In some cases, the molecular mechanisms underlying developmental potential are not strongly conserved. Should the RNA silencing mechanisms underlying oocyte-to-zygote transition and pluripotency in mammals be viewed as unique solutions superimposed on broader general principles?
Keywords: pluripotency, RNAi, oocyte, Dicer, Dgcr8, miR-290
Abstract
RNA silencing is a complex of mechanisms that regulate gene expression through small RNA molecules. The microRNA (miRNA) pathway is the most common of these in mammals. Genome-encoded miRNAs suppress translation in a sequence-specific manner and facilitate shifts in gene expression during developmental transitions. Here, we discuss the role of miRNAs in oocyte-to-zygote transition and in the control of pluripotency. Existing data suggest a common principle involving miRNAs in defining pluripotent and differentiated cells. RNA silencing pathways also rapidly evolve, resulting in many unique features of RNA silencing in different taxonomic groups. This is exemplified in the mouse model of oocyte-to-zygote transition, in which the endogenous RNA interference pathway has acquired a novel role in regulating protein-coding genes, while the miRNA pathway has become transiently suppressed.
See Glossary for abbreviations used in this article.
Glossary.
- DAZAP2
DAZ (deleted in azoospermia)-associated protein 2
- DGCR8
Di George Critical Region 8
- DCR
DICER, an RNAse III family endoribonuclease
- DNMT
de novo DNA methyltransferase
- dpc
days post-coitum
- dsRNA
double-stranded RNA
- EB
embryoid bodies
- EEmiRC
early embryonic microRNA cluster
- ELAVL2
(HuB) A+U-rich sequences binding translational repressor
- ESCs
embryonic stem cells
- iPSCs
induced pluripotent stem cells
- ESCC
ES cell-specific cell cycle-regulating miRNAs
- miRNA
microRNA, a small RNA encoded by the genome
- OZT
oocyte-to-zygote transition
- OCT4
octamer 4, also known as POU class 5 homeobox
- oncomiRs
miRNAs with oncogenic potential
- piRNA
Piwi-interacting RNA
- RBL2
retinoblastoma-like protein 2
- RNAi
RNA interference, one of the RNA silencing pathways that uses short RNAs generated from long dsRNA to induce sequence-specific cleavage of cognate mRNAs
- RT-PCR
reverse transcription PCR
- siRNA
small interfering RNA
- SOX2
(sex determining region Y)-Box 2 transcription factor
- UTR
untranslated region
- ZGA
zygotic genome activation
Introduction
Pluripotency is the ability of cells to differentiate into any fetal or adult cell type. Pluripotency is formed during early development and self-renewing pluripotent cells can be maintained in culture under specific conditions. Pluripotency in mammals is most often studied in embryonic stem cells (ESCs), which are derived from the inner cell mass of the blastocyst (Smith, 2001), and induced pluripotent stem cells (iPSCs), which are formed after reprogramming gene expression in somatic cells with specific pluripotency factors (Takahashi & Yamanaka, 2006). The OCT4 (POU5F1), SOX2 and NANOG transcription factors form the core of a network responsible for the transcriptional control of ESC renewal and pluripotency (Boyer et al, 2005; Loh et al, 2006). Microarray data indicate that a similar transcription factor network also forms in a stepwise manner upon mouse zygotic genome activation (ZGA; Zeng et al, 2004; Zeng & Schultz, 2005), which starts around 1.0 dpc during the early two-cell stage (the blastocyst, which is the source of ESCs, forms at 3.5 dpc). Thus, reprogramming of an oocyte into pluripotent blastomeres of an early embryo—here referred to as oocyte-to-zygote transition (OZT)—is a physiological analogy of the experimental reprogramming of somatic cells into iPSCs. In fact, the cytoplasm of an enucleated oocyte can induce pluripotency in the nuclei of somatic cells during nuclear transfer (reviewed in Gurdon & Melton, 2008), which is the basis of mammalian cloning (Wilmut et al, 1997). As there is no mRNA synthesis between the end of the mouse oocyte growth phase and the first zygotic cleavage, post-transcriptional mechanisms are essential for the natural formation of pluripotency.
Here, we review the role of mammalian miRNA and RNAi during OZT and in the control of pluripotency. MicroRNA and RNAi pathways use small RNA molecules as sequence-specific guides for translational repression and/or mRNA degradation (reviewed in Chapman & Carrington, 2007). Briefly, siRNAs are produced from long dsRNA by cleavage with RNase III Dicer. MicroRNAs originate as long primary transcripts with local hairpin structures, that are cleaved by the Microprocessor complex composed of DGCR8 and RNase III Drosha. MicroRNAs are produced from small hairpin precursors upon cleavage by Dicer. Small interfering RNAs and miRNAs are loaded on Argonaute (AGO) proteins, which mediate silencing effects (reviewed in Kim et al, 2009).
Animal miRNAs typically base-pair imperfectly with the 3′-UTR of target mRNAs and induce translational repression and/or mRNA degradation. MicroRNAs are the dominant class of short RNAs in mammalian cells—more than a thousand miRNAs with defined sequences have been identified and implicated in the regulation of many cellular processes (Kloosterman & Plasterk, 2006). It has been estimated that miRNAs could directly target more than half of all mammalian genes (Friedman et al, 2009).
Target mRNA recognition involves the nearly perfect hybridization of the miRNA 5′-end, which nucleates the miRNA–mRNA interaction (Brennecke et al, 2005; Lai, 2002). Perfect base-pairing of a miRNA or siRNA results in the AGO2-mediated sequence-specific endonucleolytic cleavage of the target in the middle of the base-pairing sequence (Liu et al, 2004; Meister et al, 2004). However, miRNAs can induce substantial mRNA degradation even in the absence of extensive base-pairing to their targets (Bagga et al, 2005; Lim et al, 2005). This is a likely consequence of translational repression and the relocation of repressed mRNAs to cytoplasmic foci known as P-bodies (reviewed in Eulalio et al, 2007a). P-bodies harbour mRNAs, translational repressors, mRNA-binding proteins and components of the mRNA degradation machinery. It has been proposed that their formation is a consequence of miRNA activity (Eulalio et al, 2007b).
The role of microRNAs in pluripotent stem cells
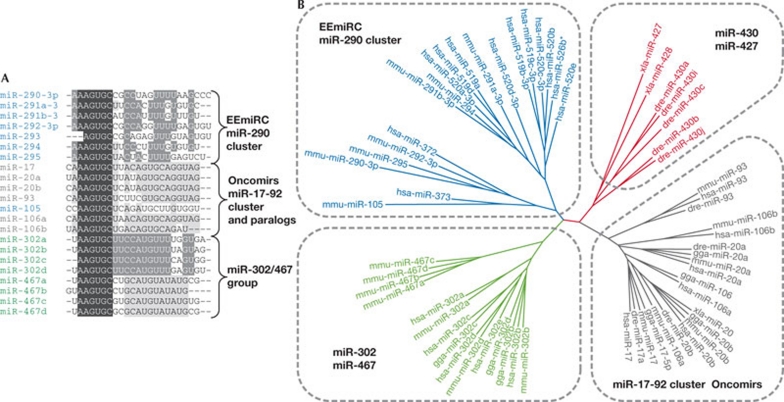
The well-characterized ESC miRNome is dominated by miRNAs sharing a 5′-proximal AAGUGC motif (Babiarz et al, 2008; Houbaviy et al, 2003; Landgraf et al, 2007; Suh et al, 2004). These miRNAs (referred to hereafter as AAGUGC miRNAs) can be divided into three groups (Fig 1): (i) EEmiRC miRNAs, found in placental mammals (Houbaviy et al, 2005); (ii) the miR-17-92 cluster (and its paralogues), which is conserved across vertebrates and carries onco-miRs (reviewed in Mendell, 2008); and (iii) the miR-302/miR-467 group, including the miR-302 family in tetrapods and the miR-467 family in the mouse. In addition, there is a closely related group of miRNAs (Fig 1B) that are found in fish (miR-430 cluster) and amphibian embryos (miR-427), where they contribute to the degradation of maternal mRNAs (Giraldez et al, 2006; Lund et al, 2009). The miR-17-92 cluster group probably evolved independently as these miRNAs—in contrast to the other two groups—derive from the 5′ arm of small hairpin and are not as GU-rich in the 3′ end.
Figure 1.
MicroRNAs sharing the proximal AAGUGC motif. (A) Alignment of mouse AAGUGC miRNAs. The conserved 5′end is shaded in black. Grey shading indicates nucleotides conserved in the major groups of AAGUGC miRNAs. (B) Unrooted neighbour-joining tree showing divergence of animal AAGUGC miRNAs. hsa, Homo sapiens; mmu, Mus musculus; xla, Xenopus laevis; gga, Gallus gallus; dre, Danio rerio.
Another remarkable feature of the ESC miRNome is the minimal expression of the let-7 miRNA family. The highly conserved let-7 family is expressed in adult and differentiated animal tissues; it is upregulated during the differentiation of ESCs and is thought to regulate ‘stemness' by suppressing self-renewal and promoting differentiation (reviewed in Bussing et al, 2008). The opposing activities of let-7 and pluripotent miRNAs represent one of the features that distinguish pluripotent and differentiated cells (Melton et al, 2010).
The role of ESC miRNAs has been investigated using cells lacking Dgcr8 and Dcr1, both of which are required for miRNA biogenesis. While DGCR8 is involved in the production of canonical miRNAs, DICER1 is involved in producing both siRNAs and miRNAs. The loss of Dcr1 in mouse ESCs results in the depletion of miRNAs and causes slower proliferation and differentiation defects in vivo and in vitro (Kanellopoulou et al, 2005; Murchison et al, 2005). In vivo, Dcr1−/− ESCs do not contribute to chimeric mice and fail to generate teratomas. In vitro, Dcr1−/− ESCs form EB-like structures, but there is little morphological evidence of differentiation. Expression of Oct4 is only partly decreased in mutant EBs after day 5 of differentiation, and expression of endodermal and mesodermal markers is not detectable (Kanellopoulou et al, 2005). Similarly, the loss of Dgcr8 causes partial downregulation of pluripotency markers during the retinoic-acid-induced differentiation (Wang et al, 2007). In contrast to the Dcr1−/− ESCs, Dgcr8−/− cells show weaker differentiation defects that include largely undifferentiated teratomas and abnormal expression (but not an absolute loss) of differentiation markers. This indicates that a part of the Dcr1−/− phenotype is caused by the loss of endogenous siRNAs (endo-siRNAs).
The analysis of Dcr1−/− ESCs has also revealed defects in the centromeric chromatin, manifested as a loss of DNA methylation and histone H3K9 trimethylation, and an increased abundance of RNAs derived from centromeric repeats (Fukagawa et al, 2004; Kanellopoulou et al, 2005). However, Dcr1−/− ESCs produced independently did not show the same phenotype (Murchison et al, 2005). Further analysis has revealed that the chromatin defects reported from Dcr1−/− ESCs can be at least partly explained by miRNA-mediated regulations (Benetti et al, 2008; Sinkkonen et al, 2008).
Two approaches have been taken to address the specific roles of miRNAs in mouse ESCs. Sinkkonen et al undertook a bioinformatic analysis of the transcriptome of Dcr1−/− ESCs (Sinkkonen et al, 2008), while Wang et al screened rescue effects of individual miRNAs in Dgcr8−/− ESCs (Wang et al, 2007). Dcr1−/− analysis has revealed that the most significantly enriched motifs in the 3′UTRs of transcripts upregulated in Dcr1−/− ESCs are sequences complementary to AAGUGC motifs. This implicates a dominant function for EEmiRC miRNAs among ESC miRNAs. Subsequent analysis has identified around 250 primary targets of EEmiRC miRNAs in ESCs (Sinkkonen et al, 2008), and Rbl2 (p130) and Cdkn1a (p21) were verified as primary targets of EEmiRC miRNAs (Fig 2). RBL2 is a transcriptional repressor that suppresses expression of de novo DNA methyltransferases. Thus, in the absence of EEmiRC miRNAs, elevated levels of RBL2 reduce levels of DNMT3A and DNMT3B. This causes DNA methylation defects during differentiation, whereby the Oct4 promoter does not accumulate de novo DNA methylation (Benetti et al, 2008; Sinkkonen et al, 2008). Defective de novo DNA methylation probably contributes to differentiation defects in ESCs, similarly to a Dnmt3a and Dnmt3b double knockout (Chen et al, 2003). However, differentiation defects are not rescued by EEmiRC miRNAs (Wang et al, 2008), suggesting that the cause of differentiation defects includes the loss of other ESC miRNAs or perhaps miRNAs that appear and function during differentiation, such as miR-145 (Xu et al, 2009).
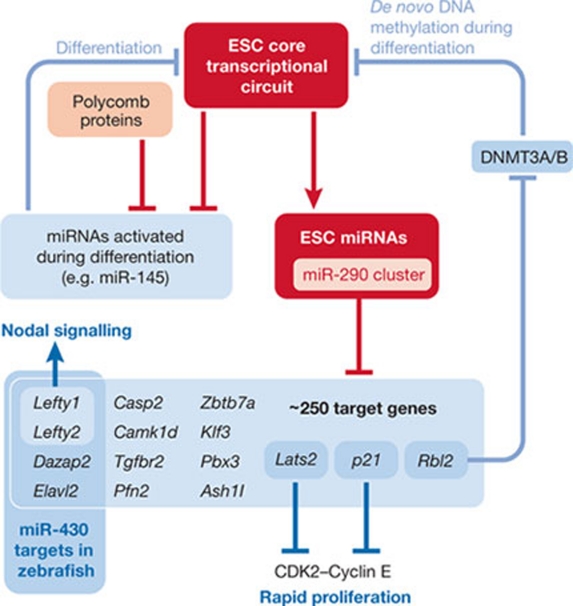
Figure 2.
Roles of the miR-290 cluster in undifferentiated mouse embryonic stem cells. Red frames highlight two important components of control of gene expression in ESCs: the core pluripotency transcription factor network (ESC core transcriptional circuit), which also controls expression of the second component, ESC miRNAs. The miR-290 cluster dominates miRNA function in ESCs and regulates ∼250 primary targets, some of which are shown here. Murine miR-290 have a role in several processes including cell-cycle regulation, nodal signalling and transcriptional regulation. The miR-290 cluster indirectly regulates de novo DNA methylation by targeting RBL2, a transcriptional repressor acting on Dnmt3 genes. MicroRNAs—including miR-145 and others—also contribute to suppression of the ESC core transcriptional circuit during differentiation. The scheme and listed miR-290 targets are based on published data (Marson et al, 2008; Melton et al, 2010; Sinkkonen et al, 2008; Tay et al, 2008; Xu et al, 2009). ESC, embryonic stem cell.
Studies of Dcr1−/− and Dgcr8−/− have also revealed that cell-cycle progression in ESCs is under the control of miRNAs. Sinkkonen et al reported that EEmiRC miRNAs regulate p21 and that transfection of EEmiRC miRNAs partly rescues growth defects in Dcr1−/− cells (Sinkkonen et al, 2008). Wang et al subsequently showed, in Dgcr8−/− cells, that several ESC miRNAs designated ESCC contribute to the rescue of proliferation by the regulation of CDK2–cyclin E-dependent control of G1–S progression (Wang et al, 2008). ESCC miRNAs include EEmiRC miRNAs, members of the miR-17-92 cluster and an unrelated miR-223. MicroRNA-mediated control of proliferation also involves cyclin D1, which is targeted by miR-302 in human ESCs (Card et al, 2008).
Interestingly, despite the massive transcriptome remodeling in Dcr1−/− ESCs, the loss of pluripotency is not accompanied by notable changes in the expression of the core pluripotency circuit factors (Sinkkonen et al, 2008). This implies that ESC miRNAs, the transcription of which is largely controlled by core pluripotency factors (Marson et al, 2008), do not form a feedback loop to maintain the core pluripotency factors and that the core pluripotency network is insufficient to maintain pluripotency in the absence of miRNAs. In addition to their role in the maintenance of pluripotency, EEmiRC miRNAs can also contribute to the establishment of pluripotency during the formation of iPSCs (Judson et al, 2009). miR291-3p, miR-294 and miR-295 increase the efficiency of reprogramming by pluripotency factors Oct4, Sox2 and Klf4. Thus, AAGUGC miRNAs dominate miRNA activities in ESCs and have a broader relationship to the control of cell growth and differentiation.
The role of microRNAs and endo-siRNAs in OZT
Mouse ovarian oocytes are highly specialized cells that are arrested in the prophase of the first meiotic division. The fully grown mouse oocyte can resume meiosis during ovulation but arrests again at metaphase II and dies unless fertilized. Even if diploidized and parthenogenetically activated, a zygotic genome of entirely maternal origin cannot support normal embryonic development (Surani et al, 1984). The fertilized egg, however, transforms into a cleaving zygote, the cells of which exhibit pluripotency. The factors required for genome reprogramming during OZT are present in the oocyte cytoplasm. OZT has two essential components: (i) control of stability and translation of maternal mRNAs and (ii) control of the transcriptional activation of zygotic genes. As no new mRNAs are produced before the two-cell stage (Zeng & Schultz, 2005), gene expression during OZT is mostly governed by post-transcriptional mechanisms such as reversible deadenylation coupled with transcriptional repression—which is the basis for controlling storage and recruitment of specific mRNAs for translation during meiosis and later (reviewed in Richter, 2007). RNA silencing pathways—that have essential roles elsewhere—are also involved in the regulation of mammalian OZT, but in a rather unexpected way.
MicroRNAs during OZT
The cloning of small RNAs from mouse oocytes has revealed three classes of small RNA: miRNAs, endo-siRNAs and piRNAs (Tam et al, 2008; Watanabe et al, 2008). Mammalian piRNAs are small RNAs produced by a Dcr1-independent mechanism that are essential for the male but not the female germline (reviewed in Klattenhoff & Theurkauf, 2008). The presence of all three known mammalian RNA silencing pathways in one cell type is unique. Interestingly, mouse oocytes do not contain any abundant oocyte-specific miRNAs and, due to a high abundance of members of the let-7 family and low levels of the miR-290 cluster miRNAs, the miRNA profile appears somatic-like. This is counterintuitive, considering the reprogramming ability of oocyte cytoplasm.
RT-PCR analysis of mouse miRNA expression during OZT (Tang et al, 2007) has revealed typical maternal, maternal-zygotic and zygotic expression patterns (Fig 3B). MicroRNA expression patterns are relatively unvarying when compared with mRNA expression patterns during OZT, and essentially depend on the ‘entry' level of maternal miRNAs and the intensity of zygotic transcription. Maternal miRNAs undergo a common degradation on fertilization, and zygotic miRNAs increase their abundance from the two-cell stage onwards (Tang et al, 2007). This common degradation probably reflects the fact that miRNAs are too short to be selectively degraded on the basis of sequence motifs, but the common transcriptional activation during ZGA is peculiar because miRNA expression is controlled by polymerase II, which allows for variable expression patterns. This is in contrast to maternal mRNAs, which have diverse individual patterns of expression (Zeng et al, 2004; Zeng & Schultz, 2005).
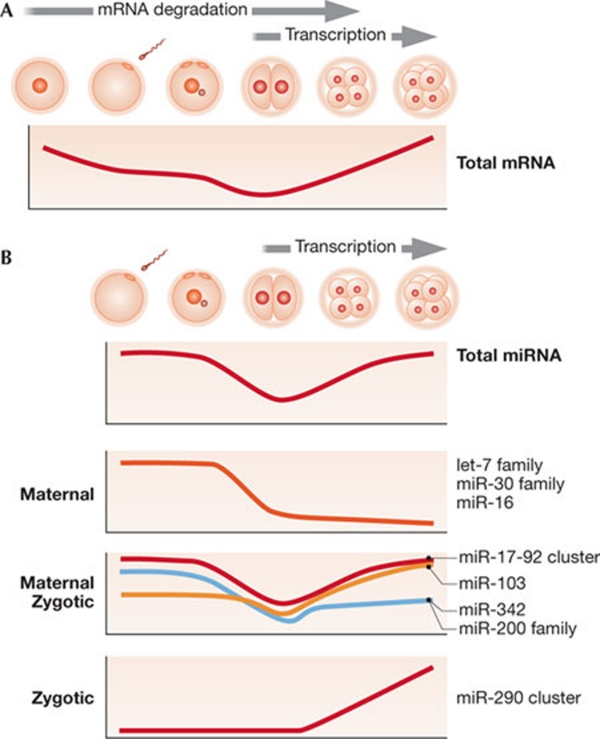
Figure 3.
miRNAs during mouse oocyte-to-zygote transition. (A) Total mRNA (poly(A) RNA) amount changes during OZT (based on Piko & Clegg, 1982). From left to right the following developmental stages are shown: fully grown oocyte, mature MII egg, one-cell, two-cell, four-cell and eight-cell. (B) Schematic miRNA expression profiles during OZT. From left to right the following developmental stages are shown: mature MII egg, one-cell, two-cell, four-cell and eight-cell. Graphs are based on previously published data (Tang et al, 2007). miRNA, microRNA; OZT, oocyte-to-zygote transition.
The most abundant maternal miRNAs are members of the let-7 family, which account for about one-third of miRNAs in the oocyte (Tam et al, 2008; Tang et al, 2007; Watanabe et al, 2008). Such a high quantity of mature let-7 miRNAs is surprising considering that let-7 is typically found in adult and differentiated tissues, and its cloning frequency in Xenopus eggs is relatively low (Armisen et al, 2009). let-7 levels rapidly decline during OZT and let-7 expression is not restored in the early embryo, which is consistent with very low let-7 levels in ESCs. The most pronounced zygotic miRNAs are EEmiRC members (the miR-290 cluster), which are transcribed during the two-cell stage (Zeng & Schultz, 2005) and accumulate throughout preimplantation development (Tang et al, 2007). There are also maternal-zygotic miRNAs (for example, the miR-17-92 cluster), which are present at considerably high levels in oocytes and embryos, with a transiently lower expression at the two-cell stage (Fig 3B). These data imply that there is a switch between maternal and zygotic miRNAs that involves the rapid degradation of maternal miRNAs between the fertilized egg and the two-cell embryo stages. The active degradation must cease at times during the two-cell stage to allow for expression of zygotic miRNAs. Presumably, the remaining maternal miRNAs that survive the degradation disappear as they are replaced with zygotic miRNAs.
Idling microRNAs
The loss of Dcr1-dependent small RNAs during oocyte growth causes defects in meiotic spindle formation and infertility (Murchison et al, 2007; Tang et al, 2007). This phenotype was originally attributed to the loss of maternal miRNAs, because mature miRNAs were abundant in the wild-type oocyte and it seemed unlikely that the loss of endo-siRNAs would cause a spindle defect (Murchison et al, 2007; Tang et al, 2007). More recent data, however, questions the activity and role of maternal miRNAs, and changes the view of RNA silencing in mouse oocytes and early embryos. A study of the maternal loss of Dgcr8—which is involved in the biogenesis of canonical miRNA—found that Dgcr8−/− oocytes develop and ovulate normally, can be fertilized and give rise to viable mice (Suh et al, 2010). Although the loss of maternal miRNAs slightly reduced developmental competence, it became clear that oocytes have an impressive tolerance to the loss of miRNAs. This tolerance appears to extend into early development because Dgcr8−/− oocytes fertilized with Dgcr8−/− sperm develop to the blastocyst stage (Suh et al, 2010).
In parallel, a loss of P-bodies in fully grown mouse oocytes was discovered by indirect immunofluorescence and by visualizing mRNAs targeted to P-bodies (Flemr et al, 2010; Ma et al, 2010). During OZT, P-bodies disappear as meiotically incompetent small postnatal oocytes start to grow in size, and appear again at the morula and blastocyst stages (Fig 4; Flemr et al, 2010). Although P-bodies are absent in growing oocytes, their components—TNRC6, DDX6, CPEB—and other proteins are found in subcortical ribonucleoprotein aggregates, which are thought to be a storage place for maternal mRNAs (Flemr et al, 2010).
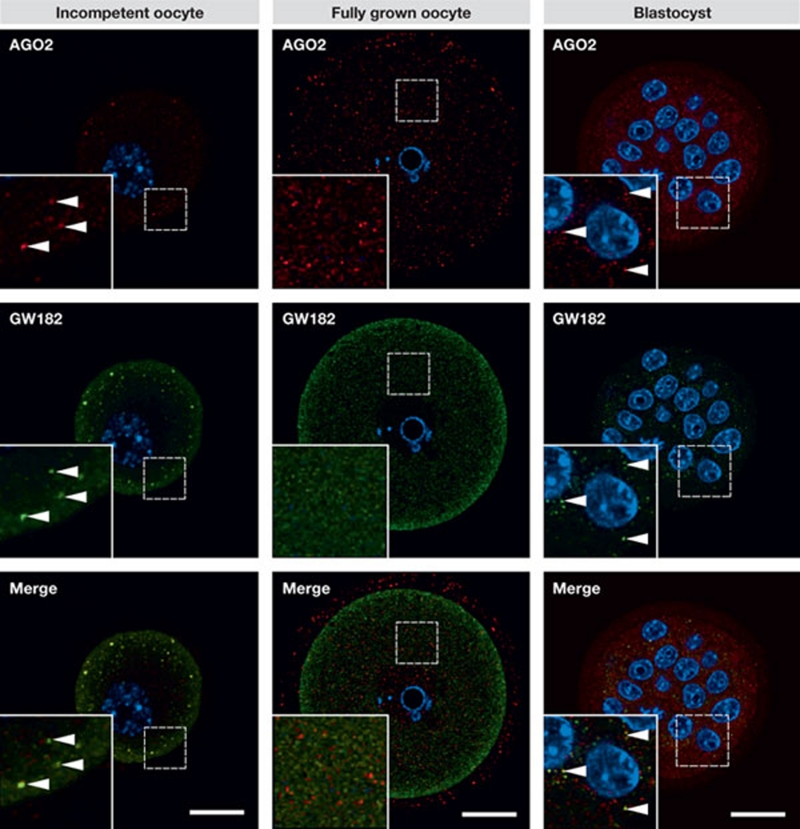
Figure 4.
Loss of P-bodies in fully grown oocytes. Confocal images of GW182 (green) and AGO2 (red) in meiotically incompetent mouse oocytes (from 12-day-old mice), fully grown preovulatory oocytes and blastocysts. The inset shows a detail of cytoplasmic staining, the part of the specimen covered by the insert is not any different from the rest of the image. Immunofluorescent staining of GW182 and AGO2 was carried out as previously described (Flemr et al, 2010). DNA is shown in blue (DAPI). Colocalization yields a yellow colour. Arrowheads depict P-bodies in blastocysts and somatic cells. Note the absence of GW182 and AGO2 colocalization and enhanced subcortical staining of GW182 in the subcortical region in fully grown oocytes. Scale bars, 20 μm.
The absence of P-bodies provoked a more detailed examination of maternal miRNA activities using luciferase-based reporters, which can distinguish between the abilities of endogenous miRNAs to induce RNAi-like cleavage and translational repression (Schmitter et al, 2006). Reporters targeted by endogenous let-7 and miR-30 showed that RNAi-like cleavage (that is, loading of miRNA on AGO2 and accessibility of cognate 3′ UTRs) is relatively normal, but the ability of miRNAs to repress translation is strongly reduced during oocyte growth and maturation (Ma et al, 2010). Interestingly, the less abundant miR-30 apparently retained more silencing activity, suggesting that an additional mechanism or mechanisms suppresses let-7 function in the oocyte (Ma et al, 2010). Suppression of miRNA activity is further supported by microarray data from Dgcr8−/− and Dcr1−/− oocytes (Suh et al, 2010). Taken together, this suggests that miRNAs are not only non-essential for OZT, but also that their function is suppressed by the oocyte. Therefore, uncoupling miRNAs from the repression of translation is the earliest known event in OZT in the mouse.
The mechanism and benefits of idling miRNAs are unknown. Suppression of miRNA function seems to take place after AGO2 loading and possibly involves uncoupling of AGO2 and TNRC6 (GW182), the latter being necessary for miRNA-mediated translational repression (Eulalio et al, 2008). This model is supported by the loss of colocalization of AGO2 and GW182 in oocytes (Flemr et al, 2010). Interestingly, overexpression of AGO2 in early embryos induces formation of P-bodies (Lykke-Andersen et al, 2008). At the moment, it is unclear why the miRNA pathway is turned off during the largest genome reprogramming event in the mammalian life cycle. We can only speculate that it might contribute to OZT by facilitating the switch between maternal and zygotic miRNAs. Idling miRNA might be seen as a waste of resources, but such a gearshift mechanism is a simple solution to disengaging maternal miRNAs during oocyte growth and engaging zygotic miRNAs later in preimplantation development.
As mentioned above, members of the let-7 family constitute about one-third of maternal miRNome, and are negative regulators of pluripotency. Although Lin-28, an inhibitor of let-7 (Hagan et al, 2009), has high transcript levels in the oocyte (Zeng et al, 2004), it does not appear to prevent high levels of mature let-7 from accumulating. A global suppression of activity of maternal miRNAs might, therefore, be able to relieve let-7-mediated repression, and allow the expression of maternal factors, which will promote pluripotency in the zygote. Furthermore, as a considerable amount of let-7 survives maternal miRNA degradation between fertilization and the two-cell stage, continual suppression of the miRNA pathway into preimplantation development would further facilitate replacement of let-7 with zygotic miRNAs, particularly with the miR-290 cluster, which has a let-7 opposing role (Melton et al, 2010). In this situation, LIN28 would prevent zygotic expression of let-7, which could cause problems when the miRNA pathway becomes functional again.
Mammalian RNA interference in the spotlight
One important output of recent studies of maternal miRNAs is new data concerning endogenous RNAi in mammals. The mammalian RNAi pathway has so far been a mystery, as endo-siRNAs were found only in oocytes and ESCs (Babiarz et al, 2008; Tam et al, 2008; Watanabe et al, 2008). Oocyte endo-siRNAs derived from processed pseudogenes suggest that mammalian RNAi, in addition to roles in the suppression of mobile and repetitive sequences known from invertebrates, might also regulate endogenous genes (Tam et al, 2008; Watanabe et al, 2008). This hypothesis is now supported by the defective spindle phenotype of Dcr1−/− and Ago2−/− oocytes, which is absent in Dgcr8−/− oocytes. Furthermore, the bioinformatic analysis of the Dcr1−/− transcriptome shows that many upregulated transcripts have complementary sequences to endo-siRNAs found in the oocyte (Ma et al, 2010). Thus, data support a model in which the miRNA pathway becomes disengaged early during oocyte growth and RNAi becomes the dominant RNA silencing pathway essential for OZT. Here, it presumably targets genes that would interfere with proper meiotic spindle assembly. The requirement for endogenous RNAi might extend into early development, because Ago2 knockdown in early embryos causes arrests at the two-cell stage (Lykke-Andersen et al, 2008). The role of endogenous RNAi still needs to be validated by evidence that the ‘slicer' activity of AGO2 is required for normal oocyte development. At present, it implies that the Dcr1−/− and Ago2−/− phenotypes might be unique to the mouse and that the role of mammalian RNAi in the oocyte might not be well conserved because it relies on endo-siRNAs produced from transcribed, processed pseudogenes, which evolve rapidly.
MicroRNAs in the zygote
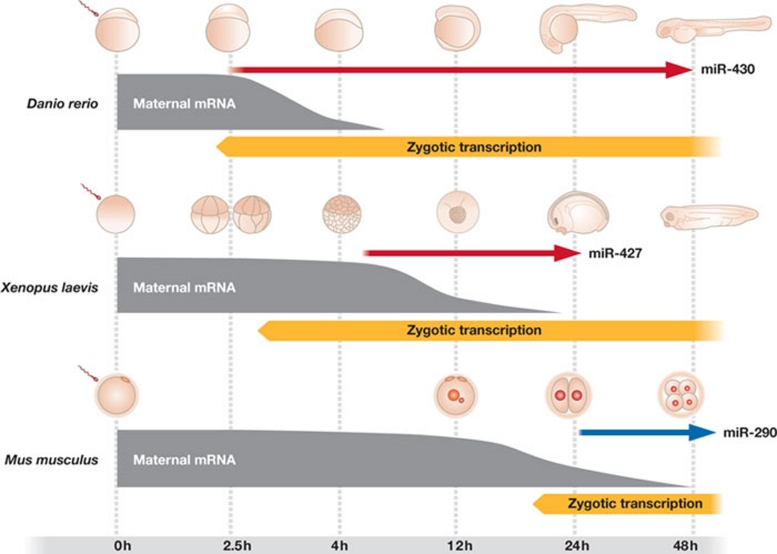
Although we have argued above that mammalian miRNAs are not needed for OZT, it is necessary to put the function of zygotic miRNAs into the context of other vertebrate systems. When such a comparison is made (Fig 5), it becomes apparent that vertebrate zygotes express AAGUGC miRNAs, but use them in a different spatiotemporal context.
Figure 5.
Oocyte-to-zygote transition in the mouse, zebrafish and frog. Specific developmental stages, the course of maternal mRNA degradation, ZGA and expression of zygotic AAGUGC miRNAs in mouse, zebrafish and frog embryos is shown on the same timescale.
In the mouse, the major reprogramming of the zygote by newly expressed genes initiates at the two-cell stage. Although the one-cell embryo is transcriptionally active, it does not seem to produce functional mRNAs (Zeng & Schultz, 2005). The final stage of degradation of maternal mRNAs is superimposed on the activation of the zygotic genome. It is initiated by the resumption of meiosis and is an integral part of the switch from the maternal to the zygotic programme, although a direct link between maternal mRNA degradation and establishment of pluripotency has not been documented. Maternal mRNA degradation is substantial and, by the two-cell stage, eliminates 75% of the poly(A) RNA that is originally found in the pre-ovulatory oocyte (Piko & Clegg, 1982). The zygotic miR-290 family transcription starts during the two-cell stage, but mature miRNA accumulation is first observed at the four-cell stage (Tang et al, 2007; Zeng & Schultz, 2005). Zygotic miRNAs are, therefore, unlikely to contribute significantly to maternal mRNA degradation in the mouse: first, because their activity is suppressed at the early stages of preimplantation development; and second, even if the zygotic miRNAs were active, they would appear when the majority of maternal mRNAs are already degraded.
This situation is different from that in the fish and the frog, in which development is very fast and zygotic miRNAs appear when maternal mRNAs are still abundant (Fig 5). Due to this fast development, there is likely to be greater pressure to eliminate maternal mRNAs so that they do not interfere with differentiation, and zygotic miRNAs offer a way to deal with this problem. A contribution of miRNAs to degradation of maternal mRNA was reported in Drosophila (Bushati et al, 2008). In the zebrafish, the miR-430 family becomes strongly upregulated a few hours after fertilization and targets up to an estimated 40% of maternal mRNAs (Giraldez et al, 2006). This suggests that the miR-430 family significantly contributes to maternal mRNA degradation. A similar role has also been proposed for miR-427 during Xenopus embryo development (Lund et al, 2009).
Despite the differences detailed above, there are also notable similarities. One example is a partly conserved role in Nodal signalling, which is important for germ layer specification (Choi et al, 2007; Rosa et al, 2009). Early vertebrate embryos express a set of related AAGUGC miRNAs during ZGA. This suggests that AAGUGC miRNAs have common and evolving roles in the regulation of development. In addition to Lefty1 and Lefty2 in Nodal signalling, only a small number of genes—including Dazap2 and Elavl2 (HuB)—are common among miR-290 and miR-430 targets (Giraldez et al, 2006; Sinkkonen et al, 2008). However, the general role of EEmiRCs and the miR-430 cluster is the same: they support development by targeting genes the expression of which should be kept low in the embryo. From this perspective, the difference is in the context: the miR-290 cluster targets mostly zygotic transcripts, whereas the miR-430 cluster deals also with a large amount of maternal mRNAs. Variability among targets and specific roles of AAGUGC miRNAs across vertebrates should not be surprising, because early post-zygotic regulations might evolve fast during speciation.
Conclusions
It is remarkable that the molecular mechanisms underlying developmental potential are not strongly conserved. This can be contrasted with the example of let-7 miRNA, which is found in differentiated cells from Caenorhabditis elegans to mammals, and zygotic AAGUGC miRNAs, which are found in vertebrates with considerable variation in conservation at the genomic level. RNA silencing mechanisms underlying OZT and pluripotency in mammals should be viewed as unique solutions superimposed on broader general principles. In the mouse, miRNA function is suppressed in oocytes: miRNAs do not contribute to zygotic genome activation, maternal mRNA degradation or establishment of the core pluripotency transcriptional network. However, the importance of endogenous RNAi in the oocyte has recently become apparent. MicroRNAs are essential for pluripotency, but whether this reflects the activity of ESC miRNAs or the requirement for other miRNAs during differentiation remains to be revealed.
Sidebar A | In need of answers.
Which miRNAs are essential for pluripotency in embryonic stem cells?
How do miRNAs support pluripotency in embryonic stem cells?
What is the role of RNAi in embryonic stem cells?
How are miRNAs suppressed in the oocyte?
Would normal miRNA activity interfere with OZT?
Is mammalian RNAi-mediated control of gene expression essential for meiosis?
How conserved is the function of RNAi in the germline in mammals?
Acknowledgments
The authors thank Richard M. Schultz, Robert Blelloch and Witold Filipowicz for fruitful discussions. P.S. is a holder of the J.E. Purkyne Fellowship and his lab is supported by the following grants: EMBO SDIG project 1483, GACR 204/09/0085, GACR P305/10/2215 and Kontakt ME09039.
Footnotes
The authors declare that they have no conflict of interest.
References
- Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA (2009) Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res 19: 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563 [DOI] [PubMed] [Google Scholar]
- Benetti R et al. (2008) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA et al. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM (2008) Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol 18: 501–506 [DOI] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H (2008) let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 14: 400–409 [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK (2008) Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28: 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E (2003) Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23: 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF (2007) Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318: 271–274 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007a) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E (2007b) P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Flemr M, Ma J, Schultz RM, Svoboda P (2010) P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod 8: 1008–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6: 784–791 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Melton DA (2008) Nuclear reprogramming in cells. Science 322: 1811–1815 [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI (2009) Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA (2003) Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Dennis L, Jaenisch R, Sharp PA (2005) Characterization of a highly variable eutherian microRNA gene. RNA 11: 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W (2008) Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450 [DOI] [PubMed] [Google Scholar]
- Lai EC (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30: 363–364 [DOI] [PubMed] [Google Scholar]
- Landgraf P et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Loh YH et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE (2009) Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15: 2351–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, Zernicka-Goetz M (2008) Maternal Argonaute 2 is essential for early mouse development at the maternal–zygotic transition. Mol Biol Cell 19: 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM (2010) MicroRNA activity is suppressed in mouse oocytes. Curr Biol 20: 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A et al. (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT (2008) miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ (2007) Critical roles for Dicer in the female germline. Genes Dev 21: 682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piko L, Clegg KB (1982) Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol 89: 362–378 [DOI] [PubMed] [Google Scholar]
- Richter JD (2007) CPEB: a life in translation. Trends Biochem Sci 32: 279–285 [DOI] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH (2009) The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16: 517–527 [DOI] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W (2006) Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 34: 4801–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15: 259–267 [DOI] [PubMed] [Google Scholar]
- Smith AG (2001) Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol 17: 435–462 [DOI] [PubMed] [Google Scholar]
- Suh MR et al. (2004) Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498 [DOI] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R (2010) MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML (1984) Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tam OH et al. (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA (2007) Maternal microRNAs are essential for mouse zygotic development. Genes Dev 21: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1–S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T et al. (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813 [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS (2009) MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM (2005) RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 283: 40–57 [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM (2004) Transcript profiling during preimplantation mouse development. Dev Biol 272: 483–496 [DOI] [PubMed] [Google Scholar]