High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA
Legionella pneumophila uses DrrA to recruit ER-derived vesicles to the Legionella-containing vacuole. This study demonstrates the structural basis for DrrA-binding to PtdIns(4)P, and describes an unexpected high affinity of DrrA to PtdIns(4)P.
Keywords: DrrA, Legionella, phosphatidylinositol 4-phosphate, Rab1, SidM
Abstract
The DrrA protein of Legionella pneumophila is involved in mistargeting of endoplasmic reticulum-derived vesicles to Legionella-containing vacuoles through recruitment of the small GTPase Rab1. To this effect, DrrA binds specifically to phosphatidylinositol 4-phosphate (PtdIns(4)P) lipids on the cytosolic surface of the phagosomal membrane shortly after infection. In this study, we present the atomic structure of the PtdIns(4)P-binding domain of a protein (DrrA) from a human pathogen. A detailed kinetic investigation of its interaction with PtdIns(4)P reveals that DrrA binds to this phospholipid with, as yet unprecedented, high affinity, suggesting that DrrA can sense a very low abundance of the lipid.
Introduction
The pathogen Legionella pneumophila causes Legionnaires' disease by infecting human lung macrophages (Muder et al, 1986). Shortly after uptake, the bacterium establishes the Legionella-containing vacuole (LCV) as an intracellular replicative organelle, from which it injects several proteins through its type IV secretion system into the cytosol of the host cell (Isberg et al, 2009). One of the substrates of the type IV secretion system is DrrA, also known as SidM, a 647-amino-acid protein that is involved in redirecting endoplasmic reticulum (ER)-derived vesicles to the LCV. DrrA consists of three domains, the central domain (amino acids 340–533) having guanine nucleotide exchange factor (GEF) activity towards the GTPase Rab1 (Machner & Isberg, 2007; Schoebel et al, 2009; Suh et al, 2010), a carboxy-terminal lipid-binding domain and an amino-terminal domain of unknown function. Immediately after transfer from the Legionella bacterium into the host cytosol, DrrA localizes to the cytosolic surface of the LCV through its C-terminal domain by binding specifically to phosphatidylinositol 4-phosphate (PtdIns(4)P; Brombacher et al, 2009). Once bound to the LCV, the GEF domain of DrrA mediates redirection of ER-derived vesicles through the recruitment and activation of Rab1 (Derre & Isberg, 2004; Kagan et al, 2004; Schoebel et al, 2009). As Rab proteins, such as Rab1, can exert the function of regulating vesicular transport only when they are attached to their respective target membrane, the correct membrane localization of the Rab-activating GEFs is crucial. Thus, Legionella ensures the correct targeting and activation of Rab1 during infection to the LCV by attaching DrrA through lipid binding to the LCV.
The PtdIns(4)P-binding domain of SidM/DrrA (P4M) has been identified recently (Brombacher et al, 2009). To understand the structural basis for PtdIns(4)P recognition, we have determined the crystal structure of P4M. A subsequent quantification revealed an unusually high binding affinity for the P4M–PtdIns(4)P interaction, which has not been observed for other phosphatidylinositol phosphate (PtdInsP)-binding proteins.
Results And Discussion
Structure of DrrA340–647
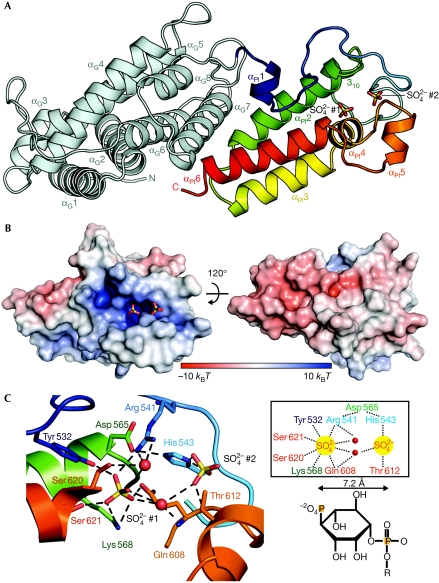
The minimal fragment of P4M is small, consisting of about 100 amino acids, and has no sequence homology to any known lipid-binding domain. As constructs containing only P4M (amino acids 544–647) seemed to reduce PtdIns(4)P binding to some degree (Brombacher et al, 2009), we determined the crystal structure of a fragment of DrrA comprising the GEF domain and P4M (amino acids 340–647) at 2.5 Å resolution (Fig 1A; for data collection, phasing and refinement statistics, see Table 1). As reported previously for the GEF domain (Schoebel et al, 2009; Suh et al, 2010), P4M has a new protein fold with no homologous structures found in the Protein Data Bank when running a search using the Dali server (Holm et al, 2008). P4M consists of approximately 50% of α-helices (six α-helices and one 310-helix) and 50% of ordered loops. The three central helices αPI2, αPI3 and αPI6 are arranged perpendicularly to α-helices αG6–αG8 of the GEF domain. At the tip of these helices, two sulphate ions from the crystallization buffer are found in a positively charged pocket (Fig 1B), surrounded mainly by the αPI1–αPI2 loop and the αPI4–αPI5 loop (Fig 1C). The sulphate ions are spaced by 7.2 Å, in accordance with the distance between the phosphate groups in PtdIns(4)P (Fig 1C, right). This mimicking property of sulphate ions for binding of the phosphate head groups of PtdInsP has been observed in another instance (p47phox-PX; Karathanassis et al, 2002). It is therefore probable that this positively charged pocket constitutes the binding site for the PtdIns(4)P head group (supplementary Fig S1 online). Furthermore, a negatively charged surface patch is observed on the opposite side of the presumed PtdIns(4)P-binding cavity, which is likely to be repelled by the negatively charged cytosolic surface of intracellular membranes (Fig 1B). This effect will presumably help to orient the protein during the LCV-binding process such that the supposed PtdIns(4)P-binding pocket faces towards intracellular PtdIns(4)P-containing membranes (Weber et al, 2006).
Figure 1.
DrrA reveals a new fold for phosphatidylinositol-4-phosphate binding. (A) Representation of the crystal structure of DrrA340–647. The GEF domain is shown in light blue, P4M in the spectrum from blue to red. Two sulphate ions (SO42− #1 and SO42− #2) found in the crystal structure indicate the presumed PtdIns(4)P-binding pocket and are drawn in stick representation. (B) Surface representations of P4M coloured by its electrostatic potential with two sulphate ions (stick representation) occupying the PtdIns(4)P-binding pocket. (C) Left: polar contacts between sulphate ions (sticks), the sulphate surrounding water molecules (red spheres) and selected residues of P4M. Right: schematic depiction of the polar contacts between DrrA and sulphate ions. The PtdIns(4)P head group is depicted schematically below to illustrate the spacing between 1- and 4-phosphates (sulphates are numbered according to Fig 1A). GEF, guanine nucleotide exchange factor; P4M, PtdIns(4)P-binding domain of SidM/DrrA; PtdIns(4)P, phosphatidylinositol 4-phosphate.
Table 1. Data collection, phasing and refinement statistics.
| DrrA340–647 | DrrA340–647 SeMet* | |
|---|---|---|
| Data collection‡ | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 74.9, 75.4, 131.0 | 74.1, 75.3, 131.0 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)§ | 20–2.5 (2.6–2.5) | 20–2.7 (2.8–2.7) |
| Rmean | 11.1 (41.2) | 15.6 (46.2) |
| I/σI | 15.8 (4.2) | 10.6 (4.0) |
| Completeness (%) | 100 (100) | 100 (100) |
| Redundancy | 8.1 (8.2) | 7.3 (7.1) |
| Refinement | ||
| Resolution (Å) | 2.5 | |
| No. of reflections | 26,298 | |
| Rwork/Rfree | 18.9/26.2 | |
| No. of atoms | ||
| Protein | 4,752 | |
| Ligand/ion | 40 | |
| Water | 174 | |
| B-factors | ||
| Protein | 35 | |
| Ligand/ion | 42 | |
| Water | 40 | |
| r.m.s.d.∥ | ||
| Bond lengths (Å) | 0.019 | |
| Bond angles (°) | 1.658 | |
| *Data collection statistics for single-wavelength anomalous dispersion data refer to unmerged Friedel pairs; | ||
| ‡data sets were collected from a single crystal; | ||
| §values in parentheses refer to the highest resolution shell; | ||
| ∥r.m.s.d. values. | ||
The interactions between the P4M and GEF domains are mainly polar (supplementary Fig S2 online), which leads to a defined relative orientation that is probably also stable in solution or when DrrA is bound at the membrane, as is corroborated by the crystal structure of a similar DrrA fragment that was published while this paper was under review (Zhu et al, 2010). This structure is nearly identical to the one described here (supplementary Fig S3A online), despite having been obtained at markedly different pH and from crystals that diffract to only 3.5 Å, whereas our model was refined at 2.5 Å. The higher resolution allowed us to identify the position of a second sulphate ion, which leads to the modelling of the presumed position and orientation of PtdIns(4)P, thereby providing a reasonable model for the mechanism of PtdIns(4)P recognition (supplementary Fig S1 online) and membrane binding (see below). There is no structural similarity between P4M and other PtdInsP-binding domains. However, the Epsin N-terminal homology domain of Epsin is also an α-helical protein, and harbours an amphiphatic α-helix (α0) close to the PtdInsP-binding pocket that is implicated in membrane binding (Ford et al, 2002). This is also seen in our structure (supplementary Fig S3B online), supporting the idea that α-helix αPI5 could be involved in membrane binding.
Interaction of DrrA340–647 with PtdIns(4)P
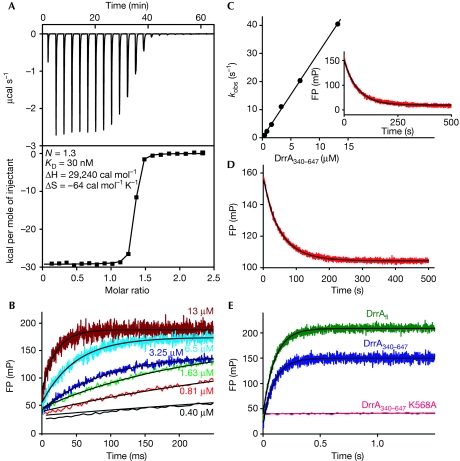
To determine the affinity between DrrA and PtdIns(4)P, we performed isothermal titration calorimetry measurements with a water-soluble analogue of PtdIns(4)P, namely, di-C4-PtdIns(4)P. The experiment revealed an unexpectedly high affinity of DrrA towards di-C4-PtdIns(4)P, with a dissociation equilibrium constant (KD) of approximately 30 nM (Fig 2A). In further experiments, it was possible to determine the association and dissociation rate constants (kon and koff, respectively) of DrrA340–647 by using a fluorescent analogue of di-C4-PtdIns(4)P labelled with boron-dipyrromethene (BODIPY; BODIPY–PtdIns(4)P), exploiting the change in fluorescence polarization on binding. Fitting the association traces to single exponentials (Fig 2B) and plotting the observed pseudo-first order rate constants against the DrrA concentration resulted in kon=3.2 × 106 M−1 s−1 (Fig 2C). In a separate experiment, the displacement of BODIPY–PtdIns(4)P from DrrA with non-fluorescent di-C4-PtdIns(4)P resulted in a slow dissociation rate constant (koff) of 0.016 s−1 (Fig 2C, inset). Together, the KD value for the DrrA–(BODIPY–PtdIns(4)P) interaction is calculated to be 5 nM.
Figure 2.
DrrA binds phosphatidylinositol 4-phosphate selectively with unexpectedly high affinity. (A) Isothermal titration calorimetry experiment of 50 μM di-C4-PtdIns(4)P titrated into 5 μM DrrA340–647. (B) Individual stopped-flow time traces of the association between 166 nM BODIPY–PtdIns(4)P and indicated concentrations of DrrA340–647 monitored by the change in fluorescence polarization. (C) Linear fit of the observed DrrA340–647–BODIPY–PtdIns(4)P association rates against DrrA340–647 concentration. Inset: BODIPY–PtdIns(4)P displacement from a DrrA340–647:BODIPY–PtdIns(4)P complex (0.4 μM) by 5 μM di-C4-PtdIns(4)P. (D) Stopped-flow competition binding experiment (red line) using 200 nM DrrA340–647, 200 nM BODIPY–PtdIns(4)P and 400 nM di-C4-PtdIns(4)P. The data were fitted to a simple competition model for the determination of KD, kon and koff of di-C4-PtdIns(4)P to DrrA340–647. (E) Association of 166 nM BODIPY–PtdIns(4)P with 6.5 μM DrrAfl, DrrA340–647 or DrrA340–647 K568A. BODIPY, boron-dipyrromethene; FP, fluorescence polarization; PtdIns(4)P, phosphatidylinositol 4-phosphate.
As it is possible that the high binding affinity between DrrA and BODIPY–PtdIns(4)P is in part due to the presence of the fluorescence group, we performed competition experiments with unlabelled di-C4-PtdIns(4)P (Fig 2D). The displacement of 200 nM BODIPY–PtdIns(4)P from a mixture with 200 nM DrrA340–647 was monitored by the addition of 400 nM di-C4-PtdIns(4)P and was fitted to a simple competition model. From the end point of the reaction, the dissociation constant for di-C4-PtdIns(4)P was determined to be KD=18.2 nM. Kinetic constants were calculated from the time dependence of the change in fluorescence polarization (kon=4.1 × 106 M−1 s−1, koff=0.079 s−1). Thus, the absence of the fluorescence reporter group resulted in a fourfold weaker di-C4-PtdIns(4)P binding compared with BODIPY–PtdIns(4)P. Nevertheless, to the best of our knowledge, the affinity of P4M-PtdIns(4)P is at least an order of magnitude higher than that of most PtdInsP–protein interactions reported so far. The strongest interaction with a PtdInsP known to date is between the PH domains of oxysterol-binding protein and PtdIns(4)P and has a KD value of 40–100 nM (Stahelin et al, 2007).
Owing to the close proximity of the P4M and GEF domains of DrrA in the crystal structure, we investigated whether Rab1b can modulate the affinity of DrrA towards PtdIns(4)P. For this purpose, we measured the time-dependent displacement of BODIPY–PtdIns(4)P from DrrA340–647 with unlabelled PtdIns(4)P in the presence of Rab1b:GDP and Rab1b:GTP (supplementary Fig S4 online). The rate of dissociation of BODIPY–PtdIns(4)P from the complex was unchanged, regardless of the presence of Rab1b:GDP, Rab1b:GTP or the nucleotide-free Rab1b:DrrA340–647 complex, indicating that Rab1b has no modulatory role on PtdIns(4)P affinity for P4M.
Using the fluorescent binding assay, we also investigated the relevance of the positively charged pocket of P4M of DrrA for PtdIns(4)P binding. As the sulphate ions could potentially mimic the position of the 4-phosphate head group of PtdIns(4)P, we mutated the sulphate-interacting residue Lys568 to alanine (Fig 1C). Melting point analysis by circular dichroism showed no effect on protein stability (supplementary Fig S5 online), but the mutation completely abolished the DrrA–(BODIPY–PtdIns(4)P) interaction (Fig 2E). This result confirms that the positively charged pocket of P4M containing sulphates is indeed the binding cavity for PtdIns(4)P.
Biological relevance of the high affinity of PtdIns(4)P
To recruit Rab1, DrrA must associate with LCV after it has been released into the cytosol of the host cell. PtdIns(4)P is found mainly in the Golgi and to a lesser extent in the plasma membrane (di Paolo & de Camilli, 2006), the latter being the primary origin of LCV. Owing to the presumably low abundance of PtdIns(4)P in the LCV immediately after Legionella phagocytosis and because of possible competition with other PtdIns(4)P-binding proteins, DrrA seems to have evolved to bind to PtdIns(4)P with exceptionally high affinity. DrrA is released from the LCV in the course of the Legionella infectious cycle, so that membrane binding is only transient (Ingmundson et al, 2007). As binding towards PtdIns(4)P is exceptionally strong, release from the membrane most probably occurs as a result of a reduction of the amount of PtdIns(4)P in the course of the maturation of LCVs. Despite the high affinity, the DrrA:PtdIns(4)P complex is moderately dynamic (koff=0.079 s−1; that is, half-life=ln2/koff=8.7 s) and would therefore allow for PtdIns(4)P modifications on a physiologically relevant time scale.
The high specificity of DrrA for PtdIns(4)P (Brombacher et al, 2009), together with its exceptionally high binding affinity, makes P4M a potentially valuable molecular tool for the specific and sensitive labelling of intracellular membranes for the presence of PtdIns(4)P. As proof of principle, it was shown that the PH domains of oxysterol-binding protein and four-phosphate adaptor protein 1 can be used to monitor the cellular activity and spatial distribution of the PtdIns(4)P-generating enzyme PtdIns(4) kinase (Balla et al, 2005). However, the tighter interaction of DrrA with PtdIns(4)P makes the protein potentially even better suited to this purpose.
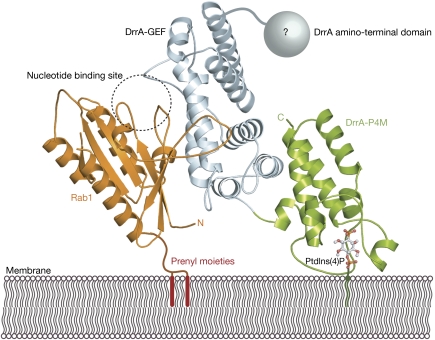
The structure of DrrA340–647, comprising the GEF domain and the P4M, allows us to propose a structural model for DrrA membrane binding and Rab1 membrane anchoring (Fig 3). DrrA presumably binds to PtdIns(4)P-containing membranes in a manner that orients the GEF domain away from the membrane surface. In this model, P4M can potentially interact with a negatively charged membrane through an arginine residue of the αPI1–αPI2 loop (Arg544) and through hydrophobic interactions of two leucines of the α-helix αPI5 (Leu 610, Leu 614) with the lipid bilayer. The GEF domain extends into the cytosol and is able to recruit Rab1 from a pool of cytosolic Rab1:GDI (GDP dissociation inhibitor) complexes. Binding of Rab1 and the concomitant nucleotide exchange from GDP to GTP are necessary for effective GDP dissociation inhibitor displacement (Schoebel et al, 2009). In our model, the binding of Rab1 to membrane-bound DrrA would orient the GTPase with the nucleotide-binding pocket facing the cytosol, thus allowing direct access for GTP to the Rab1:DrrA complex. In addition, the C-terminus of Rab1 will be positioned in close proximity to the membrane and could thus facilitate the incorporation of the C-terminal geranylgeranyl moieties of Rab1 into the lipid bilayer. Thus, the P4M of DrrA achieves three goals simultaneously: first, it binds specifically and tightly to PtdIns(4)P and hence localizes DrrA to the LCV. Second, it orients the GEF domain of DrrA in a manner that promotes undisturbed nucleotide exchange on Rab1. Third, P4M brings the prenylated C-terminus of Rab1 in close proximity to the membrane, thereby supporting the insertion of geranylgeranyl moieties and leading to stable attachment of Rab1 to the LCV for subsequent recruitment of ER-derived vesicles.
Figure 3.
Structural model of DrrA membrane binding and simultaneous Rab1 interaction. The model demonstrates a possible orientation of P4M towards a membrane and the implications for Rab1 binding to the DrrA–GEF domain. In this view, P4M orients the preceding GEF domain such that the GDP/GTP-binding pocket of Rab1 points to the cytosol. Concomitantly, the carboxyl terminus of the GTPase is positioned in close proximity to the membrane, thereby presumably facilitating the incorporation of the prenyl moieties into the LCV membrane. The position of Rab1 on DrrA340–647 was modelled by superimposing DrrA340–647 with the Rab13–174:DrrA340–533 complex structure (Schoebel et al, 2009). Green, DrrA-P4M; light blue, DrrA-GEF domain; grey sphere, DrrA amino-terminal domain; orange, Rab1; ball and sticks, PtdIns(4)P model; dashed circle, GDP/GTP bindings site of Rab1; red bars, schematic for Rab1 C-terminal prenyl moieties. GEF, guanine nucleotide exchange factor; LCV, Legionella-containing vacuole; P4M, PtdIns(4)P-binding domain of SidM/DrrA; PtdIns(4)P, phosphatidylinositol 4-phosphate.
Methods
Materials. Di-C4-PtdIns(4)P (di-C4-PtdIns(4)P, P-4004) and BODIPY tetramethylrhodamine (TMR) PtdIns(4)P (BODIPY–PtdIns(4)P, C-04M6) were obtained from Echelon.
Purification of Legionella DrrA340–647 and mutants. DrrA340–647 was subcloned into a modified pET19 vector (Novagen) that contained an N-terminal hexa-histidine tag and a tobacco etch virus (TEV) protease cleavage sequence. The site-specific mutant DrrA340–647 K568A was created by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Proteins were expressed in Escherichia coli (BL21-CodonPlus(DE3)-RIL) at 37°C after induction with 1 mM isopropyl-β-dithiogalactopyranoside. Selenomethionine-labelled DrrA340–647 was expressed using the methionine biosynthesis inhibition method (van Duyne et al, 1993). After bacterial cell lysis, the supernatant was applied to nickel–nitrilotriacetic acid (Ni–NTA) chromatography and proteins were eluted with a linear Imidazole gradient (5–500 mM) in buffer A (20 mM HEPES (pH 8.0), 50 mM NaCl and 1 mM β-mercaptoethanol). Fractions containing DrrA340–647 or the mutant protein were pooled and digested with His10-tagged TEV protease while dialysing against buffer A to remove imidazole. Uncleaved protein and TEV protease were removed by passing over the Ni–NTA column again, and the concentrated sample was subjected to size exclusion chromatography (Superdex75 16/60, GE Healthcare) in buffer containing 20 mM HEPES (pH 8.0), 50 mM NaCl and 2 mM dithiothreitol. The protein was concentrated to 20 mg/ml and flash-frozen in liquid nitrogen. Rab1b purification has been described previously (Schoebel et al, 2009).
Crystallization and structure determination of DrrA340–647. Native and selenomethionine-labelled crystals of DrrA340–647 were obtained by the hanging-drop vapour diffusion method by mixing 1 μl of protein (20 mg/ml) in gel filtration buffer with 1 μl of a reservoir solution containing 12% (w/v) polyethylene glycol 4000, 20% (v/v) glycerol, 0.16 M ammonium sulphate and 0.1 M sodium acetate at pH 4.8. Diffraction data were collected on beamline X10SA of the Swiss Light Source and processed with XDS (Kabsch, 1993). Selenium atoms were located and a model was obtained with autoSHARP (Vonrhein et al, 2007) in MIR(AS) mode. Structure refinement was completed by using phenix.refine (Adams et al, 2007) and manual rebuilding in Coot (Emsley & Cowtan, 2004) with alternating rounds of refinement in REFMAC5 (Murshudov et al, 1997). The protein crystallizes in space group P212121 with two independent copies of DrrA340–647 in the asymmetric unit. No electron density is visible for C-terminal residues 639–647. Molecular presentations were prepared with PyMOL (Delano, 2002).
Isothermal titration calorimetry of DrrA–PtdIns(4)P interaction. Di-C4-PtdIns(4)P (50 μM) in running buffer (20 mM HEPES (pH 8.0), 50 mM NaCl, 1 mM tris (2-carboxyethyl) phosphine) was titrated automatically into a 5 μM solution of DrrA340–647 dialysed in the same buffer, using an iTC200 Micro Calorimeter (MicroCal). Titrations were performed at 25°C, with an injection volume of 6 μl at time intervals of 5 min to ensure that the titration peak returned to baseline. Data were collected using iTC200 control software and analysed with Origin (Version 7.0, MicroCal). Data were corrected for heat of dilution of Di-C4-PtdIns(4)P into buffer, which was determined in a separate titration.
Kinetic analysis of PtdIns(4)P interaction with DrrA. All measurements were carried out at 25°C in buffer that contained 20 mM HEPES (pH 8.0), 50 mM NaCl and 2 mM dithiothreitol. Fast kinetic measurements were performed with a stopped-flow apparatus (Applied Photophysics). For BODIPY TMR PtdIns(4)P binding or release, time-dependent changes in fluorescence polarization were monitored with a 570nm cutoff filter in the stopped-flow machine, excited at 546 nm. The competition experiment to determine the affinity of non-fluorescent di-C4-PtdIns(4)P with DrrA (200 nM) was carried out in the presence of 400 nM di-C4-PtdIns(4)P and 200 nM BODIPY–PtdIns(4)P. Data were fitted to a simple competition model yielding the kon and koff values for di-C4-PtdIns(4)P by using the program KinTek explorer (Johnson et al, 2009).
Accession codes. Protein Data Bank: coordinates for the DrrA340–647 crystal structure have been deposited under accession code 3N6O.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
C. Herrmann and M. Wehner from the University of Bochum are acknowledged for help in isothermal titration calorimetry measurements. N. Bleimling is acknowledged for invaluable technical assistance. We thank the staff of Beamline X10SA at the Paul Scherrer Institute (Villingen, Switzerland) for access to their facilities and the X-ray communities at the Max Planck Institute of Molecular Physiology (Dortmund, Germany) and the Max Planck Institute for Medical Research (Heidelberg, Germany) for help with data collection.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T (2005) A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell 16: 1282–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H (2009) Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem 284: 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Derre I, Isberg RR (2004) Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect Immun 72: 3048–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Paolo G, de Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366 [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR (2007) Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450: 365–369 [DOI] [PubMed] [Google Scholar]
- Isberg RR, O'Connor TJ, Heidtman M (2009) The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Simpson ZB, Blom T (2009) Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem 387: 20–29 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kagan JC, Stein MP, Pypaert M, Roy CR (2004) Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med 199: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL (2002) Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J 21: 5057–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR (2007) A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318: 974–977 [DOI] [PubMed] [Google Scholar]
- Muder RR, Yu VL, Woo AH (1986) Mode of transmission of Legionella pneumophila. A critical review. Arch Intern Med 146: 1607–1612 [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A (2009) RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell 36: 1060–1072 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W (2007) Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J Biol Chem 282: 25737–25747 [DOI] [PubMed] [Google Scholar]
- Suh HY, Lee DW, Lee KH, Ku B, Choi SJ, Woo JS, Kim YG, Oh BH (2010) Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J 29: 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol 229: 105–124 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H (2006) Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu L, Zhou Y, Yao Q, Liu L, Shao F (2010) Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA. Proc Natl Acad Sci USA 107: 4699–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.