The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties
The SCA7 domain of ATXN7/Sgf73 is involved in nucleosome binding through the H2A.H2B dimer, providing an additional chromatin anchoring point for SAGA.
Keywords: SAGA, ATXN7, ubiquitin, zinc-finger, nucleosome
Abstract
SAGA (Spt–Ada–Gcn5 acetyltransferase), a coactivator complex involved in chromatin remodelling, harbours both histone acetylation and deubiquitination activities. ATXN7/Sgf73 and ATXN7L3, two subunits of the SAGA deubiquitination module, contain an SCA7 domain characterized by an atypical zinc-finger. We show that the yeast Sgf73–SCA7 domain is not required to recruit Sgf73 into SAGA. Instead, it binds to nucleosomes, a property that is conserved in the human ATXN7–SCA7 domain but is lost in the ATXN7L3 domain. The solution structures of the SCA7 domain of both ATXN7 and ATXN7L3 reveal a new, common zinc-finger motif at the heart of two distinct folds, providing a molecular basis for the observed functional differences.
Introduction
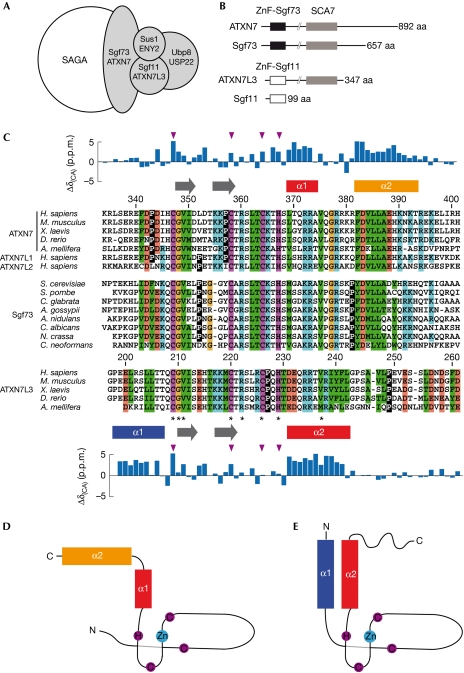
Transcription initiation is regulated by the diverse functions of multiprotein coactivator complexes that include histone modification and nucleosome remodelling activities, as well as interactions with gene-specific activators and general transcription factors (Naar et al, 2001; Martinez, 2002). SAGA (Spt–Ada–Gcn5 acetyltransferase) is one such coactivator complex that is highly conserved throughout evolution, with the histone acetyltransferase GCN5 being the catalytic subunit (Baker & Grant, 2007; Nagy & Tora, 2007). The structural organization of the SAGA complex revealed that its activities are localized in distinct functional modules (Wu et al, 2004). This complex hosts a second enzymatic activity, as it was shown to remove ubiquitin from monoubiquitinated histone H2B (Henry et al, 2003; Daniel et al, 2004). In agreement with this observation, the human ubiquitin protease USP22 (or its yeast orthologue Ubp8), together with ATXN7L3 (Sgf11), ENY2 (Sus1) and ATXN7 (Sgf73), forms a deubiquitination module (DUBm; Fig 1A; Powell et al, 2004; Rodriguez-Navarro et al, 2004; Ingvarsdottir et al, 2005; Lee et al, 2005; Zhang et al, 2008; Zhao et al, 2008). Analysis of ATXN7/Sgf73 protein sequences reveals the presence of two domains showing a strict conservation of cysteine and histidine residues (Fig 1B). The first amino-terminal domain contains a C2H2 zinc-finger (ZnF) and its deletion dissociates the DUBm from SAGA (Kohler et al, 2008). This ZnF has an important structural role in the DUBm, mediating interactions with the three other components (Kohler et al, 2010; Samara et al, 2010). The second domain, hereafter referred to as the SCA7 domain (InterPro: IPR013243), shows an atypical sequence pattern with a Cys-X9–10–Cys-X5–Cys-X2-His motif, which is characterized by a long sequence insertion between the first two zinc-coordinating residues (Fig 1C; Helmlinger et al, 2004). An SCA7 domain is also found in ATXN7L3, but not in its yeast orthologue Sgf11, suggesting that the ATXN7L3–SCA7 domain might have evolved to achieve a function distinct from that of ATXN7–SCA7 (Zhao et al, 2008). Both the structure and the function of the two distinct SCA7 domains remain unknown.
Figure 1.
Conservation of the SCA7 domain in ATXN7 and ATXN7L3 orthologues. (A) Schematic representation of the SAGA deubiquitination module. (B) Domain organization of ATXN7 homologues. The highly conserved SCA7 domain (grey box) is shared by all ATXN7 orthologues, as well as a subset of ATXN7L3 orthologues. Sequence conservation of the amino-terminal ZnF domain distinguishes ATXN7L3/Sgf11 orthologues (ZnF-Sgf11, white box) from ATXN7/Sgf73 orthologues (ZnF-Sgf73, black box). (C) Multiple sequence alignment of the SCA7 domain from selected species reveals the sequence similarities and differences that allow the SCA7 domains of ATXN7/Sgf73 orthologues to be distinguished from those of ATXN7L3 orthologues. Invariant residues are indicated by asterisks and zinc-chelating residues are coloured in magenta. Hydrophobic, positive, negative and glycine residues are coloured in green, blue, red and yellow, respectively. Prolines are shown in inverted contrast. α-Helices are indicated as rectangles and extended segments as arrows, and the deviations of Cα chemical shifts from random coil values are represented above and below the ATXN7 and ATXN7L3 sequences. The positions of zinc-coordinating residues are indicated with magenta triangles. (D,E) Topologies of ATXN7–SCA7 (D) and ATXN7L3–SCA7 domains (E). SAGA, Spt–Ada–Gcn5 acetyltransferase; ZnF, zinc-finger.
To provide structural and functional insights into the SCA7 domains, we undertook a structure–function analysis of these atypical ZnFs. Our results unveil a new zinc-binding fold, and we link the striking structural differences observed between the two SCA7 domains to their ability to bind to nucleosomes.
Results And Discussion
ATXN7 and ATXN7L3 contain distinct SCA7 domains
The SCA7 domain is found exclusively in members of the ATXN7 gene family, which includes two distinct subunits of SAGA complexes: ATXN7 and ATXN7L3 orthologues (Fig 1B). To examine the functional divergence of the SCA7 domains between ATXN7 and ATXN7L3 proteins, we performed multiple alignments of the corresponding sequences from different genomes. This analysis highlights the consensus signature for the SCA7 domain (Fig 1C, asterisks), encompassing the putative zinc-coordinating residues, but also reveals the distinct features of the two proteins (Fig 1C). Marked differences are found mostly in the carboxy-terminal of the domain, suggesting that divergent evolution of the SCA7 ZnF domain occured in order to achieve specific functions in the SAGA complex.
SCA7 domains contain a new type of ZnF
On the basis of both sequence conservation analysis and biochemical screening, the SCA7 domains of ATXN7 and ATXN7L3, encompassing residues Lys 330–His 401 of ATXN7 (ATXN7–SCA7) and Gly 197–Gly 276 of ATXN7L3 (ATXN7L3–SCA7), were produced and analysed using NMR spectroscopy. Notably, analysis of Cα chemical shifts revealed the presence of disordered regions in both SCA7 domains, albeit not in the same region (Fig 1C). Whereas the first and last 10 residues of ATXN7–SCA7 are not folded, the N-terminal region of ATXN7L3–SCA7 is well structured and the last 30 residues of this domain are not folded.
The structures of the two proteins were determined by using nuclear Overhauser enhancement spectroscopy (NOESY) spectra recorded at 950 MHz, allowing efficient automated analysis by ATNOS/CANDID software (Table 1 and supplementary Table S1 online). Both sets of calculations resulted in a well-defined fold as shown by the superimposition of the lowest energy models of ATXN7–SCA7 and ATXN7L3–SCA7 (supplementary Fig S1 online). The larger number of NOEs assigned for ATXN7–SCA7 reflects the greater number of folded residues that form an extended core, as compared with ATXN7L3–SCA7.
Table 1. NMR and refinement statistics for protein structures.
| ATXN7 | ATXN7L3 | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 1589 | 970 |
| Intra-residue | 387 | 289 |
| Inter-residue | ||
| Sequential (∣i–j∣=1) | 474 | 383 |
| Medium-range (∣i–j∣<4) | 376 | 167 |
| Long-range (∣i–j∣>5) | 352 | 131 |
| Protein–zinc restraints | 10 | 10 |
| Hydrogen bonds | 0 | 0 |
| Total dihedral angle restraints | ||
| φ | 29 | 23 |
| ψ | 27 | 23 |
| Structure statistics | ||
| Violations (mean±s.d.) | ||
| Distance constraints (Å) | 0.0664±0.0034 | 0.0725±0.0040 |
| Dihedral angle constraints (°) | 0.1706±0.2998 | 0.1293±0.1373 |
| Max. dihedral angle violation (°) | 4.935 | 2.441 |
| Max. distance constraint violation (Å) | 0.488 | 0.356 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.0177±0.0009 | 0.0147±0.0007 |
| Bond angles (°) | 2.0232±0.0400 | 1.8581±0.0471 |
| Impropers (°) | 1.9413±0.0777 | 1.8466±0.0865 |
| Average pairwise r.m.s. deviation* (Å) | ||
| Heavy | 1.17±0.16 | 1.44±0.17 |
| Backbone | 0.60±0.19 | 0.54±0.12 |
| NOE, nuclear Overhauser enhancement. | ||
| *Pairwise r.m.s. deviations were calculated among 20 refined structures by using residues 342–394 for ATXN7–SCA7 and residues 199–211 and 220–244 for ATXN7L3–SCA7. | ||
In both ATXN7–SCA7 and ATXN7L3–SCA7, the large sequence insertion between the first and second zinc-coordinating cysteines corresponds to a protruding extended hairpin structure (Fig 2A,B). The core of the zinc-binding site shows a conserved structure formed by two short adjacent loops (Fig 2A,B, L1 and L2) located at the bottom of the hairpin. This particular three-dimensional organization of zinc-coordinating residues has not been observed previously and represents a new fold, which seems to be specific to SCA7 domains as no homologous fold was found in the Protein Data Bank. The zinc-binding scaffold contains two conserved hydrophobic residues that hook onto the remaining parts of the domain. In particular, the valine residue of the SCA7-specific Cys-Gly-Val motif (Val 349 in ATXN7, Val 211 in ATXN7L3) interacts with the α-helix located immediately downstream from the zinc-binding site and contributes to its orientation (Fig 2A,B). A second hook is provided by the Leu 362 located at the tip of the first loop of the zinc-coordination cage that anchors the second α-helix in the ATXN7–SCA7 domain (Fig 2A).
Figure 2.
Solution structures of the SCA7 domains of human ATXN7 and ATXN7L3. (A,B) Ribbon representation of (A) ATXN7–SCA7 and (B) ATXN7L3–SCA7 domains showing the hydrophobic contacts that hook the two α-helices onto the zinc-finger. (C,D) Conserved interactions define the relative orientations of the two α-helices within (C) ATXN7–SCA7 and (D) ATXN7L3–SCA7 domains. The side chains of conserved hydrophobic, positive or negative residues are shown by using green, blue or red sticks, respectively. The figures were prepared using PyMOL (DeLano Scientific LLC).
SCA7 domains differ in their helical structures
Although the SCA7 domains of both ATXN7 and ATXN7L3 contain two α-helices, these are not located at similar positions in the sequences. In ATXN7–SCA7, the two α-helices (encompassing residues Leu 369–Ala 374 and Phe 382–Thr 393) are located downstream from the zinc-binding site and are separated by a loop containing a large number of positively charged residues (Fig 1C,D). In ATXN7L3–SCA7, the two α-helices (encompassing residues Pro 198–Thr 207 and Asp 231–Leu 242) lie to either side of the zinc-binding site (Fig 1C,E), leading to a different packing of the two helices (Fig 2C,D).
In ATXN7–SCA7, the two helices have an almost perpendicular orientation, the α2 helix being anchored to the zinc-binding site by the hydrophobic contact between Leu 362 and Leu 385 (Fig 2A). The closed conformation of the elbow formed by the two helices is due to the tight packing of several conserved residues, namely Phe 382 and Leu 386 in helix α2 and Arg 372 in helix α1 (Fig 2C). In addition, a conserved salt bridge is observed between Arg 373 in helix α1 and Asp 383 in helix α2. Notably, the side chain of Arg 378 is buried inside the elbow and might contribute to the closed conformation through a cation–pi interaction with the side chain of Phe 382.
In ATXN7L3–SCA7, the helices α1 and α2 adopt an anti-parallel orientation defined by hydrophobic interactions involving residues Leu 201 and Leu 205 in helix α1, and Val 237, Phe 241 and Leu 242 in helix α2 (Fig 2D). Interestingly, sequence analysis of the Apis mellifera ATXN7L3 orthologue suggests that the hydrophobic interaction between Leu 201 and Leu 242 is replaced by an electrostatic interaction between a lysine and a glutamate residue (Fig 1C).
Analysis of the solution structures of ATXN7–SCA7 and ATXN7L3–SCA7 shows that sequence divergence between the two SCA7 domains translates into a major structural rearrangement of helical structures around a conserved zinc-coordination scaffold.
The ATXN7–SCA7 domain binds to nucleosomes
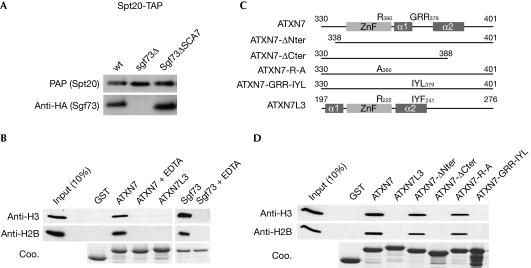
We then asked whether these structural differences account for potential distinct functions of the SCA7 domains of ATXN7 and ATXN7L3. Although the function of the ATXN7L3–SCA7 domain has never been addressed, it has been suggested that SCA7 domains from both Sgf73 and ATXN7 are involved in the incorporation of these proteins into SAGA (Helmlinger et al, 2004; Kohler et al, 2008). To confirm this observation, we analysed a yeast strain in which the SCA7 domain of Sgf73 was deleted (Sgf73ΔSCA7). Tandem affinity purification (TAP) of TAP-tagged Spt20 from this mutant strain revealed normal incorporation of Sgf73ΔSCA7 (Fig 3A; supplementary Fig S2 online), indicating that Sgf73–SCA7 is dispensable for Sgf73-mediated incorporation of the DUBm into SAGA.
Figure 3.
The SCA7 domains of human ATXN7 and yeast Sgf73 bind to nucleosomes. (A) Tandem affinity purifications (TAP) of SAGA using TAP-tagged Spt20 from the indicated strains were analysed by western blot using PAP or HA antibody. (B) SCA7 domains from ATXN7, ATXN7L3 or Sgf73 fused to GST or GST alone were immobilized on glutathione-Sepharose beads and incubated with mononucleosomes. The retained material was analysed by Coomassie blue staining (Coo.) and western blotting using H2B or H3 antibodies, as indicated. All interactions were lost when the fusion proteins were pre-incubated with EDTA solution. (C) Schematic representation of ATXN7–SCA7 mutant domains used in GST-pulldown experiments. (D) Mononucleosomes retained by immobilized SCA7 domains were analysed by western blotting using H3 and H2B antibodies. GST, glutathione S-transferase; HA, haemagglutinin; PAP, peroxidase anti-peroxidase; SAGA, Spt–Ada–Gcn5 acetyltransferase; ZnF, zinc-finger.
We then asked whether the ATXN7/Sgf73 SCA7 domain helps recruit nucleosomes to the DUBm of SAGA, as described for the SAGA histone acetyltransferase module that contains several chromatin-binding domains (Lee & Workman, 2007). In glutathione S-transferase (GST)-pulldown assays, nucleosomes were retained specifically by the GST–ATXN7–SCA7 fusion protein, but not by GST alone, nor by the fusion protein pretreated with EDTA solution (Fig 3B). Under the same experimental conditions, we observed the nucleosome-binding properties of the SCA7 domain of Sgf73, whereas the ATXN7L3–SCA7 domain did not interact with nucleosomes (Fig 3B). Evidence of the nucleosome-binding property of the ATXN7–SCA7 domain was also obtained by NMR, as the addition of an increasing amount of mononucleosomes to a solution of 15N-labelled ATXN7–SCA7 led to the progressive loss of free ATXN7–SCA7 resonances (supplementary Fig S3 online). The observation of a subset of correlation peaks, the intensity of which increases on addition of nucleosomes, suggests slow exchange between the bound and free states of ATXN7–SCA7. By assuming a diffusion-limited on-rate, an upper limit of 10 μM was obtained for the Kd, suggesting a fairly strong interaction (supplementary Fig S3 online).
Role of the C-terminal helix in nucleosome binding
The different organizations of the two helices found in the SCA7 domains of ATXN7 and ATXN7L3 suggested that the nucleosome-binding properties depend on sequence elements located outside the zinc-binding site. To confirm this, we generated and expressed mutant forms of the GST–ATXN7–SCA7 fusion protein, in which either the N-terminal or the C-terminal residues of the domain were deleted (ATXN7-ΔNter and ATXN7-ΔCter, respectively; Fig 3C).
GST-pulldown experiments revealed that the N-terminal deletion mutant interacted with nucleosomes as efficiently as the full-length domain. By contrast, deletion of the C-terminal residues of the domain completely abolished the interaction (Fig 3D). Superposition of the 1H–15N heteronuclear single quantum coherence (HSQC) NMR spectrum of ATXN7-ΔCter on that of ATXN7–SCA7 revealed that the mutation disrupts α2 helix but leaves the rest of the structure unaffected, thus emphasizing the role of this helix in nucleosome binding (supplementary Fig S4 online). This result was reinforced by further mutagenesis experiments in which the Gly-Arg-Arg motif, found specifically in ATXN7 homologues (positions 377–379) in the elbow between the two helices (Fig 1C), was mutated to Ile-Tyr-Leu, reminiscent of the Ile-Tyr-Phe motif found in the ATXN7L3 domain. This mutation abolished nucleosome binding, whereas the mutation of the strictly conserved Arg 360—located in the ZnF fold and exposed to solvent—to an alanine did not affect nucleosome binding, again suggesting that the core SCA7 ZnF is not involved directly in the interaction (Fig 3D). Together, these observations suggest that both the orientation and composition of the C-terminal helix in the SCA7 domain of ATXN7, which are strikingly different from those of ATXN7L3, have a role in nucleosome binding.
Regulation of SAGA deubiquitination activity
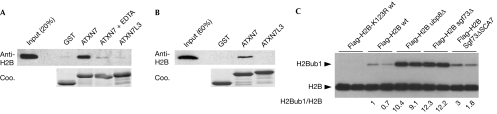
We further demonstrated that the ATXN7–SCA7 domain interacted with H2A–H2B dimers (Fig 4A) and with octamers reconstituted from histones of which the N-terminal tails had been deleted (Fig 4B), but not with H3-H4 tetramers (supplementary Fig S5 online). Our in vitro binding experiments indicate that the ATXN7–SCA7 domain binds to the core or the C-terminal ends of the histone H2A and H2B dimer, a region located on the lateral face of the nucleosome that contains the ubiquitinated Lys 120 of H2B (Luger et al, 1997; Zhang, 2003). The ATXN7–SCA7 domain could therefore facilitate the recruitment of the SAGA DUBm to its substrate. To test this hypothesis, we analysed whether the deletion of the Sgf73–SCA7 domain affected the deubiquitination activity of SAGA. As reported previously, we observed a strong accumulation of monoubiquitinated H2B (H2Bub1) in ubp8Δ and sgf73Δ strains in which the SAGA deubiquitination activity is completely lost (Kohler et al, 2008; Fig 4C). We further analysed a mutant form of Sgf73 lacking the SCA7 domain (Sgf73ΔSCA7) but containing the N-terminal region that interacts with other subunits of the DUBm. In the Sgf73ΔSCA7 strain, the mutant form of Sgf73 remained associated with SAGA (Fig 2A) and the global levels of H2Bub1 were reproducibly increased by 2.5-fold as compared with the wild-type strain (Fig 4C). Together, our results suggest that the nucleosome-binding properties of the ATXN7/Sgf73–SCA7 domain provide a further regulatory mechanism for USP22/Ubp8. Indeed, the first ZnF of Sgf73, together with other subunits of the DUBm, mediates the allosteric activation of Ubp8 (Bonnet et al, 2008; Kohler et al, 2008, 2010). In addition, we propose that the interaction of Sgf73–SCA7 with nucleosomes has a role in fine-tuning the deubiquitination activity of SAGA through optimal positioning of the DUBm relative to its substrate.
Figure 4.
The yeast Sgf73–SCA7 domain regulates H2B deubiquitination. (A,B) GST-pulldown experiments using H2A–H2B dimers or tail-less histone octamers were analysed by Coomassie blue staining (Coo.) or western blotting using H2B antibodies. (C) Extracts of the indicated yeast strains carrying a Flag–HTB1 plasmid were separated on a 15% SDS–PAGE gel and subjected to western blot analysis using Flag antibodies. GST, glutathione S-transferase; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
The structures of the SCA7 domains of ATXN7 and ATXN7L3 provide striking examples of the diversity and plasticity of ZnFs as protein interaction mediators. In contrast to previous observations, the nucleosome-binding ability that distinguishes the SCA7 domains of ATXN7 and ATXN7L3 is related to a significant change in the position of the helical structural elements around an otherwise conserved zinc-binding core. Indeed, whereas a unique prototypal structure can usually be associated with a given ZnF sequence signature, we found two distinct folds for the SCA7 sequence signature, providing the first example of a class of ZnF domains for which distinct interaction properties are associated with different folds.
Methods
Plasmids. Plasmids for the expression of GST–SCA7 fusion proteins (ATXN7: 330–401, ATXN7-ΔNter: 338–401 and ATXN7-ΔCter: 330–388) were described previously (Helmlinger et al, 2004). The DNA coding for SCA7 domains of Sgf73 (211–283) and of ATXN7L3 (197–276) were amplified by PCR and inserted into pGEX-4T1 (GE Healthcare). The sgf73 genomic locus (ChrVII: 377258, 379763) was amplified by PCR from genomic DNA of a BY4742 strain and cloned into a pRS315 vector. This plasmid was modified to introduce the sequence coding for 3-haemagglutinin and a Myc Tag upstream from the Sgf73 stop codon. Mutants were generated by using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene).
Expression and purification of SCA7 domains. Expression of all proteins was achieved by using Escherichia coli BL21 [DE3] cells (Novagen) as described earlier (Nomine et al, 2005). The final purification step involved size exclusion chromatography using a Superdex G75 (HiLoad 16/60) gel filtration column equilibrated with phosphate buffer (50 mM phosphate, pH 7.0, 200 mM NaCl, 2 mM dithiothreitol) before concentration.
NMR spectroscopy. NMR samples contained 0.2 mM of 13C,15N-labelled or unlabelled proteins. Most NMR experiments were recorded at 298 K on a Bruker DRX600 spectrometer. For structure calculations, homonuclear 2D NOESY spectra were recorded with a mixing time of 150 ms on a Bruker 950 MHz spectrometer. The protonation state of the zinc-coordinating His367 of ATXN7 was set to Nɛ2, based on the frequencies of the 15N nuclei of the imidazole ring, which were measured using a long-range 1H–15N HSQC spectrum.
Structure calculation. 950 MHz 2D NOESY and 600 MHz 3D 15N- and 13C-edited NOESY spectra, together with backbone Φ and Ψ angles derived from resonance assignments using the TALOS program (Cornilescu et al, 1999), were used as input data to the semi-automatic ATNOS/CANDID procedure (Herrmann et al, 2002). The final set of structures was refined in explicit water using the RECOORD protocol (Nederveen et al, 2005). The final ensemble of the 20 lowest energy structures contained no distance violations greater than 5 Å, nor dihedral angle violations greater than 5°. Three-dimensional structures of ATXN7–SCA7 and ATXN7L3–SCA7 have been deposited in the Protein Data Bank under accession numbers 2kkr and 2kkt, respectively.
Generation of yeast strains. The deletion of sgf73 gene and the genomic integration of a TAP-Tag in the C-terminal of Spt20 were performed using standard methods involving the transformation of yeast with PCR fragments for homologous recombination. The constructs inserted in pRS315 and pRS413 were transformed, selected and grown on complete synthetic medium minus leucine and minus histidine, respectively. All strains were generated in a BY4742 background and are listed in supplementary Table S2 online.
Analysis of the level of ubiquitinated H2B. Two independent clones of each tested strain were grown in complete synthetic medium minus leucine and histidine to the midlog phase. A volume of 1 ml of each culture was resuspended in 100 μl of 200 mM NaOH for 5 min at room temperature (20°C). Pellets of cells were then boiled for 5 min at 100°C in 100 μl Laemmli buffer. In all, 10% of the whole-cell extract was analysed by western blot with the Flag antibody (M2, Sigma) to detect Flag-H2B. Quantification of the H2Bub1/H2B ratio was done by using ImageJ software.
GST-pulldown assay and tandem affinity purifications. GST-pulldown assay and tandem affinity purifications were performed according to standard procedures, with minor modifications (see the supplementary information online).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank J. Koffler, G. Lang, E. Guittet, C. Ling, M. Stierle, M. Vitorino, X. Gao and M. Oulad-Abdelghani for their contribution to this work. Research in the authors' laboratories is supported by grants from the European Transcriptome, Regulome and Cellular Commitment Consortium, Institut National du Cancer, Agence Nationale de la Recherche, Structural Proteomics in Europe (SPINE-2), Ligue contre le cancer and NWO-Horizon. Access to high-field NMR spectrometers was granted by the CNRS through the Très Grands Equipements Résonance Magnétique Nucléaire programme.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baker SP, Grant PA (2007) The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26: 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Romier C, Tora L, Devys D (2008) Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem Sci 33: 369–375 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR III, Grant PA (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871 [DOI] [PubMed] [Google Scholar]
- Helmlinger D et al. (2004) Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet 13: 1257–1265 [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24: 171–189 [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL (2005) H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol 25: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E (2008) Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10: 707–715 [DOI] [PubMed] [Google Scholar]
- Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N (2010) Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL (2007) Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 8: 284–295 [DOI] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL (2005) The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol 25: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Martinez E (2002) Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol 50: 925–947 [DOI] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70: 475–501 [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26: 5341–5357 [DOI] [PubMed] [Google Scholar]
- Nederveen AJ et al. (2005) RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59: 662–672 [DOI] [PubMed] [Google Scholar]
- Nomine Y, Charbonnier S, Miguet L, Potier N, Van Dorsselaer A, Atkinson RA, Trave G, Kieffer B (2005) 1H and 15N resonance assignment, secondary structure and dynamic behaviour of the C-terminal domain of human papillomavirus oncoprotein E6. J Biomol NMR 31: 129–141 [DOI] [PubMed] [Google Scholar]
- Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ (2004) Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol 24: 7249–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C (2010) Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P (2004) Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15: 199–208 [DOI] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB (2008) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y (2003) Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev 17: 2733–2740 [DOI] [PubMed] [Google Scholar]
- Zhao Y et al. (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29: 92–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.