) | |||
| SB0673 | 264073777777600 | 9 | 51 | 58 | 118 (23.8) | Yes (SB0673) |
| SB0121 | 676773677777600 | 29 | 15 | 4 | 48 (9.7) | No |
| SB0140 | 664073777777600 | 10 | 18 | 6 | 34 (6.9) | Yes (SB0140) |
| SB0669 | 264063777777600 | 5 | 8 | 18 | 31 (6.3) | No |
| SB0145 | 640013777777600 | 4 | 16 | 10 | 30 (6.0) | Yes (SB0145) |
| SB0971 | 664073777700600 | 0 | 9 | 9 | 18 (3.6) | Yes (SB0971) |
| SB0269 | 664063777777600 | 2 | 2 | 10 | 14 (2.8) | No |
| SB0327 | 676573777077600 | 3 | 9 | 2 | 14 (2.8) | No |
| SB1165 | 640033777777600 | 0 | 3 | 6 | 9 (1.8) | No |
| SB0986 | 264071777777600 | 4 | 2 | 2 | 8 (1.6) | No |
| SB0663 | 640003777777600 | 4 | 0 | 4 | 8 (1.6) | No |
| SB1040 | 640013377777600 | 1 | 5 | 2 | 8 (1.6) | Yes (SB1040) |
| SB1116 | 664063777700600 | 2 | 2 | 4 | 8 (1.6) | No |
| SB0119 | 676763677777600 | 5 | 0 | 0 | 5 (1.0) | No |
| SB1044 | 064063777777600 | 4 | 0 | 0 | 4 (0.8) | No |
| SB1496 | 640023777777600 | 0 | 1 | 3 | 4 (0.8) | No |
| SB0272 | 664073777757600 | 3 | 1 | 0 | 4 (0.8) | No |
| SB0130 | 676573777777600 | 0 | 1 | 3 | 4 (0.8) | No |
| SB1111 | 064061777777600 | 3 | 0 | 0 | 3 (0.6) | No |
| SB1114 | 440003777777600 | 3 | 0 | 0 | 3 (0.6) | No |
| SB0987 | 624073777700600 | 3 | 0 | 0 | 3 (0.6) | No |
| SB1118 | 640003777776600 | 0 | 0 | 3 | 3 (0.6) | No |
| SB1216 | 076573777077600 | 0 | 3 | 0 | 3 (0.6) | No |
| SB1110 | 064263777777600 | 2 | 0 | 0 | 2 (0.4) | No |
| SB1696 | 444063777746600 | 1 | 1 | 0 | 2 (0.4) | No |
| SB1125 | 464063735546600 | 0 | 0 | 2 | 2 (0.4) | No |
| SB1113 | 464063777756600 | 0 | 0 | 2 | 2 (0.4) | No |
| SB1129 | 674563777077600 | 0 | 0 | 2 | 2 (0.4) | No |
| SB1503 | 674573777077600 | 0 | 1 | 1 | 2 (0.4) | No |
| SB0807 | 676773675777600 | 2 | 0 | 0 | 2 (0.4) | No |
| SB1757 | 644073777700600 | 0 | 2 | 0 | 2 (0.4) | No |
| SB1755 | 666073777777600 | 0 | 1 | 1 | 2 (0.4) | No |
| SB0152 | 400000077777600 | 0 | 1 | 0 | 1 (0.2) | Yes (SB0152) |
| Mutliplea | Multiple | 47 | 15 | 31 | 93 | No |
| Total | 146 | 167 | 183 | 496 | ||
There are 94 isolates each represented by a single cow. Ninety-three did not match the human isolates and were grouped together in this table as ‘Multiple’. SB0152 was listed separately in this table since it was a match to human spoligotype SB0152.
3.3. Human/cattle spoligotype matches
Over 91% (97/106) of the human M. bovis TB isolates from San Diego County had spoligotypes that were identical to those found in Mexican cattle (Table 1). Four human cases had M. bovis spoligotypes that matched cattle originating from both Mexico and the USA, and nine human cases had spoligotypes that did not match cattle in Mexico or the USA. Eleven of the 13 Mexican states sampled yielded cattle M. bovis isolates with spoligotypes that matched human M. bovis spoligotypes in San Diego (Table 2).
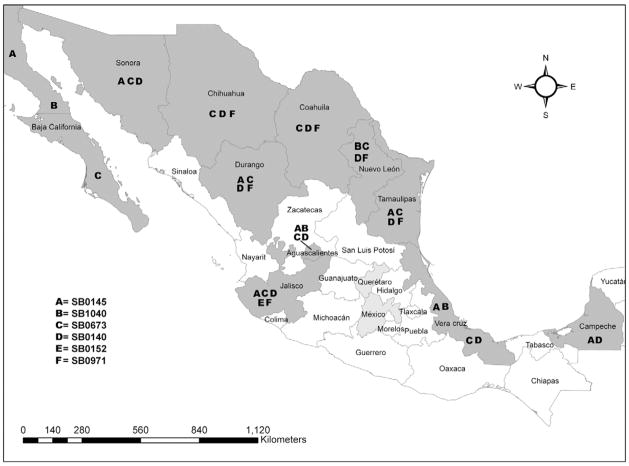
Human M. bovis spoligotypes SB0145, SB1040, SB0152, and SB0971, which accounted for 88% (93/106) of all the human cases, were found in only 11.5% of the Mexican cattle sampled (Table 3), but were found widely distributed in all but two of the Mexican states sampled (Figure 1). Human M. bovis spoligotypes SB0673 and SB0140, found in both Mexican and US cattle, accounted for only 3.8% of the human cases, but were found in 31% (152/496) of M. bovis isolates obtained from Mexican cattle (Table 3), and in 45% (99/219) of US cattle tested (Table 4).
Figure 1.
Mexican states where cattle were sampled for Mycobacterium bovis (gray shading). Letters represent the San Diego human M. bovis spoligotypes that matched cattle in each state sampled. States shaded in light gray indicate Mexican states in which M. bovis was found in cattle but their spoligotypes did not match San Diego human cases.
Table 4.
Mycobacterium bovis spoligotypes collected from US cattle, 1997–2008, that matched human M. bovis spoligotypes in San Diego
| Spoligotype ID | Spoligotype/state | Number of isolates (%) |
|---|---|---|
| SB0673 | 264073777777600 | |
| New Mexico | 78 (78.8) | |
| California | 6 (6.1) | |
| Texas | 5 (5.1) | |
| North Dakota | 3 (3.0) | |
| Arizona | 2 (2.0) | |
| SB0140 | 664073777777600 | |
| Texas | 2 (2.0) | |
| Colorado | 2 (2.0) | |
| California | 1 (1.0) | |
| Total | 99 (100) |
4. Discussion
Our analyses indicate that the majority of human M. bovis pathogen strains identified in San Diego from 2004 through 2007 had the same spoligotype as strains found only in cattle in Mexico, supporting existing epidemiological evidence7–14 that the population of M. bovis strains causing human M. bovis disease in this region most likely originated in Mexican cattle. Most of the human M. bovis cases with spoligotypes matching Mexican cattle spoligotypes were clustered into two closely-related patterns (SB0145 and SB1040). These spoligotypes were rare among the sampled Mexican cattle, but widely distributed across 11 of the 13 Mexican states sampled, and were not detected among the US cattle surveillance cohort. This lack of concordance in the frequency of M. bovis strains circulating in the suspected animal source population relative to the strains circulating in humans has been documented in other countries where M. bovis is a zoonotic disease.33–35 It is possible that the discordance observed is a sampling artifact as the Mexican cattle spoligotypes represent only a small proportion of the approximately 23 million head of cattle in Mexico, but it might also reflect changes in the Mexican cattle population that occurred between the time the human infections were acquired and the time the infections reactivated as TB disease, as has been documented in the UK.36 It is also possible that human M. bovis strains SB0145 and SB1040, which differ by only a single spoligotype spacer, have been spread from person-to-person, which would increase their frequency.
While person-to-person transmission of M. bovis has recently been demonstrated in France37 and the UK38 and has been hypothesized to be occurring in San Diego,11,12 there is considerable epidemiological evidence in San Diego that indicates person-to-person transmission is probably rare, despite the fact that approximately 54% of M. bovis cases in this region have pulmonary disease.13 This evidence includes the absence of M. bovis TB cases in children not yet exposed to dairy products (these children do get M. tuberculosis TB which is only transmitted person-to-person);10 and the evidence that M. bovis incidence in San Diego, which increased significantly from 1994 through 2005, has not followed the same trends as M. tuberculosis incidence, which has steadily decreased since 1994. This suggests that M. bovis transmission mechanisms are different from the person-to-person transmission of M. tuberculosis.13
The two spoligotypes from four human cases that matched strains from Mexican and US cattle (SB0673 and SB0140), were common in Mexican cattle, and were also found in cattle from six different states in the USA, including California. It is not surprising that some of the spoligotypes examined were present in cattle from both sides of the border. Restrictions on cattle trade between Mexico and the USA have only been in place for 10 years, and only prevent the importation of dairy cattle into the USA.20 Beef cattle can still be exported from Mexico into the USA if they are demonstrated to be tuberculin skin test (TST)-negative at the time of export.
Since the US bovine TB eradication program began in 1917, M. bovis infection in US cattle has been extremely rare.4 Less than 0.02% of US cattle tested positive for TB by TST in 2002,39 and less than 0.002% of 377 000 cattle tested in a 2008 California M. bovis investigation were positive for TB.40 Localized M. bovis outbreaks, low prevalence of M. bovis infection in US cattle herds, and strict national pasteurization requirements in the USA make it unlikely that the San Diego human cases with SB0673 and SB0140 spoligotypes are associated with US-born cattle with these spoligotypes.
In contrast, there have been several reports of widespread M. bovis infection in Mexican cattle.18–21,23,32,41 In some regions of Mexico, up to 13% of dairy herds are reported to be infected with M. bovis42 and up to 30% of milk produced in Mexico is not pasteurized.19 Additionally, unpasteurized soft fresh cheeses, such as queso fresco, which is very popular within the Mexican community, are brought into the USA from Mexico for personal use and have been shown to be contaminated with M. bovis,14,15 making it much more likely that human cases with SB0673 and SB0140 spoligotypes are associated with Mexican cattle with those spoligotypes.
4.1. Limitations
While spoligotyping is a practical approach for genotyping in large scale, population-level M. bovis studies such as this one, it is not a complete tool for molecular epidemiological analyses.43 Spoligotyping requires less DNA than other methods, is easily replicated in low-resource settings, and results can be digitally expressed,25,30,44,45 but, it can have limited discriminatory power if used in isolation.46
While we were able to obtain MIRU data for all of the human M. bovis cases, which demonstrated some heterogeneity amongst the human spoligotype clusters, it was not possible, due to the retrospective nature of the study and the lack of access to the cattle isolates, to obtain MIRU data for any of the cattle cases. However, we do not believe that the lack of MIRU data limited our conclusion that the population of human M. bovis strains from San Diego likely originated from the population of M. bovis strains found in Mexican cattle, as that is the most parsimonious conclusion from all of the available molecular and epidemiological data. We could not exclude the possibility of homoplasy, where similar mutations in the human and cattle M. bovis strains could have arisen in phylogenetically unrelated strains. But given the clonal nature of M. bovis, the approximately 10–20 year stability of spoligotypes and the unidirectionality of mutations in the DR region (spacers can only be lost not re-acquired),36 the likelihood of significant homoplasy in our binational pathogen populations is remote in the time-period under study. Additional MIRU and restriction fragment length polymorphism sub-typing, based on a systematic cattle sample, to further distinguish strains, are needed to determine the contribution of specific dairy sources to specific human cases in the USA.
Little can be inferred from the nine human M. bovis cases that did not match any of the cattle cases from Mexico or the USA. It is very unlikely that these cases match undetected cattle cases in the USA as the US cattle sample represents all known cattle TB suspects from 1997 through 2008. The same cannot be said for the Mexican sample. It represents only a small fraction of incident M. bovis cases among cattle in Mexico and additional matching strains may have gone undetected. Furthermore, the use of a convenience sample of cattle isolates from Mexico may have introduced a selection bias.
5. Conclusions
Our genotyping data indicate that the population of M. bovis strains causing human M. bovis TB disease in Southern California is closely related to the M. bovis strain population found in Mexican cattle and supports existing epidemiological evidence10,13 that human M. bovis TB in San Diego likely originates in Mexican cattle. This suggests that elimination of M. bovis TB in San Diego will require a reduction in M. bovis prevalence in Mexican cattle, increasing pasteurization of milk products in Mexico, and reducing the flow of unpasteurized milk products between Mexico and the USA.
The California State and San Diego County Health Departments have long standing enforcement programs that target the illegal sale of dairy products, in addition to providing educational resources regarding the risks of unpasteurized dairy products. Furthermore, in 2008, the San Diego Health Department initiated a media campaign to educate the Hispanic community on both sides of the border about prevention of TB from M. bovis; however, studies to evaluate the effectiveness of these efforts have not been completed.
Restriction of importation of unpasteurized cheeses for personal use is also being tested at the region’s USA–Mexico border,47 and may assist in reducing exposures in the USA if found feasible to implement. However, since exposure will likely continue via consumption of dairy products in Mexico, it is critical that binational cooperation and resources for collaboration be made available in the USA and Mexico in order to eradicate M. bovis from Mexican dairy herds. Our analysis indicates that most M. bovis strain clusters are found throughout Mexico, suggesting that unrestricted cattle movement in the past has played a role in M. bovis transmission in Mexico. In order to fully address M. bovis TB infections in Mexican cattle, prevention measures that incorporate a nationwide approach will be required. Increased emphasis on pasteurization of most dairy products could also assist in prevention efforts.
Acknowledgments
The authors thank Dr Benjamin Sanchez for his assistance with dataset preparation and Dr Edward Desmond for his assistance with the human M. bovis genotyping. Dr Rodwell received financial support from the California HIV/AIDS Research Program at the University of California, Fellowship No. CF07-SD-302 and the National Institutes of Health: T32 #DA023356 and K01AI083784-01.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Conflict of interest: No conflict of interest to declare.
References
- 1.World Health Organization. [(accessed March 2009)];Who report 2009: global tuberculosis control, surveillance, planning, financing. Available at: http://www.who.int/tb/publications/global_report/2009/pdf/full_report.pdf.
- 2.O’Reilly LM, Daborn CJ. The epidemiology of Mycobacterium bovis infections in animals and man. A review. Tuber Lung Dis. 1995;76(Suppl 1):1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 3.Thoen C, Lobue P, De Kantor I. The importance of Mycobacterium bovis as a zoonosis. Vet Microbiol. 2006;112:339–45. doi: 10.1016/j.vetmic.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 4.Kaneene JB, Miller R, Meyer RM. Abattoir surveillance: the US experience. Vet Microbiol. 2006;112:273–82. doi: 10.1016/j.vetmic.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Hlavsa MC, Moonan PK, Cowan LS, Navin TR, Kammerer JS, Morlock GP, et al. Human tuberculosis due to Mycobacterium bovis in the United States, 1995–2005. Clin Infect Dis. 2008;47:168–75. doi: 10.1086/589240. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Human tuberculosis caused by Mycobacterium bovis—New York City, 2001–2004. MMWR Morb Mortal Wkly Rep. 2005;52:605–8. [PubMed] [Google Scholar]
- 7.Dankner WM. Mycobacterium bovis: a significant cause of childhood tuberculous disease in San Diego, California. Program and Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy 33rd Interscience Conference on Antimicrobial Agents and Chemotherapy; October 17–20, 1993; New Orleans, Louisiana, USA. 1993. p. 367. [Google Scholar]
- 8.Dankner WM, Davis CE. Mycobacterium bovis as a significant cause of tuberculosis in children residing along the United States–Mexico border in the Baja California region. Pediatrics. 2000;105:79. doi: 10.1542/peds.105.6.e79. [DOI] [PubMed] [Google Scholar]
- 9.Dankner WM, Waecker NJ, Essey MA, Moser K, Thompson M, Davis CE. Mycobacterium bovis infections in San Diego: a clinicoepidemiologic study of 73 patients and a historical review of a forgotten pathogen. Medicine (Baltimore) 1993;72:11–37. [PubMed] [Google Scholar]
- 10.Lobue PA, Betacourt W, Peter C, Moser KS. Epidemiology of Mycobacterium bovis disease in San Diego County, 1994–2000. Int J Tuberc Lung Dis. 2003;7:180–5. [PubMed] [Google Scholar]
- 11.Lobue PA, Betancourt W, Cowan L, Seli L, Peter C, Moser KS. Identification of a familial cluster of pulmonary Mycobacterium bovis disease. Int J Tuberc Lung Dis. 2004;8:1142–6. [PubMed] [Google Scholar]
- 12.Lobue PA, Leclair JJ, Moser KS. Contact investigation for cases of pulmonary Mycobacterium bovis. Int J Tuberc Lung Dis. 2004;8:868–72. [PubMed] [Google Scholar]
- 13.Rodwell TC, Moore M, Moser KS, Brodine SK, Strathdee SA. Tuberculosis from Mycobacterium bovis in binational communities, United States. Emerg Infect Dis. 2008;14:909–16. doi: 10.3201/eid1406.071485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris NB, Payeur J, Bravo D, Osorio R, Stuber T, Farrell D, et al. Recovery of Mycobacterium bovis from soft fresh cheese originating in Mexico. Appl Environ Microbiol. 2007;73:1025–8. doi: 10.1128/AEM.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinde H, Mikolon A, Rodriguez-Lainz A, Adams C, Walker RL, Cernek-Hoskins S, et al. Recovery of Salmonella, Listeria monocytogenes, and Mycobacterium bovis from cheese entering the United States through a noncommercial land port of entry. J Food Prot. 2007;70:47–52. doi: 10.4315/0362-028x-70.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Grange JM, Yates MD, De Kantor IN. Guidelines for speciation within the Mycobacterium tuberculosis complex: WHO/EMC/ZOO/96.4. Geneva, Switzerland: World Health Organization; 1996. pp. 1–23. [Google Scholar]
- 17.Streicher EM, Victor TC, Van Der Spuy G, Sola C, Rastogi N, Van Helden PD, et al. Spoligotype signatures in the Mycobacterium tuberculosis complex. J Clin Microbiol. 2007;45:237–40. doi: 10.1128/JCM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobos-Marin L, Montes-Vargas J, Zumarraga M, Cataldi A, Romano MI, Estrada-Garcia I, et al. Spoligotype analysis of Mycobacterium bovis isolates from Northern Mexico. Can J Microbiol. 2005;51:996–1000. doi: 10.1139/w05-083. [DOI] [PubMed] [Google Scholar]
- 19.Milian-Suazo F, Banda-Ruiz V, Ramirez-Casillas C, Arriaga-Diaz C. Genotyping of Mycobacterium bovis by geographic location within Mexico. Prev Vet Med. 2002;55:255–64. doi: 10.1016/s0167-5877(02)00015-6. [DOI] [PubMed] [Google Scholar]
- 20.Milian-Suazo F, Harris B, Diaz CA, Torres CR, Stuber T, Ojeda GA, et al. Molecular epidemiology of Mycobacterium bovis: usefulness in international trade. Prev Vet Med. 2008;87:261–71. doi: 10.1016/j.prevetmed.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Milian-Suazo F, Salman MD, Black WC, Triantis JM, Ramirez C, Payeur JB, et al. Molecular epidemiologic analysis of Mycobacterium bovis isolates from Mexico. Am J Vet Res. 2000;61:90–5. doi: 10.2460/ajvr.2000.61.90. [DOI] [PubMed] [Google Scholar]
- 22.Samper S, Martin C, Pinedo A, Rivero A, Blazquez J, Baquero F, et al. Transmission between HIV-infected patients of multidrug-resistant tuberculosis caused by Mycobacterium bovis. AIDS. 1997;11:1237–42. doi: 10.1097/00002030-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Santillan-Flores MA, Flores J, Arriaga-Diaz C, Romero-Torres C, Suarez-Guemes F, Espitia C. Polymorphism of the PE domain of PE/PE_PGRS sequences in clinical isolates of Mycobacterium bovis in Mexico. Vet Microbiol. 2006;115:364–9. doi: 10.1016/j.vetmic.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Zumarraga MJ, Martin C, Samper S, Alito A, Latini O, Bigi F, et al. Usefulness of spoligotyping in molecular epidemiology of Mycobacterium bovis-related infections in South America. J Clin Microbiol. 1999;37:296–303. doi: 10.1128/jcm.37.2.296-303.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frothingham R, Meeker-O’Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144(Pt 5):1189–96. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 27.Roring S, Scott A, Brittain D, Walker I, Hewinson G, Neill S, et al. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J Clin Microbiol. 2002;40:2126–33. doi: 10.1128/JCM.40.6.2126-2133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skuce RA, McCorry TP, McCarroll JF, Roring SM, Scott AN, Brittain D, et al. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology. 2002;148(Pt 2):519–28. doi: 10.1099/00221287-148-2-519. [DOI] [PubMed] [Google Scholar]
- 29.Gibson AL, Hewinson G, Goodchild T, Watt B, Story A, Inwald J, et al. Molecular epidemiology of disease due to Mycobacterium bovis in humans in the United Kingdom. J Clin Microbiol. 2004;42:431–4. doi: 10.1128/JCM.42.1.431-434.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aranaz A, Liebana E, Mateos A, Dominguez L, Vidal D, Domingo M, et al. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–40. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costello E, O’Grady D, Flynn O, O’Brien R, Rogers M, Quigley F, et al. Study of restriction fragment length polymorphism analysis and spoligotyping for epidemiological investigation of Mycobacterium bovis infection. J Clin Microbiol. 1999;37:3217–22. doi: 10.1128/jcm.37.10.3217-3222.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Guerrero L, Milian-Suazo F, Arriaga-Diaz C, Romero-Torres C, Escartin-Chavez M. Molecular epidemiology of cattle and human tuberculosis in Mexico. Salud Publica Mex. 2008;50:286–91. doi: 10.1590/s0036-36342008000400006. [DOI] [PubMed] [Google Scholar]
- 33.Muller B, Hilty M, Berg S, Garcia-Pelayo MC, Dale J, Boschiroli ML, et al. African 1, an epidemiologically important clonal complex of Mycobacterium bovis dominant in Mali, Nigeria, Cameroon, and Chad. J Bacteriol. 2009;191:1951–60. doi: 10.1128/JB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cadmus S, Palmer S, Okker M, Dale J, Gover K, Smith N, et al. Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J Clin Microbiol. 2006;44:29–34. doi: 10.1128/JCM.44.1.29-34.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sales MP, Taylor GM, Hughes S, Yates M, Hewinson G, Young DB, et al. Genetic diversity among Mycobacterium bovis isolates: a preliminary study of strains from animal and human sources. J Clin Microbiol. 2001;39:4558–62. doi: 10.1128/JCM.39.12.4558-4562.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith NH, Gordon SV, De La Rua-Domenech R, Clifton-Hadley RS, Hewinson RG. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat Rev Microbiol. 2006;4:670–81. doi: 10.1038/nrmicro1472. [DOI] [PubMed] [Google Scholar]
- 37.Sunder S, Lanotte P, Godreuil S, Martin C, Boschiroli ML, Besnier JM. Human-to-human transmission of tuberculosis caused by Mycobacterium bovis in immunocompetent patients. J Clin Microbiol. 2009;47:1249–51. doi: 10.1128/JCM.02042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans JT, Smith EG, Banerjee A, Smith RM, Dale J, Innes JA, et al. Cluster of human tuberculosis caused by Mycobacterium bovis: evidence for person-to-person transmission in the UK. Lancet. 2007;369:1270–6. doi: 10.1016/S0140-6736(07)60598-4. [DOI] [PubMed] [Google Scholar]
- 39.Aphis Veterinary Services Fact Sheet. [(accessed March 2009)];Bovine tuberculosis. Available at: http://www.aphis.usda.gov/publications/animal_health/content/printable_version/fs_ahtb.pdf.
- 40.California Department of Food and Agriculture (CDFA) [(accessed March 2009)];California update: bovine tuberculosis (TB) update. Available at: http://www.cdfa.ca.gov/ahfss/animal_health/pdfs/tb/bovine_tb_update_021309.pdf.
- 41.Perumaalla VS, Adams LG, Payeur JB, Jarnagin JL, Baca DR, Guemes FS, et al. Molecular epidemiology of Mycobacterium bovis in Texas and Mexico. J Clin Microbiol. 1996;34:2066–71. doi: 10.1128/jcm.34.9.2066-2071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Kantor IN, Ritacco V. An update on bovine tuberculosis programmes in Latin American and Caribbean countries. Vet Microbiol. 2006;112:111–8. doi: 10.1016/j.vetmic.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 43.Allix C, Walravens K, Saegerman C, Godfroid J, Supply P, Fauville-Dufaux M. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J Clin Microbiol. 2006;44:1951–62. doi: 10.1128/JCM.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durr PA, Hewinson RG, Clifton-Hadley RS. Molecular epidemiology of bovine tuberculosis. I. Mycobacterium bovis genotyping. Rev Sci Tech. 2000;19:675–88. doi: 10.20506/rst.19.3.1241. [DOI] [PubMed] [Google Scholar]
- 45.Caimi K, Romano Mi, Alito A, Zumarraga M, Bigi F, Cataldi A. Sequence analysis of the direct repeat region in Mycobacterium bovis. J Clin Microbiol. 2001;39:1067–72. doi: 10.1128/JCM.39.3.1067-1072.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–56. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 47. [(accessed March 2010)];FDA Issues Health Advisory About Certain Soft Cheese Made From Raw Milk. Available at: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2005/ucm108420.htm.