Abstract
Polarized epithelial cells contain apical and basolateral surfaces with distinct protein compositions. To establish and maintain this asymmetry, newly made plasma membrane proteins are sorted in the trans-Golgi network for delivery to apical or basolateral surfaces. Signals for basolateral sorting are generally located in the cytoplasmic domain of the protein, while signals for apical sorting can be in any part of the protein and can depend on N-linked glycosylation of the protein. Signals for constitutive transcytosis to the apical surface have not been reported. Here we utilized the polymeric immunoglobulin receptor (pIgR), which is biosynthetically delivered to the basolateral surface. There the pIgR can bind a ligand and, with or without bound ligand, the pIgR can then be transcytosed to the apical surface. We found that the glycosylation of the pIgR did not affect the biosynthetic transport of the pIgR. However, glycosylation had an effect on pIgR apical transcytosis. Importantly, analysis of the cytoplasmic tail of the pIgR suggested that a short peptide segment was sufficient to transcytose the pIgR or a neutral reporter from the basolateral to the apical surface. This apical transcytosis sorting signal was not involved in polarized biosynthetic traffic of the pIgR.
Keywords: Epithelial polarity, apical sorting, transcytosis, glycosylation, polymeric immunoglobulin receptor
Introduction
The plasma membrane of polarized epithelial cells is divided into apical and basolateral domains, which have very different protein compositions. Proteins reach these surfaces by two classes of pathways (1). Newly made proteins can be sorted in biosynthetic pathways into carriers that deliver them to the apical or basolateral surface. Once reaching a surface, proteins can be endocytosed and then sorted in endocytic pathways for recycling to the original surface, degradation or transcytosis to the opposite surface. A typical long-lived plasma membrane protein may be endocytosed many times during its lifetime and so the accuracy of its post-endocytic sorting to the recycling or transcytotic pathway is crucial to the maintenance of the distinct compositions of the apical and basolateral surfaces.
Sorting signals within the plasma membrane protein determine its apical or basolateral destination as the protein travels this pathway. Such sorting signals have been primarily studied in the biosynthetic pathway. For basolateral sorting, most signals have been found to reside in relatively short amino acid sequences in the cytoplasmic portion of the protein (2). This was first shown in the case of the polymeric immunoglobulin receptor (pIgR), where a 14 residue portion of its cytoplasmic domain could be transplanted to a heterologous reporter, redirecting its biosynthetic delivery to the basolateral surface (3). Many other cytoplasmically localized, basolateral sorting signals have been identified, some of which (but notably not the pIgR) interact with the AP1b clathrin adaptor complex, which is responsible for directing these proteins to the basolateral surface (4, 5). In some cases, these basolateral sorting signals also operate in the endocytic pathway to recycle proteins back to the basolateral surface (2).
Signals for biosynthetic apical sorting are more heterogeneous and less well understood (6). Some apical sorting signals are in the cytoplasmic domain. This has been best studied for rhodopsin (7), whose cytoplasmic signal has been demonstrated to redirect biosynthetic delivery of a heterologous reporter to the apical surface, as shown by metabolic pulse chase analysis. In other cases, apical sorting depends on a membrane-spanning polypeptide segment of a transmembrane protein (8); this segment may cause association of the protein with specific lipid microdmains or “rafts”, which may promote apical sorting (9). Many apical proteins are not transmembrane, but are instead anchored to the outer leaflet of the membrane by a glycosyl phosphatidyl inositol (GPI) lipid modification, which may also lead to raft association. This modification, together with clustering of the protein, may lead to apical sorting (10-13). Finally, both N-linked and O-linked carbohydrates coupled to the extracellular portion of apical plasma membrane proteins have been proposed to act as apical sorting signals (14-17).
Signals for post-endocytic sorting to the apical surface, i.e. for transcytosis from the basolateral to the apical surface, have been less investigated; most of the research has been carried out in an experimental system consisting of the rabbit pIgR expressed in Madin-Darby canine kidney (MDCK) cells (18). The pIgR contains a C-terminal cytoplasmic domain of 103 amino acids, numbered 653-755 (numbering based on rabbit pIgR). Phosphorylation of Ser664, which is part of the 14-residue basolateral sorting signal mentioned above, promotes transcytosis. This basolateral sorting signal works in both the biosynthetic and endocytic pathways and phosphorylation of Ser664 probably acts by reducing the activity of the basolateral sorting signal in the endocytic pathway, thereby allowing transcytosis to occur. Ordinarily in the pIgR-MDCK system, much of the pIgR endocytosed from the basolateral surface is recycled to that surface and transcytosis is relatively inefficient. This relatively inefficient transcytosis is considered “constitutive.” Other than phosphorylation of Ser664, little is known about constitutive transcytosis. To our knowledge, no specific sorting signals for constitutive transcytosis of the pIgR or other molecules have been identified. It is not even clear that such signals exist.
The pIgR is capable of binding an extracellular ligand, polymeric IgA (pIgA). Binding of pIgA causes “ligand-stimulated” transcytosis, so that the complex of pIgR and pIgA is more efficiently transcytosed. Binding of pIgA causes the pIgR to initiate a signaling pathway involving the non-receptor tyrosine kinase p62Yes, the small GTPase rab3, activation of a phospholipase Cγ, activation of protein kinase C and elevation of intracellular free calcium, all of which promote transcytosis (19-24). This ligand-stimulated transcytosis occurs not only in the rabbit pIgR-MDCK system, but also in vivo in rodent liver, which transports large amounts of pIgA from blood to bile (25, 26).
Here we report the identification of a short polypeptide sequence in the cytoplasmic domain of the pIgR that promotes constitutive transcytosis both of the pIgR and of a heterologous reporter protein.
Results
Deglycosylation of pIgA does not affect its endocytosis, recycling or transcytosis
As glycosylation has been shown to function as an apical signal in the biosynthetic pathway, we investigated if glycosylation played a role in basolateral to apical transcytosis of pIgR. The principal function of pIgR is to bind its ligand, pIgA, and carry the pIgA from the basolateral surface to the apical surface. There the extracellular domain of pIgR is proteolytically cleaved off from its transmembrane anchor and released together with pIgA into secretions. This released fragment of pIgR is called secretory component (SC). Both the pIgR and its pIgA ligand are heavily glycosylated and the sugar chains from pIgR or pIgA could promote apical sorting and transcytosis. We focused first on the glycosylation of the pIgA. Human IgA is produced in two isotypes, IgA1 and IgA2. IgA2 has two allotypic variants, IgA2m(1) and IgA2m(2). One major difference between the two isotypes is the presence of a 13 amino-acid hinge region in IgA1. This region contains five O-linked sites absent from the IgA2. IgA1 has two N-glycosylation sites, while IgA2 contains an additional two (IgA2m(1)) or three (IgA2m(2)) conserved N-glycans. The oligosaccharide structures of IgA1 and IgA2 display a high degree of heterogeneity not only in the number of carbohydrate chains, but also in their composition. Finally, the IgA J chain contains one conserved N-glycan (27, 28).
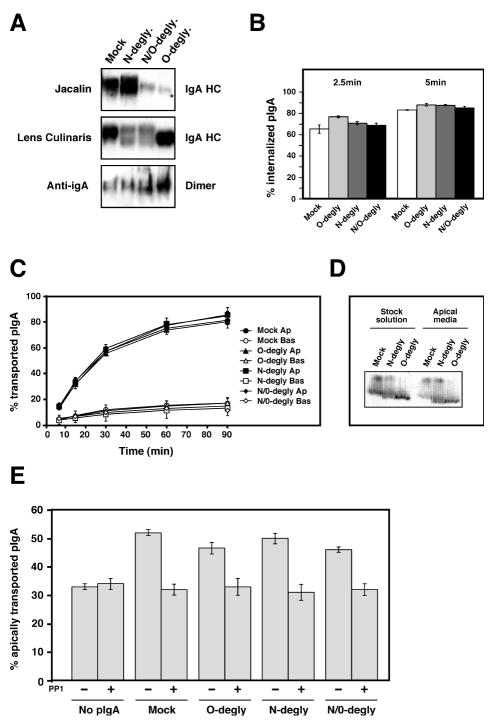
We first tested the role of pIgA glycosylation in pIgR trafficking by use of differential enzymatic deglycosylation of the N-and/or O-linked sugars as explained in the Materials and Methods section. The challenge was to find conditions whereby the pIgA would be efficiently deglycosylated while under non-denaturing conditions, which would preserve its structural and biological properties. Many different conditions were tested and the efficiency of the deglycosylation was assessed by western-blot using lectins specific for O- or N-linked sugars, Jacalin and Lens culinaris lectins respectively. Figure 1 shows the results of Nor/and O-deglycosylation performed under the optimal conditions. The top panel of Fig. 1A shows the binding of Jacalin to the N-, O- and N/O-deglycosylated pIgA compared to the control mock-deglycosylated pIgA (gels in the upper and middle panels were run under reducing conditions). As expected, the N-deglycosylated pIgA still bound Jacalin. The appearance of the two bands in the N-deglycosylated sample was due to the N-deglycosylation (see right panel) but note that both bands showed a strong binding to the O-linked sugar specific lectin, Jacalin. In contrast, the O- and N/O-deglycosylated pIgA showed a weak signal. Conversely, the binding of Lens culinaris was reduced in the N- and N/O-deglycosylated pIgA but intact in the O-deglycosylated pIgA (middle panel). The efficiency of deglycosylation is strikingly high considering that the pIgA could not be denatured to facilitate deglycosylation. The efficiency of deglycosylation was consistently over 75% for N-linked glycans and up to 95% for O-linked glycans. We next investigated whether the enzymatic deglycosylation procedure altered the structural characteristics of the pIgA.
Figure 1.
Biochemical and functional characterization of the enzymatically deglycosylated human purified pIgA.
A) pIgA was mock treated, N-deglycosylated (N-degly.), O-deglycosylated (O-degly.) or N- and O-deglycosylated (N/O-degly.) using specific glycosydases as described in the Material and Methods section. 10μg of each pIgA solution was resolved under reducing conditions by SDS-PAGE and transferred onto a nitrocellulose membrane. The glycosylation status of the IgA heavy chain (IgA HC) was analyzed by probing the membrane with the O-glycan specific lectin Jacalin (top panel) or the N-glycan specific lectin Lens Culinaris (middle panel) coupled to HRP. The oligomerization of the pIgA was analyzed by SDS-PAGE and anti-IgA immunoblot under non-reducing conditions to reveal the dimeric oligomers bottom panel). Small amounts of larger oligomers are also visible near the top of the gel. (The results shown are representative of three independent preparations of deglycosylated pIgA.
B) Each pIgA preparation was iodinated and tested for their pIgR-mediated endocytosis from the basolateral surface of polarized MDCK cells. The experiment was performed three times in triplicate, and data from all experiments were combined. Values are mean ± SD.
C) Basolateral recycling and apical transcytosis of iodinated wild-type and deglycosylated pIgA endocytosed from the basolateral surface. The experiment was performed five times in triplicate and one representative experiment is shown.
D) The radioactive pIgA collected in the apical media at the end of the transcytosis assay presented in (C) were analyzed after TCA precipitation by SDS-PAGE under reducing conditions and revealed by fluorography. The arrowhead points to glycosylated IgA heavy chain, while the bracket indicates the position of the deglycosylated heavy chain.
E) The ability of the deglycosylated pIgA to stimulate pIgR transcytosis was assessed in the absence or presence of the protein tyrosine kinase inhibitor PP1. The experiment was performed three times in triplicate, and data from all experiments were combined. Values are mean ± SD. Statistical analysis was done by ANOVA followed by Tukey’s multiple comparison test to determine the statistical significance of the differences between each sample group. Statistically significant differences (p<0.01) were found between the sample “No pIgA” versus each pIgA preparations in the absence of PP1. For each pIgA preparations, the differences were also significant (p<0.01) when comparing the samples “without PP1” versus “with PP1”. In all other cases no statistically significant difference was found.
It is well known that only polymeric IgA, and not the monomeric, binds to pIgR (29). It was therefore important to control that the deglycosylation did not affect the polymerization status of the different preparations of pIgA. The pIgA preparations were run under non-reducing conditions on a 5% SDS-PAGE and revealed by immunoblot with an anti-IgA antibody to assess the degree of polymerization compared to the non-deglycosylated pIgA. The same proportion of trimeric and dimeric forms were observed in each samples showing that deglycosylation did not affect polymerization of the pIgA (Fig. 1A, bottom panel). Finally, we directly tested the capability of each pIgA preparation to bind to the pIgR. In agreement with previous studies that had shown that recombinant IgA1 lacking the N-linked glycans was still able to bind pIgR or that IgA2, which does not have O-linked sugars, binds as well to pIgR (27, 30), we found that N- and/or O-deglycosylation did not affect the binding capabilities of our different IgA preparations (data not shown).
To follow their intracellular transport, the different pIgA preparations were radio-iodinated. Endocytosis of N-, O- and N/O-deglycosylated pIgA was compared to the control of mock-deglycosylated pIgA. We first tested the ability of the different pIgA preparations to be endocytosed by the pIgR. Radio-iodinated pIgAs were bound to the basolateral surface for 1 hr at 4°C and then unbound material removed by extensive washing at the same temperature. The cells were then warmed up to 37°C for 2.5 or 5 minutes to allow internalization to occur, cooled back down to 4°C, and pIgA remaining on the surface was removed by protease treatment at 4°C. The amount of radio-iodinated pIgA remaining associated with the cells was taken as the amount of internalized IgA. As shown in Fig. 1B, both the initial rate (2.5 min values) and the plateau (5 min values) of endocytosis were unchanged for each pIgA preparation.
Next, the ability of a cohort of pIgA that had been pre-internalized from the basolateral surface to be transcytosed to the apical surface or recycled to the basolateral surface was measured. Radio-iodinated pIgA was pre-internalized from the basolateral surface for 10 min at 37°C, which causes accumulation of the pIgA in basolateral early endosomes. After washing to remove non-bound pIgA, the cells were incubated at 37°C and the apical and basolateral media were collected and replaced with fresh warm medium at various intervals continuing for 90 min total. This assay essentially follows the post-endocytic fate of the internalized pIgA. Figure 1C plots the cumulative radio-iodinated pIgA released into the apical medium (i.e. transcytosed pIgA) or basolateral medium (i.e. recycled pIgA). Transcytosis and recycling curves for the various pIgA preparations were almost super imposable.
An additional experiment was performed to control for the possibility that in each preparation of deglycosylated pIgA the non-or partially deglycosylated fraction might be preferentially transcytosed, thus masking a possible defect due to glycosylation. In this control the total radio-iodinated pIgA transported into the apical medium over the entire 90 min chase period was immunoprecipitated and run on a SDS-PAGE in parallel with the equivalent quantity of the corresponding starting solution of radio-iodinated pIgA. As shown in Fig. 1D the proportion of deglycosylated pIgA transcytosed into the apical medium and in the starting solution is equivalent for both N- and O-deglycosylated pIgA indicating that the small proportion of non-deglycosylated pIgA was not preferentially transcytosed compared to the deglycosylated pIgA.
Finally, we analyzed the ability of the different pIgA preparations to stimulate transcytosis of the pIgR, i.e. to cause ligand-stimulated transcytosis as described in the Introduction. As shown in Fig. 1C, independent of the glycosylation state of the pIgA, it was still capable of stimulating pIgR transcytosis as efficiently as the control pIgA. In addition, this stimulation was still inhibited by the protein tyrosine kinase PP1 as previously reported (23).
Mutation of the major N-glycosylation site of a recombinant human pIgA has no effect on pIgR intracellular transport
As an alternative approach to reduce N-glycosylation of pIgA, we used a point mutant of a recombinant human pIgA where one of the two glycosylated asparagines was replaced by a glutamine. IgA1 is glycosylated on N263 and N459 (28, 31). N263 was mutated to glutamine (mutant designated “NQ”). We could not mutate N459, as this is involved in interaction with J chain and polymerization of the IgA; this interaction makes N459 less likely to be recognized by the sorting machinery. We found that NQ was indistinguishable from wild-type pIgR in its ability to bind to pIgR, and to be endocytosed, transcytosed to the apical surface and stimulate transcytosis of the pIgR (Supplementary Figure 1).
Taken together these data demonstrate that the glycosylation of the pIgA, although important for many of its biological aspects, does not play a role in pIgR-mediated transcytosis. Moreover, the glycosylation of pIgA cannot account even partially for the stimulation of pIgR transcytosis by pIgA.
Mutation of the major N-glycosylation site of the pIgR reduced transcytosis
We next tested if glycosylation of the pIgR itself could be a signal that promotes transcytosis of the pIgR. Almost all of the work in the pIgR MDCK system has utilized rabbit pIgR. Three allotypes (t61, t62 and t63) of rabbit pIgR have been described, which display slightly different amino acid sequences and N-glycosylation sites. The three allotypes possess a common glycosylation site in domain 4 at Asn400 (throughout this paper, we number pIgR residues based on the rabbit sequence). The second site is in domain 1 at Asn70 for t61 and Asn90 for t62 and t63. Moreover, biochemical analysis revealed that 75% of the molecules produced by the t62 allotype are not glycosylated at position 90 but solely at position 400. Finally, it was shown that binding of IgA to purified secretory component was not measurably affected by the variation in amino acid sequence or by the variation in degree of glycosylation (32, 33).
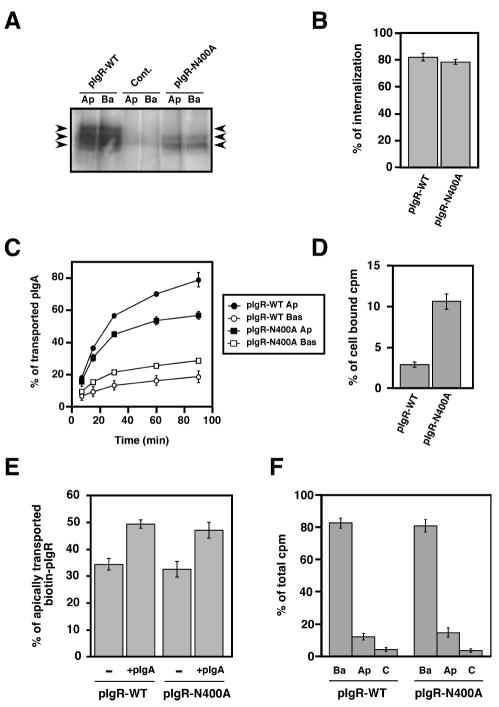
We therefore selectively mutated the major glycosylation site of the pIgR t62allotype at Asn400, converting it to Ala (pIgR-N400A). (The second site is only glycosylated in 25% of the molecules). We selectively biotinylated the apical or basolateral cell surface, precipitated the biotinylated pIgR with streptavidin-agarose and displayed it by SDS-PAGE and immunoblotting with antibodies to SC. Figure 2A shows that compared to pIgR-WT, the mutated pIgR-N400A had entirely lost its highest glycosylated form (see Figure 2A, uppermost arrow). The middle band was also significantly reduced and most likely represents the 25% of pIgR glycosylated on a second minor site. In Figure 2A, the lanes labeled Ap were from cells selectively biotinylated at the apical surface, whereas the lanes labeled Ba were selectively labeled at the basolateral surface. Note that relatively distribution of pIgR between apical and basolateral was unchanged by the N400A mutation, suggesting that this mutation did not alter the steady state polarized distribution of pIgR.
Figure 2.
Mutation of the major N-glycosylation site of pIgR hinders its constitutive apical transcytosis.
A) Apical (Ap) or basolateral (Ba) cell surface biotinylation of untransfected MDCK control cells (cont.) and cells expressing either the pIgR-WT or the mutated pIgR-N400A. The arrows point to the three major bands immunoprecipitated using the sheep anti-rabbit pIgR antibody from pIgR-WT expressing cells. The control cells were not transfected with rabbit pIgR. The pIgRN400A mutant is found as a doublet with a majority of the protein visualized as the lowest band demonstrating its weak glycosylation. The highest of the three bands is not seen with the pIgRN400A mutant, indicating that this highest band is the result of glycosylation at the N400 site.
B) Basolateral endocytosis of iodinated pIgA at 5 min mediated by the pIgR-WT and pIgR-N400A. The experiment was performed twice in triplicate, and data from both experiments were combined. Values are mean ± SD.
C) Apical transcytosis and basolateral recycling of iodinated pIgA endocytosed from the basolateral surface of MDCK cells expressing pIgR-WT or pIgRN400A. The experiment was performed three times in triplicate and one representative experiment is shown.
D) Percentage of pIgA remaining in the cells at the end of the 90 min chase of the experiment presented in C).
E) PIgA-stimulated transcytosis of pIgR-WT and pIgR-N400A. The experiment was performed four times in triplicate, and data from all experiments were combined. Values are mean ± SD. For each cell line, data were compared between +/- pIgA by using a paired Student’s t-test. We determined a statistically significant difference with p<0.01 for both pIgR-WT and pIgR-N400A.
F) Apical (Ap) and basolateral (Ba) cell surface delivery of pIgR-WT and pIgR-N400A. The experiment was performed three times in triplicate, and one representative experiment is shown.
We then compared the ability of pIgR-N400A and pIgR-WT to internalize pIgA for 5 min, using the internalization assay described above. As shown in Fig. 2B, there was no significant difference between pIgR-WT and pIgR-N400A.
Next, we assessed the post-endocytic transport of pIgA by pIgR-N400A, using the assay described in Materials and Methods. We observed reduced apical transcytosis partially compensated by increased basolateral recycling (Fig. 2C). The amount of pIgA that remained associated with the cells at the end of the 90 min chase period was increased, as shown in Fig. 2D. Thus, in sum the N400A mutation reduced pIgR constitutive post-endocytic apical transport.
We then assessed the ability of pIgR-N400A to undergo pIgA-stimulated transcytosis as described in Materials and Methods. As shown in Fig. 2E, the addition of pIgA stimulated transcytosis of pIgR, as assayed by SC release, by an equal amount in pIgR-WT and pIgR-N400A.
Finally, we examined whether N-glycosylation of pIgR could affect its biosynthetic delivery (Fig. 2F). Cells were metabolically labeled with a 15 min pulse of 35S-cysteine/methionine and then chased for 60 min. The arrival of metabolically labeled pIgR at the apical or basolateral surface was detected by its sensitivity to V8 protease, as described in Materials and Methods. We found no significant difference in polarized biosynthetic delivery between the wild type and N400A mutant suggesting that the N-glycan apical targeting signal is not involved in polarized biosynthetic sorting.
Taken together, these data indicate that N-glycosylation of the pIgR serves as a signal to promote basolateral to apical transcytosis of pIgR, even in the absence of bound pIgA. Neither N-glycosylation of the pIgR nor N/O glycosylation of the pIgA appears to play a detectable role in ligand-stimulated transcytosis.
Residues 731-741 of the pIgR are sufficient for apical transcytosis
The pIgR’s C-terminal cytoplasmic domain extends from residues 653 to 755. We have previously made a series of truncation and deletion mutants, which have been used to define signals in the pIgR for basolateral sorting, endocytosis and ligand-stimulated transcytosis. We have previously reported that the truncation of the last 30 amino acids of the cytoplasmic tail (pIgR-725t) blocks both constitutive and pIgA-stimulated apical transport of the fully glycosylated pIgR-WT (23). This observation suggested that a signal for basolateral to apical transcytosis was present in this region of the pIgR’s cytoplasmic tail. Now we have addressed this possibility by analyzing two additional truncation mutants, pIgR-737t and pIgR-747t, with truncations at residue 737 or at residue 747, respectively. We used a post-endocytic fate assay as shown in Fig. 3A. We found that the deletion of the C-terminal 8 amino-acids (pIgR-747t) had no effect on apical transcytosis, compared to pIgR-WT. In striking contrast, deletion of the last 18 amino acids (pIgR-737t) inhibited transcytosis almost as dramatically as pIgR-725t.
Figure 3.
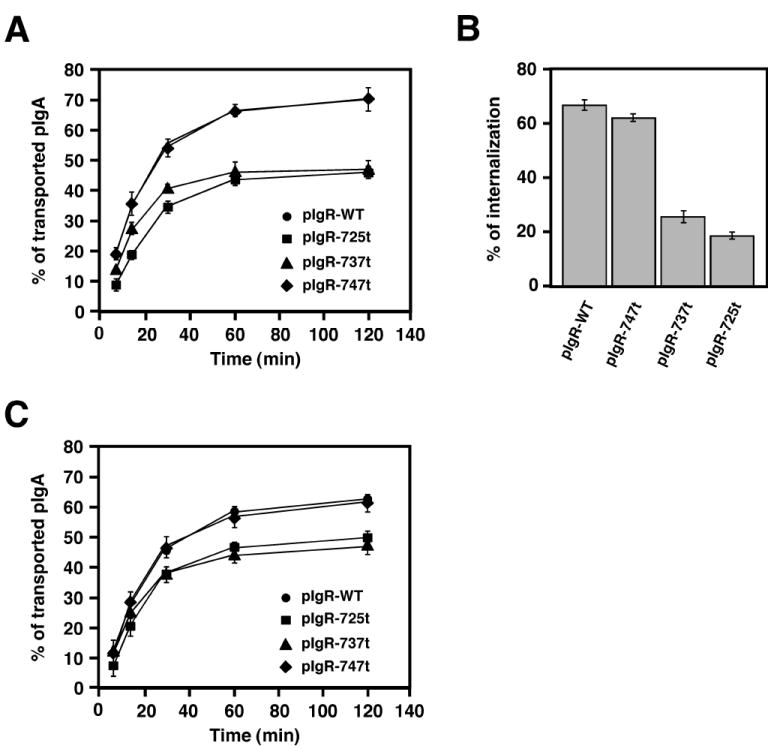
The pIgR cytoplasmic tail of pIgR contains a putative apical targeting signal.
A) Apical transcytosis of iodinated pIgA in MDCK cells expressing pIgR-WT or the C-terminal truncated mutants pIgR-725t, pIgR-737t and pIgR-747t. The experiment was repeated at least 5 times in triplicate for each mutant, and one representative experiment is shown.
B) Basolateral endocytosis of iodinated pIgA at 5 min mediated by the pIgR-WT or the C-terminal truncated mutants pIgR-725t, pIgR-737t and pIgR-747t. The experiment was performed twice in triplicate, and data from both experiments were combined. Values are mean ± SD.
C) Post-endocytic traffic (combined step 2 and 3 of pIgA apical transcytosis) of iodinated pIgA in MDCK cells expressing pIgR-WT or the C-terminal truncated mutants pIgR-725t, pIgR-737t and pIgR-747t. The experiment was repeated 3 times in triplicate for each mutant, and one representative experiment is shown.
We have previously shown that transcytosis of the pIgR can be divided into 3 steps (34). Step 1 is endocytosis from the basolateral plasma membrane and delivery to basolateral early endosomes. Step 2 is the microtubule dependent translocation from basolateral early endosomes to apical recycling endosomes. Step 3 is delivery from apical recycling endosomes to the apical plasma membrane. Only step 3 is stimulated by pIgA binding to the pIgR.
Endocytosis of the pIgR is controlled by Ser726, Tyr734 and to a lesser extent by Tyr668 (35, 36). We have previously shown that deletion at 725t or point mutation of Ser726 decreased basolateral endocytosis and consequently apical transcytosis of pIgA (37). We suspected that deletion at 737t would also affect pIgR endocytosis, i.e. step 1. We measured the endocytosis of pre-bound radio-iodinated pIgA at the basolateral surface. At 5 min, pIgA endocytosis was equivalent to control in pIgR-747t MDCK cells, but severely impaired by deletion at 737t similarly to the 725t deletion (Fig. 3B).
To determine whether the 737t deletion also affected the post-endocytic traffic of pIgA we performed a modified transcytosis assay where the cells were allowed to accumulate radio-iodinated pIgA in the basolateral endosome for 30 min at 17°C Fig. 3C. The cells were then transferred to 4°C and treated basolaterally with trypsin to remove any remaining pIgA bound at the basal surface. The cells were then washed with soybean trypsin inhibitor and MEM-BSA to eliminate the trypsin. We then placed the cells back at 37°C to chase the pre-internalized pIgA and thus measure the post-endocytic transport of the radio-iodinated pIgA. This assay measures both steps 2 and 3 of transcytosis, but measures only constitutive pIgA transcytosis. We observed a decreased in pIgA transcytosis in both 725t and 737t cells, but not in 747t cells, which behave as cells expressing full-length pIgR. We have shown previously that deletion at 737t does not affect step 3 of pIgR transcytosis (23). These data are summarized in Fig. 4A. We conclude that 737t deletion affects steps 1 and 2 of pIgR transcytosis. Taken together with our previous results, this data suggests that the segment 737-747 contains a signal for transcytosis to the apical surface.
Figure 4.
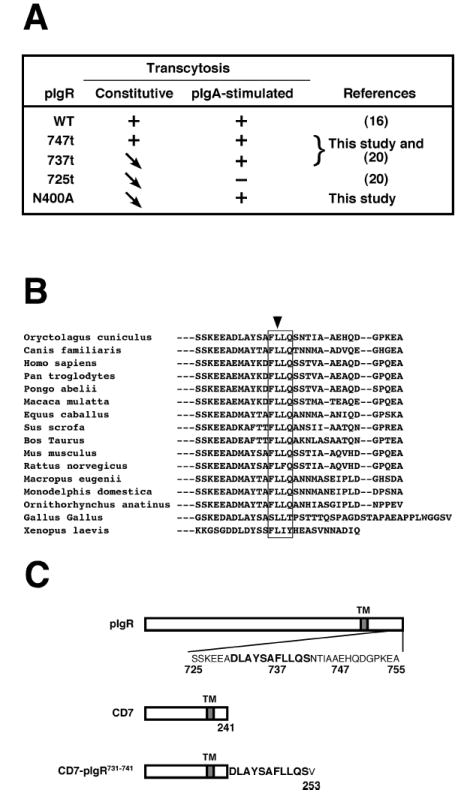
PIgR putative apical targeting signal.
A) Summary of the apical transcytotic properties of pIgR-WT and the indicated mutants suggesting the presence of a cytoplasmic apical targeting signal encompassing the region 737-747.
B) Alignment of all the known sequences of the C-terminus of pIgR cytoplasmic tail starting at serine 725. The box surrounds the highly conserved sequence FLLQ (737-740) and the arrow points to the totally conserved leucine residue 738.
C) Schematic representation of the chimeric protein CD7-pIgR731-741 construct that comprises the most conserved sequence around the FLLQ motif.
To analyze this potential apical transcytosis signal, we aligned the sequences of all of the known pIgRs from diverse species from primates to Xenopus. This revealed the presence of a highly conserved sequence FLLQ (737-740) with the first leucine being 100% conserved (Fig. 4B). The conserved upstream sequence starting at 725 is the consensus sequence for the phosphorylation of serine 726; this has been shown to be involved in regulating internalization via clathrin-coated pits (36). Thus, based on the sequence homology we transferred onto the C-terminal, cytoplasmic tail of the normally basolateral protein, CD7, which we used here as a neutral reporter, the sequence DLAYSAFLLQS (pIgR residues 731-741; Fig. 4C) to make the chimeric protein CD7-pIgR731-741.
We first measured the polarized biosynthetic delivery of CD7 and CD7-pIgR731-741. As shown in Fig. 5A, CD7 is very accurately delivered to the basal surface and the addition of the pIgR peptide did not seem to affect this polarized biosynthetic delivery. Next, we compared the steady state distribution of CD7 and CD7-pIgR731-741 at the apical and basolateral surfaces, as determined by cell surface biotinylation. While CD7 is mostly expressed at the basal surface (81% basolateral, 19% apical), the CD7-pIgR731-741 chimera was now found equally distributed between the basolateral (48%) and apical (52%) surfaces. We also examined the steady state polarized distribution of CD7 and CD7-pIgR731-741 by confocal immunofluorescent microscopy, detecting the proteins with an antibody against CD7. Fig. 5C shows reconstructed X-Z (vertical) sections through the monolayer. CD7 itself was only seen at the basolateral surface, while CD7-pIgR731-741 was observed at both the apical and basolateral surfaces of the cells in the monolayer.
Figure 5.
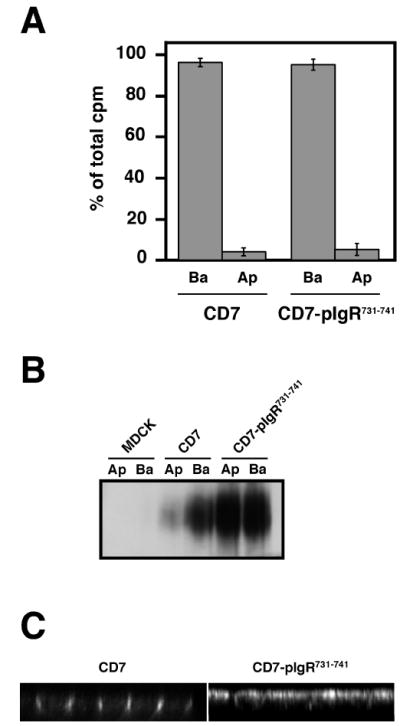
The 731-741 domain of pIgR cytoplasmic tail targets CD7 to the apical surface
A) TGN delivery to the apical (Ap) or basolateral (Ba) cell surface of CD7 and CD7-pIgR731-741. The experiment was performed three times in triplicate, and one representative experiment is shown.
B) Apical (Ap) or basolateral (Ba) cell surface biotinylation of untransfected MDCK cells, or CD7 and CD7-pIgR731-741 MDCK expressing cells.
C) Immunofluorescence analysis of the steady-state distribution of CD7 and CD7-pIgR731-741 in polarized MDCK cells.
Thus, both CD7 and CD7-pIgR731-741 are accurately delivered almost exclusively to the basolateral surface. However, at steady state CD7 is found only at the basolateral surface, while CD7-pIgR731-741 is found at both surfaces in approximately equal amounts.
In order to explain this difference, we investigated the endocytosis and post-endocytic sorting of CD7 and CD7-pIgR731-741, using an antibody directed against the extracellular domain of CD7 as a “pseudo-ligand”. This antibody was bound to CD7 or CD7-pIgR731-741 at the basolateral surface for 60 min at 4°C, unbound material washed away and the cells warmed up to 37°C for 30 min. Cells were then cooled back to 4°C and antibody remaining on the cell surface removed by incubation at 4°C with trypsin for 45 min. Figure 6A shows that there was no significant difference in internalization between CD7 and CD7-pIgR731-741 by this assay.
Figure 6.
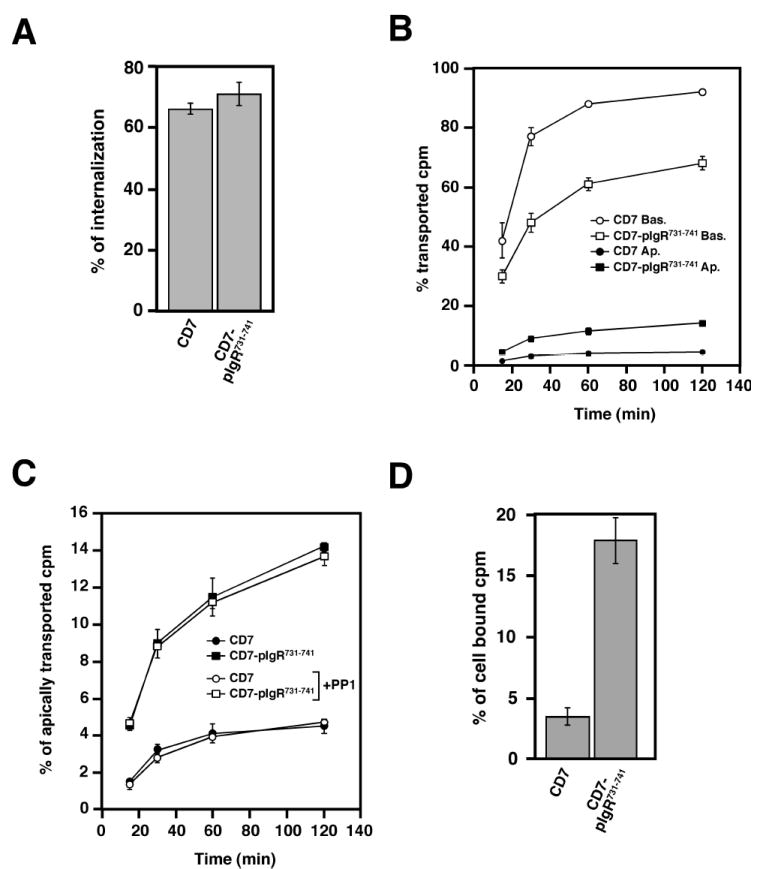
The 731-741 pIgR sequence contains an apical targeting signal.
A) Basolateral endocytosis of CD7 and CD7-pIgR731-741. The experiment was performed twice in triplicate, and data from both experiments were combined. Values are mean ± SD.
B) Apical transcytosis of iodinated anti-CD7 monoclonal antibody endocytosed from the basolateral surface of MDCK cells expressing the wild-type CD7 or the CD7-pIgR731-741. The experiment was performed four times in triplicate, and one representative experiment is shown.
In C) is presented the apical transcytosis of the same experiment shown in B).
D) Intracellular accumulation of pIgA at the end of the transport assay presented in A) and B).
Next we compared the post-endocytic fate of CD7 and CD7-pIgR731-741 using an assay similar to that employed in Fig. 3A, except that the antibody to the ectodomain of CD7 was used instead of pIgA. As shown in Fig. 6B, basolateral recycling of CD7-pIgR731-741 was reduced relative to CD7, with 68% recycled at 120 min for CD7-pIgR731-741 compared to 92% for CD7. Transcytosis to the apical surface was increased ~three fold, to 14.25% at 120 min for CD7-pIgR731-741 versus 4.5% for CD7. Transcytosis was replotted in an enlarged scale in Fig. 6C to better show this increase. Fig. 6D shows that more of the pIgR731-741 was also retained intracellularly, consistent with the increased apical transcytosis and even larger decrease in basolateral recycling. We propose that the resolution of the issue raised above is that both CD7 and CD7-pIgR731-741 are initially targeted to the basolateral surface and then endocytosed equally. However, only CD7-pIgR731-741 is then transcytosed to the apical surface. This transcytosed CD7-pIgR731-741 accounts for the material detected at the apical surface. Some of the pIgR731-741 may recycle to the basolateral surfaces for multiple cycles before eventually being transcytosed to the apical surface. These experiments suggest that the 11 amino-acid segment, residues 731-741, of pIgR cytoplasmic tail contains an apical targeting signal that specifically operates in a post-endocytic sorting pathway, rather than in a biosynthetic pathway.
As discussed above, transcytosis of the pIgR to the apical surface has two components, a constitutive component and a component that is stimulated by binding of the ligand, pIgA, to the pIgR. This latter, stimulated transcytosis is the result of a signaling pathway initiated by pIgA binding, which leads to activation of p62Yes, a Src-family non-receptor tyrosine kinase and then to downstream signaling events (38). To test if this pathway was involved in apical transcytosis of CD7-pIgR731-741, we treated cells with PP1, a specific inhibitor of the Src family of non-receptor tyrosine kinases. As shown in Fig. 6C, PP1 had no effect on transcytosis of CD7-pIgR731-741. Furthermore, it has previously been shown that brefeldin A (BFA), an inhibitor of certain ARF1 guanine nucleotide exchange factors involved in vesicular traffic, inhibits transcytosis of pIgA by the pIgR (39, 40). We tested the effect of BFA on transcytosis of CD7-pIgR731-741 and found no effect (our unpublished data). Taken together, these data suggest that the signal contained in residues 731-741 of pIgR codes for constitutive transcytosis, and not for ligand-stimulated transcytosis.
Discussion
Our most important finding is the identification of an 11 amino acid segment within the cytoplasmic domain of the pIgR (residues 731-741) that represents a novel class of sorting signal, which is sufficient basolateral to apical transcytosis. This signal acts on molecules that have been endocytosed at the basolateral surface and promotes their post-endocytic delivery to the apical surface. This sequence is highly conserved among pIgR’s from humans through birds and amphibians. Importantly, it can be transplanted to a heterologous reporter molecule, CD7. Both CD7- and CD7-pIgR731-741 are efficiently sent in the biosynthetic pathway to the basolateral surface and subsequently endocytosed. After endocytosis, CD7 is almost entirely recycled to the basolateral, whereas a significant fraction of CD7-pIgR731-741 is transcytosed to the apical surface. The novel transcytosis signal that we have identified therefore appears to work mainly in a post-endocytic pathway and not in the biosynthetic pathway.
This novel transcytotic signal can be contrasted with previously identified signals for biosynthetic sorting to the apical surface. It is largely unknown whether or not these signals also function in the post-endocytic pathway(s). Many signals for apical sorting in the biosynthetic pathway have been proposed to be weaker than basolateral sorting signals, so that apical sorting may only become apparent when the basolateral sorting signal is removed from a protein that contains both types of signals (41, 42). This does not appear to be the case for the transcytotic sorting signal described here, in that it directs the post-endocytic apical sorting of proteins that contain signals for biosynthetic delivery to the basolateral surface.
It is also interesting to compare the transcytotic signal identified here to signals for basolateral sorting, which are largely the best understood polarized sorting signals. In several cases, such basolateral sorting signals cause basolateral sorting in both the biosynthetic and post-endocytic sorting pathways (1). This originally led to the idea that the same machinery is responsible for basolateral sorting in both the biosynthetic and post-endocytic pathways. One possible reason for this is that in some cases, there is evidence that the biosynthetic pathway passes through endocytic compartments, at least in part, on the way to the basolateral surface. Furthermore, the AP1b clathrin adaptor, which is responsible for basolateral sorting of many proteins, is present in and functions in both the biosynthetic and post-endocytic pathways (43). In contrast, the transcytotic signal identified here apparently works only after endocytosis. We do not know what machinery recognizes this transcytotic signal, but presumably it is only present or at least only active in some portion of the endocytic pathway and not in the biosynthetic pathway. Another possibility is that transcytotic sorting signal is activated (e.g. by covalent modification or association with another protein) after delivery to the basolateral surface. The ability of this transcytotic signal to function even when transplanted to a completely heterologous reporter, CD7, makes this possibility seem less likely, though it cannot be ruled out.
We have previously described (34) that transcytosis of the pIgR involves three distinguishable steps, i.e. endocytosis and delivery to basolateral early endosomes (step 1), microtubule dependent movement from basolateral early endosomes to apical recycling endosomes (step 2) and movement from apical recycling endosomes to apical plasma membrane (step 3). We found that truncation of pIgR at residue 725 and 737 blocked steps 1 and 2 but not step 3. This raises the possibility that the region of pIgR residues 731-741 may contain two adjacent or overlapping determinants. We were unable to analyze this further using the CD7-pIgR731-741 fusion, due to technical reasons. It is worth noting that some basolateral sorting signals also seem to consist of two closely adjacent regions (44). The apical sorting mechanisms are poorly understood. However, there is accumulating evidence that microtubule motors might promote the directional apical transport (6). We and others have shown that step 2 of pIgR transcytosis is sensitive to microtubule disrupting agents (34, 45). The cytoplasmic tail of rhodopsin that contains the apical determinant binds to the microtubule motor dynein, which mediates the translocation of rhodopsin to the apical membrane in MDCK cells (46). Similarly, one could speculate that the determinant identified in pIgR mediates binding to microtubule motors for its apical transport
The pIgR is constitutively transcytosed from the basolateral to the apical plasma membrane by a mechanism that utilizes the novel transcytotic sorting signal described above. Binding of the ligand, pIgA, to the pIgR, activates a signaling network, which further stimulates transcytosis. This ligand-stimulated transcytosis mechanism does not seem to involve or depend on the transcytotic sorting signal that we describe here, as indicated by the lack of effect of PP1 or BFA, both of which are known to selectively block pIgA-stimulated transcytosis.
Apical sorting signals in the biosynthetic pathway have been shown to involve carbohydrates in a number of cases. We investigated if the glycosylation of either the pIgR itself or the pIgA ligand of the pIgR played any role in polarized sorting. We mutated a major site of Asn-linked glycosylation in the pIgR (pIgR-N400AP). Transcytosis of pIgR-N400A was reduced relative to wild-type pIgR, suggesting that N-linked glycosylation of the pIgR plays a role in its constitutive transcytosis. Ligand-stimulated transcytosis was not affected. It may be that this N-linked glycosylation and the transcytotic signal that we have identified cooperate and/or operate at different steps to promote transcytosis. However, it is difficult to rule out that the effect of N-glycosylation is only indirect, e.g., on protein folding (14).
We also tested if glycosylation on the pIgA ligand might be involved in ligand-stimulated transcytosis, acting as an apical sorting signal. We used pIgA lacking N- and/or O-linked carbohydrates. We found no evidence that glycosylation of the pIgA played any role in transcytosis. Altered glycosylation of pIgA has been implicated in several major disease processes, including IgA nephropathy, Henoch-Schonlein Purpura, primary Sjögren’s syndrome and complement activation (47, 48). Our data suggest that the altered glycosylation of pIgR does not act through stimulation of pIgR transcytosis to cause these disease processes.
In conclusion, we have identified a novel class of sorting signal involved in transcytosis from the basolateral to the apical surface. This pathway therefore cannot be assumed to operate by default, but rather like most other pathways is signal mediated.
Materials and Methods
Cells, antibodies and reagents
MDCK cell strain II cell line and its transfectants were maintained in MEM supplemented with 5% fetal calf serum, penicillin (50 U/ml) and streptomycin (50 μg/ml). For cell polarity studies, MDCK cells were grown on 0.4-μm pore polycarbonate Transwell filters (Corning-Costar, Cambridge, MA) as previously described for a minimum of 3 days before assays (49). MDCK cells expressing the rabbit pIgR allotype t62 and the truncated mutants 725t, 737t and 747t have been described elsewhere (23). The pIgR-N400A, CD7 and CD7- pIgR731-741 constructs were transfected into MDCK cells by calcium precipitation and individual clones screened by immunoblot. For each cell line, several clones were isolated and analyzed. The results shown are for one representative clone. The sheep anti-SC pIgR antibody was previously described (50) and the monoclonal antibody (clone SC166) directed against the cytoplasmic tail of pIgR was a kind gift from Prof. Anne Hubbard, Johns Hopkins Univ, Baltimore, MD. Note that the SC166 antibody recognizes the 11 amino-acid fragment 731-741 of pIgR (see below). The anti-CD7 monoclonal antibody (clone CD7-6B7) used for transport assays was from Caltag Laboratories (Burlingame, CA) and the anti-CD7 monoclonal antibody (clone T3-3A1) used for immunofluorescence was a gift from Dr. C. -H. Sung. The glycosidases were from Boehringer Ingelheim (Germany) and the biotinylated lectins from Vector Laboratories (Burlingame, CA). The sulfo-NHS-biotin and biotin-LC-hydrazide-were obtained from Pierce (Rockford, IL), and the nocodazole from Calbiochem (La Jolla, CA). EXPR35S35S protein labeling mix was purchased from NEN™ Life Science Products (Boston, MA). All chemicals and other reagents were from Sigma (St Louis, MO).
DNA constructs
PIgRN400A
The substitution of the residue asparagine by an alanine at position 400 was done by PCR mutating the asparagine AAT codon into the GCT alanine codon. The 5’ forward oligonucleotide was AGGCTGGCCCTGTTCGAAGAGCCTGGCGCTGGCACC and the 3’ reverse oligonucleotide was TGTGTGCCTAGGCCTCCTTGGGGCCATCTTGGTGCTCAGC. Underlined are the restriction sites BstBI and AvrII in the forward and reverse oligonucleotides, respectively. In the forward oligonucleotide the mutated codon is indicated in bold. The sequence AvrII is placed at the extremity of the pIgR coding region and contains the stop codon TAG indicated in bold in the reverse oligonucleotide. The last nucleotides coding for pIgR are in italic in the reverse oligonucleotide. The amplified PCR fragment was digested with BstBI and AvrII, and was subcloned into the digested BstBI/AvrII mammalian expression vector pCB7-pIgR containing the hygromycin selective marker.
CD7-pIgR731-741
The 11 amino-acids 731 to 741 from pIgR cytoplasmic tail have been fused to CD7 C-terminal extremity by PCR amplification of the last 78 nucleotides of CD7 from nucleotide 642 to 720 using a 3’ oligonucleotide containing the sequence encoding for pIgR731-741. The plasmid CD7BB1 containing the CD7 transcript was obtained from Dr. C.-H. Sung and described elsewhere (7, 51). The 5’ forward oligonucleotide was GGGTGGCGTGTGTGCTGGCGAGGACACAGATAAAGAAACTGTGCTCGTGGCGGGATAAGAATTCGG and the 3’ reverse oligonucleotide was TCTAGATCTAGACGGATTGGAGCAGGAAAGCTGAGTAGGCCAGGTCCTGGTACTGGTTGGGGGAGG. Underlined are the restriction sites EcoRI within CD7 in the 5’ oligonucleotide and XbaI in the 3’ oligonucleotide. The sequence XbaI is present in the 3’ untranslated region of the CD7 insert. In bold is the sequence coding for the 11 amino acids of pIgR (731-741) with an extra codon overlapping the XbaI restriction site and coding for a valine at the most C-terminal position. The stop codon TAG is present within the XbaI restriction site in frame with the CD7-pIgR731-741 insert. In italics is the sequence of the last 21 nucleotides of CD7 in frame with the sequence encoding for pIgR731-741 fragment. The amplified PCR fragment was digested with EcoRI and XbaI and inserted into digested EcoRI/XbaI CD7BB1 plasmid containing the geneticin selective marker.
Deglycosylation and characterization of the pIgA preparations
The human pIgA was purified as previously described (52). N-deglycosylation was completed using N-glycosidase F. Briefly, 1mg of human pIgA was incubated 16 h at 37°C on a head-to-head rotator in 20mM sodium phosphate buffer pH 7.2 with 80 U of N-glycosidase F and 2 mM PMSF in a total volume of 3 ml. O-deglycosylation was performed by incubating 1 mg of pIgA 16 h at 37°C on a head-to-head rotator in 20 mM sodium phosphate buffer pH 7.2 with 100 mU of neuraminidase, 85 mU of O-glycosidase and 2 mM PMSF in a total volume of 5 ml. Deglycosylation of both O- and N-linked sugars was achieved by successive O- and N-deglycosylation. These procedures yield approximately 90% of O-deglycosylation and 80% N-deglycosylation. A mock-treated pIgA was prepared under the same conditions but the enzymes were omitted. At the end of each treatment the pIgA, preparations were concentrated using a centricon-100, 100,000 MW cut-off (Amicon, Beverly, MA) and analyzed after SDS-PAGE by western-blots using biotinylated lectins revealed by streptavidin-HRP (Jackson ImmunoResearch, West Grove, PA) and ECL (Amersham, Arlington Heights, IL). The anti-IgA antibodies used in immunoblot were from Cappel (Durham, NC) and revealed with an HRP-conjugated secondary antibody (Jackson ImmunoResearch).
Ligand transcytosis and endocytosis
PIgA transcytosis and endocytosis were assayed as previously described (23). The pIgA preparations were iodinated by the ICl method and stored at a concentration of 75 μg/ml and 1 × 106 cpm/μl. For transcytosis assay, MDCK cells cultured on 12-mm Transwell filters were allowed to internalize iodinated pIgA (1 × 106 cpm in a 10 μl drop of MEM-BSA) from the basolateral surface for 10 min at 37°C. The filters were rapidly washed four times with MEM-BSA, transferred into a 12-well culture plate and fresh 37°C MEM-BSA was added to both the apical (300 μl) and basolateral (500 μl) chambers. The medium was collected and replaced after 7.5, 15, 30, 60 and 90 or 120 min. At the end of the chase at 37°C, the filters were cut out from the holders and the radioactivity in all fractions (cells, apical and basolateral media) was counted in a Packard γ-counter (Packard Instrument, Downers Grove, IL). The percentage of transcytosis or basolateral recycling was the percent of counts released into the apical or basolateral medium relative to total counts present in all fractions, respectively. Where indicated the media collected from the apical chamber at the indicated times were pooled and precipitated in TCA 15% at 4°C for 1 h. The TCA insoluble iodinated pIgA was collected after centrifugation at 14,000 × g for 15 min at 4°C and resuspended by 10-min boiling in Laemmli buffer. The TCA-insoluble iodinated pIgA was analyzed by SDS-PAGE and the radioactivity visualized by fluorography. To analyze the transcytosis of the CD7 proteins a slightly modified procedure was adapted. Similarly to the pIgA, the monoclonal anti-CD7 antibody (clone CD7-6B7) was iodinated by the ICl method. Cells cultured on 12-mm Transwell filters were allowed to bind the iodinated antibody (0.2 × 106 cpm in a 10 μl drop of MEM-BSA) from the basolateral surface for 1 h at 4°C. The filters were rapidly washed four times with ice-cold MEM-BSA, transferred into a 12-well culture plate and fresh 37°C MEM-BSA was added to both the apical (300 μl) and basolateral (500 μl) chambers. In addition, trypsin (10 μg/ml) was present in the apical medium during the chase to release any apically transcytosed antibody. The rest of the assay was conducted as described above for pIgA.
For endocytosis assay, cells grown on 12-mm Transwell filters were incubated at 4°C for 1 h on a 30μl-drop containing the iodinated pIgA (1 × 106 cpm in MEM-BSA) or CD7-6B7 antibody (0.2 × 106 cpm in MEM-BSA). The unbound ligand was washed away by four 5-min washes in ice-cold MEM-BSA and endocytosis followed by incubating the cells 1, 2.5, 5 min (pIgA) or 5, 15, 30 min (CD7) at 37°C. After cooling down the cells in ice-cold MEM-BSA, the basolateral surface was trypsinized to remove non-internalized ligand. The basolateral trypsin washes and the intracellular counts were determined in a Packard γ-counter. The amount of internalized ligand was plotted as the percentage of total initial binding of the iodinated ligand to the basolateral surface.
Domain selective cell surface biotinylation
To determine the steady-state cell surface distribution on the apical and basolateral domain of the pIgR and CD7 proteins, a domain selective biotinylation followed by immunoprecipitation was performed. Briefly, the MDCK cells grown on 12-mm Transwell filter for three days were quickly washed in ice-cold PBS. The basolateral or apical cell surface was biotinylated for 30 min at 4°C with 1 ml (basal) or 0.3 ml (apical) of a solution containing either 0.2 mg/ml of sulfo-NHS-biotin (pIgR) or 0.2 mg/ml of biotin-LC-hydrazide (CD7) freshly dissolved in PBS. Biotin-LC-hydrazide was used instead of sulfo-NHS-biotin to biotinylate the carbohydrates of the extracellular domain of CD7 that does not possess lysine residues. During the basal or apical biotinylation, 0.3 ml or 1 ml of MEM-BSA (MEM, 0.35g/liter NaHCO3, 20mM Hepes-Na, 6mg/ml BSA, and antibiotics) were present on the opposite side, respectively. After biotinylation, the cells were quickly washed three times in MEM-BSA and twice in PBS before solubilization in 1 ml of lysis buffer (NP-40 1%, 150mM NaCl, Hepes 20mM pH 7.4, protease inhibitors). The lysates were pre-cleared and the pIgR or CD7 proteins immunoprecipitated using a sheep anti-rabbit SC antiserum or the monoclonal anti-CD7 antibody T3-3A1. The immunoprecipitates were resolved by SDS-PAGE transferred onto a PVDF membrane and the biotinylated proteins revealed by probing the membrane with streptavidin-HRP and ECL.
Domain selective cell surface delivery assay
Filter grown MDCK cells were incubated in MEM-cys/met (MEM without cysteine and methionine) for 15 min and then pulse-labeled in MEM-cyst/met containing 5% dialyzed FCS with 50μCi of EXPRE35S35S on a 25-μl drop for 15 min. After four quick washes the cells were incubated at 37°C in MEM-BSA supplemented with non-radioactive methionine and cysteine.
PIgR proteins
The pulse-chase analysis of pIgR proteins was performed as previously described (53). Following four washes, the cells were chased in MEM-BSA supplemented with non-radioactive methionine and cysteine for 60 min with or without V8 protease (25 μg/ml) in the basolateral medium. At the end of the chase, the apical and basal media were collected and the cells quickly washed three times in MEM containing 10% horse serum and twice in PBS. The filters were cut out, transferred into an Eppendorf tube and the cells boiled 5 min in 1 ml SDS lysis buffer (0.5% SDS, 150 mM NaCl, 5 mM EDTA, 20 mM triethanolamine pH 8.1, 100 U/ml Trasylol). The tubes were then vigorously vortexed for 20 min and 0.5 ml of Triton dilution buffer (5% Triton X-100, 100 mM NaCl, 50 mM triethanolamine pH 8.1, 5 mM EDTA, 2 mM PMSF, 100 U/ml Trasylol) was added. The SC was immunoprecipitated from the apical and basolateral medium, and the pIgR was immunoprecipitated from the cells using a sheep anti-rabbit SC antiserum coupled to protein G sepharose beads. After four washes, the immuno-complexes were resolved by SDS-PAGE, revealed by fluorography and quantitated using a PhosphorImager.
CD7 proteins
A detailed procedure was described elsewhere (7). In brief, at various times of the chase the cells were cooled-down, quickly washed in ice-cold PBS and biotinylated with biotin-LC-hydrazide from the basolateral or apical surface as described above. After biotinylation, the cells were solubilized in Triton lysis buffer (1% Triton X-100, 50 mM Tris pH 7.5, 150mM NaCl, 2mM EDTA and protease inhibitors), centrifuged 20 min at 14,000 × g, the lysates pre-cleared and the CD7 proteins immunoprecipitated using the anti-CD7 monoclonal antibody (clone T3-3A1). The immuno-complexes were recovered by boiling in 40 μl 5% SDS for 10 min and diluted in 460 μl lysis buffer. A 25-μl aliquot was saved to estimate the total initial radioactivity in CD7 proteins. The biotinylated CD7 proteins were then recovered by streptavidin agarose beads, resolved by SDS-PAGE and revealed by fluorography. The cumulative radioactivity in biotinylated CD7 quantitated using a PhosphorImager was plotted as a percentage of total initial radioactivity in CD7.
PIgA-stimulated transcytosis assay
In this assay, the third step of pIgR transcytosis, i.e. from the apical recycling endosomes (ARE) to the apical plasma membrane, the only one stimulated by pIgA, is measured (49). This assay has been described in detail elsewhere (34, 54). MDCK cells grown on 12-mm Transwell filter for three days were quickly washed in ice-cold PBS. The basolateral cell surface was biotinylated for 30 min at 17°C with 1 ml of a solution containing 0.2 mg/ml of sulfo-NHS-biotin freshly dissolved in PBS. During the biotinylation 300 μl of MEM-BSA were present on the apical surface. The cells were then quickly washed three times in MEM-BSA to quench the excess of biotin. The filter units were then placed onto a 30-μl drop containing or not 0.3 mg/ml of pIgA and incubated at 17°C for 10 min. At 17°C the basolateral-to-apical transcytosis of the pIgR was shown to be largely blocked so that the empty or pIgA-bound biotinylated pIgR accumulates in the basolateral early endosomal compartment (55). The cells were then further incubated for 15 min at 37°C to accumulate the biotinylated pIgR, bound or not to pIgA, into the ARE. During this chase, 300 μl of MEM-BSA containing trypsin (25 μg/ml) were present in the apical chamber to cleave off any biotinylated pIgR reaching the apical surface. The chase was stopped by transferring the filter units into ice-cold MEM-BSA. The cells were then quickly washed three times in cold MEM-BSA containing 15% horse serum to neutralize the apical trypsin. The cells were then incubated for 1 h at 4°C in MEM-BSA containing 33 μM nocodazole. During this period, the microtubules were depolymerized to prevent further basolateral-to-apical transcytosis, a step that was shown to be microtubule-dependent and to be efficiently blocked by nocodazole treatment (49). The cells are then placed in MEM-BSA plus nocodazole (33 μM) at 37°C for 20 min to allow for the transport of the biotinylated pIgR from the ARE to the apical plasma membrane. During this chase, soybean trypsin inhibitor (125 μg/ml) was added in the basolateral medium and trypsin (25 μg/ml) in the apical medium to cleave off the apically transported pIgR. The rest of the experiment was carried out at 4°C. At the end of the chase, the cells were washed twice in MEM-BSA 15% horse serum and incubated 30 min in MEM-BSA containing 125 μg/ml soybean trypsin inhibitor to quench the trypsin. The cells were washed three more times in MEM-BSA 15% horse serum and twice in PBS before solubilization in 1 ml of lysis buffer (NP-40 1%, 150 mM NaCl, Hepes 20mM pH 7.4, protease inhibitors). The lysates were pre-cleared and the pIgR immunoprecipitated using a sheep anti-rabbit SC antiserum. The immunoprecipitates were resolved by SDS-PAGE, transferred onto a PVDF membrane and the biotinylated pIgR was revealed by probing the membrane with streptavidin-HRP and ECL. The amount of pIgR was quantitated using a Molecular Dynamics densitometer. A set of filters was used as a standard and lysed after the nocodazole treatment. The amount of biotinylated pIgR in these samples was considered as 100%. In the samples subjected to the final chase, the percentage of pIgR transported to the apical surface was estimated by subtracting the percentage of remaining biotinylated pIgR. Each experiment was repeated 3 to 7 times in either triplicates or quadruplicates. The results represent the mean of all experiments ± SD. Statistical significance was evaluated by the Student’s t test.
Immunofluorescence
The procedure followed was previously described except that we omitted the permeabilization step (55). MDCK cells grown on 12-mm Transwell filters for 5 days were fixed in 4% paraformaldehyde after 3 quick washes in PBS-CM (PBS supplemented with 1 mM CaCl2, 0.5 mM MgCl2). The cells were then exposed to the indicated primary antibodies diluted in PBS-CMF (PBS-CM containing 0.7% fish skin gelatin) for 1 h at 37°C in a humid chamber. The cells were then washed four times 5 min in PBS-CMF and incubated with the fluorescent secondary antibodies in PBS-CMF for 45 min at 37°C. The cells were then washed four times 5 min in PBS-CMF and twice in PBS-CM. The filters were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and analyzed with a Bio-Rad MRC1024 laser scanning confocal microscope.
Supplementary Material
Mutation of the major site of N-glycosylation of a recombinant human pIgA does not affect its trafficking in polarized MDCK cells.
A) The glycosylation status of the recombinant pIgA heavy chain was analyzed by probing the membrane with the O-glycan specific lectin Jacalin (top panel) or the N-glycan specific lectin Lens culinaris (bottom panel). For each panel, the arrows point to the upper glycosylated and lower unglycosylated pIgA.
B) Wild-type and NQ mutant pIgA preparation were iodinated and tested for their capacity to bind to pIgR-expressing MDCK cells.
Basolateral endocytosis (C), recycling (D) and transcytosis (E) were assayed. All experiments were performed at least three times in triplicate, and one representative experiment is shown.
In F), the pIgA preparations were analyzed for their ability to stimulate the third step of pIgR apical transcytosis. The data represent the average values (± SD) from three independent experiments. For each pIgA preparation, data were compared to the control sample without pIgA by using a paired Student’s t-test. We determined a statistically significant difference with p<0.005 for both WT and NQ pIgA.
Acknowledgments
We thank Drs. Hakon Leffler (UCSF) for fruitful discussions, Ching-Hwa Sung (Weil Medical College of Cornell Univ, New York, NY) and Anne Hubbard (Johns Hopkins Univ, Baltimore MD) for providing us with reagents and advice related to CD7 and Tao Su (UCSF) for comments on the manuscript.
References
- 1.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9(11):833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folsch H. Regulation of membrane trafficking in polarized epithelial cells. Curr Opin Cell Biol. 2008;20(2):208–213. doi: 10.1016/j.ceb.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casanova JE, Apodaca G, Mostov KE. An autonomous signal for basolateral sorting in the cytoplasmic domain of the polymeric immunoglobulin receptor. Cell. 1991;66(1):65–75. doi: 10.1016/0092-8674(91)90139-p. [DOI] [PubMed] [Google Scholar]
- 4.Folsch H, Ohno H, Bonifacino JS, Mellman I. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 1999;99(2):189–198. doi: 10.1016/s0092-8674(00)81650-5. [DOI] [PubMed] [Google Scholar]
- 5.Gan Y, McGraw TE, Rodriguez-Boulan E. The epithelial-specific adaptor AP1B mediates post-endocytic recycling to the basolateral membrane. Nat Cell Biol. 2002;4(8):605–609. doi: 10.1038/ncb827. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Boulan E, Kreitzer G, Musch A. Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol. 2005;6(3):233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 7.Chuang JZ, Sung CH. The cytoplasmic tail of rhodopsin acts as a novel apical sorting signal in polarized MDCK cells. J Cell Biol. 1998;142(5):1245–1256. doi: 10.1083/jcb.142.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Naim HY, Rodriguez AC, Roth MG. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J Cell Biol. 1998;142(1):51–57. doi: 10.1083/jcb.142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuck S, Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J Cell Sci. 2004;117(Pt 25):5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- 10.Lisanti MP, Tang ZL, Sargiacomo M. Caveolin forms a hetero-oligomeric protein complex that interacts with an apical GPI-linked protein: implications for the biogenesis of caveolae. J Cell Biol. 1993;123(3):595–604. doi: 10.1083/jcb.123.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannan LA, Lisanti MP, Rodriguez-Boulan E, Edidin M. Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J Cell Biol. 1993;120(2):353–358. doi: 10.1083/jcb.120.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paladino S, Pocard T, Catino MA, Zurzolo C. GPI-anchored proteins are directly targeted to the apical surface in fully polarized MDCK cells. J Cell Biol. 2006;172(7):1023–1034. doi: 10.1083/jcb.200507116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paladino S, Sarnataro D, Tivodar S, Zurzolo C. Oligomerization is a specific requirement for apical sorting of glycosyl-phosphatidylinositol-anchored proteins but not for non-raft-associated apical proteins. Traffic. 2007;8(3):251–258. doi: 10.1111/j.1600-0854.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Boulan E, Gonzalez A. Glycans in post-Golgi apical targeting: sorting signals or structural props? Trends Cell Biol. 1999;9(8):291–294. doi: 10.1016/s0962-8924(99)01595-0. [DOI] [PubMed] [Google Scholar]
- 15.Ihrke G, Bruns JR, Luzio JP, Weisz OA. Competing sorting signals guide endolyn along a novel route to lysosomes in MDCK cells. Embo J. 2001;20(22):6256–6264. doi: 10.1093/emboj/20.22.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potter BA, Weixel KM, Bruns JR, Ihrke G, Weisz OA. N-glycans mediate apical recycling of the sialomucin endolyn in polarized MDCK cells. Traffic. 2006;7(2):146–154. doi: 10.1111/j.1600-0854.2005.00371.x. [DOI] [PubMed] [Google Scholar]
- 17.Yu CY, Chen JY, Lin YY, Shen KF, Lin WL, Chien CL, ter Beest MB, Jou TS. A bipartite signal regulates the faithful delivery of apical domain marker podocalyxin/Gp135. Mol Biol Cell. 2007;18(5):1710–1722. doi: 10.1091/mbc.E06-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostov KE, Altschuler Y, Chapin SJ, Enrich C, Low SH, Luton F, Richman-Eisenstat J, Singer KL, Tang K, Weimbs T. Regulation of protein traffic in polarized epithelial cells: the polymeric immunoglobulin receptor model. Cold Spring Harb Symp Quant Biol. 1995;60:775–781. doi: 10.1101/sqb.1995.060.01.083. [DOI] [PubMed] [Google Scholar]
- 19.Song W, Bomsel M, Casanova J, Vaerman JP, Mostov K. Stimulation of transcytosis of the polymeric immunoglobulin receptor by dimeric IgA. Proc Natl Acad Sci U S A. 1994;91(1):163–166. doi: 10.1073/pnas.91.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luton F, Verges M, Vaerman JP, Sudol M, Mostov KE. The SRC family protein tyrosine kinase p62yes controls polymeric IgA transcytosis in vivo. Mol Cell. 1999;4(4):627–632. doi: 10.1016/s1097-2765(00)80213-0. [DOI] [PubMed] [Google Scholar]
- 21.van ISC, Tuvim MJ, Weimbs T, Dickey BF, Mostov KE. Direct interaction between Rab3b and the polymeric immunoglobulin receptor controls ligand-stimulated transcytosis in epithelial cells. Dev Cell. 2002;2(2):219–228. doi: 10.1016/s1534-5807(02)00115-6. [DOI] [PubMed] [Google Scholar]
- 22.Evans E, Zhang W, Jerdeva G, Chen CY, Chen X, Hamm-Alvarez SF, Okamoto CT. Direct interaction between Rab3D and the polymeric immunoglobulin receptor and trafficking through regulated secretory vesicles in lacrimal gland acinar cells. Am J Physiol Cell Physiol. 2008;294(3):C662–674. doi: 10.1152/ajpcell.00623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luton F, Cardone MH, Zhang M, Mostov KE. Role of tyrosine phosphorylation in ligand-induced regulation of transcytosis of the polymeric Ig receptor. Mol Biol Cell. 1998;9(7):1787–1802. doi: 10.1091/mbc.9.7.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardone MH, Smith BL, Mennitt PA, Mochly-Rosen D, Silver RB, Mostov KE. Signal transduction by the polymeric immunoglobulin receptor suggests a role in regulation of receptor transcytosis. J Cell Biol. 1996;133(5):997–1005. doi: 10.1083/jcb.133.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giffroy D, Langendries A, Maurice M, Daniel F, Lardeux B, Courtoy PJ, Vaerman JP. In vivo stimulation of polymeric Ig receptor transcytosis by circulating polymeric IgA in rat liver. Int Immunol. 1998;10(3):347–354. doi: 10.1093/intimm/10.3.347. [DOI] [PubMed] [Google Scholar]
- 26.Giffroy D, Courtoy PJ, Vaerman JP. Polymeric IgA binding to the human pIgR elicits intracellular signalling, but fails to stimulate pIgR-transcytosis. Scand J Immunol. 2001;53(1):56–64. doi: 10.1046/j.1365-3083.2001.00843.x. [DOI] [PubMed] [Google Scholar]
- 27.Mattu TS, Pleass RJ, Willis AC, Kilian M, Wormald MR, Lellouch AC, Rudd PM, Woof JM, Dwek RA. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fc alpha receptor interactions. J Biol Chem. 1998;273(4):2260–2272. doi: 10.1074/jbc.273.4.2260. [DOI] [PubMed] [Google Scholar]
- 28.Endo T, Wright A, Morrison SL, Kobata A. Glycosylation of the variable region of immunoglobulin G--site specific maturation of the sugar chains. Mol Immunol. 1995;32(13):931–940. doi: 10.1016/0161-5890(95)00078-s. [DOI] [PubMed] [Google Scholar]
- 29.Norderhaug IN, Johansen FE, Schjerven H, Brandtzaeg P. Regulation of the formation and external transport of secretory immunoglobulins. Crit Rev Immunol. 1999;19(5-6):481–508. [PubMed] [Google Scholar]
- 30.Chuang PD, Morrison SL. Elimination of N-linked glycosylation sites from the human IgA1 constant region: effects on structure and function. J Immunol. 1997;158(2):724–732. [PubMed] [Google Scholar]
- 31.Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J Biol Chem. 1979;254(8):2865–2874. [PubMed] [Google Scholar]
- 32.Frutiger S, Hughes GJ, Hanly WC, Jaton JC. Rabbit secretory components of different allotypes vary in their carbohydrate content and their sites of N-linked glycosylation. J Biol Chem. 1988;263(17):8120–8125. [PubMed] [Google Scholar]
- 33.Piskurich JF, Blanchard MH, Youngman KR, France JA, Kaetzel CS. Molecular cloning of the mouse polymeric Ig receptor. Functional regions of the molecule are conserved among five mammalian species. J Immunol. 1995;154(4):1735–1747. [PubMed] [Google Scholar]
- 34.Song W, Apodaca G, Mostov K. Transcytosis of the polymeric immunoglobulin receptor is regulated in multiple intracellular compartments. J Biol Chem. 1994;269(47):29474–29480. [PubMed] [Google Scholar]
- 35.Okamoto CT, Shia SP, Bird C, Mostov KE, Roth MG. The cytoplasmic domain of the polymeric immunoglobulin receptor contains two internalization signals that are distinct from its basolateral sorting signal. J Biol Chem. 1992;267(14):9925–9932. [PubMed] [Google Scholar]
- 36.Okamoto CT, Song W, Bomsel M, Mostov KE. Rapid internalization of the polymeric immunoglobulin receptor requires phosphorylated serine 726. J Biol Chem. 1994;269(22):15676–15682. [PubMed] [Google Scholar]
- 37.Breitfeld PP, Casanova JE, McKinnon WC, Mostov KE. Deletions in the cytoplasmic domain of the polymeric immunoglobulin receptor differentially affect endocytotic rate and postendocytotic traffic. J Biol Chem. 1990;265(23):13750–13757. [PubMed] [Google Scholar]
- 38.Mostov K, Su T, ter Beest M. Polarized epithelial membrane traffic: conservation and plasticity. Nat Cell Biol. 2003;5(4):287–293. doi: 10.1038/ncb0403-287. [DOI] [PubMed] [Google Scholar]
- 39.Hunziker W, Whitney JA, Mellman I. Selective inhibition of transcytosis by brefeldin A in MDCK cells. Cell. 1991;67(3):617–627. doi: 10.1016/0092-8674(91)90535-7. [DOI] [PubMed] [Google Scholar]
- 40.Barroso M, Sztul ES. Basolateral to apical transcytosis in polarized cells is indirect and involves BFA and trimeric G protein sensitive passage through the apical endosome. J Cell Biol. 1994;124(1-2):83–100. doi: 10.1083/jcb.124.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matter K, Mellman I. Mechanisms of cell polarity: sorting and transport in epithelial cells. Curr Opin Cell Biol. 1994;6(4):545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 42.Purifoy DJ, Benyesh-Melnick M. DNA polymerase induction by DNA-negative temperature-sensitive mutants of herpes simplex virus type 2. Virology. 1975;68(2):374–386. doi: 10.1016/0042-6822(75)90280-9. [DOI] [PubMed] [Google Scholar]
- 43.Gravotta D, Deora A, Perret E, Oyanadel C, Soza A, Schreiner R, Gonzalez A, Rodriguez-Boulan E. AP1B sorts basolateral proteins in recycling and biosynthetic routes of MDCK cells. Proc Natl Acad Sci U S A. 2007;104(5):1564–1569. doi: 10.1073/pnas.0610700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reich V, Mostov K, Aroeti B. The basolateral sorting signal of the polymeric immunoglobulin receptor contains two functional domains. J Cell Sci. 1996;109(Pt 8):2133–2139. doi: 10.1242/jcs.109.8.2133. [DOI] [PubMed] [Google Scholar]
- 45.Maples CJ, Ruiz WG, Apodaca G. Both microtubules and actin filaments are required for efficient postendocytotic traffic of the polymeric immunoglobulin receptor in polarized Madin-Darby canine kidney cells. J Biol Chem. 1997;272(10):6741–6751. doi: 10.1074/jbc.272.10.6741. [DOI] [PubMed] [Google Scholar]
- 46.Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97(7):877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 47.Novak J, Moldoveanu Z, Renfrow MB, Yanagihara T, Suzuki H, Raska M, Hall S, Brown R, Huang WQ, Goepfert A, Kilian M, Poulsen K, Tomana M, Wyatt RJ, Julian BA, et al. IgA nephropathy and Henoch-Schoenlein purpura nephritis: aberrant glycosylation of IgA1, formation of IgA1-containing immune complexes, and activation of mesangial cells. Contrib Nephrol. 2007;157:134–138. doi: 10.1159/000102455. [DOI] [PubMed] [Google Scholar]
- 48.Cunningham-Rundles C, Brandeis WE, Safai B, O’Reilly R, Day NK, Good RA. Selective IgA deficiency and circulating immune complexes containing bovine proteins in a child with chronic graft versus host disease. Am J Med. 1979;67(5):883–890. doi: 10.1016/0002-9343(79)90749-6. [DOI] [PubMed] [Google Scholar]
- 49.Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol. 2000;150(3):567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breitfeld PP, Harris JM, Mostov KE. Postendocytotic sorting of the ligand for the polymeric immunoglobulin receptor in Madin-Darby canine kidney cells. J Cell Biol. 1989;109(2):475–486. doi: 10.1083/jcb.109.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YT, Stewart DB, Nelson WJ. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol. 1999;144(4):687–699. doi: 10.1083/jcb.144.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song W, Vaerman JP, Mostov KE. Dimeric and tetrameric IgA are transcytosed equally by the polymeric Ig receptor. J Immunol. 1995;155(2):715–721. [PubMed] [Google Scholar]
- 53.Aroeti B, Kosen PA, Kuntz ID, Cohen FE, Mostov KE. Mutational and secondary structural analysis of the basolateral sorting signal of the polymeric immunoglobulin receptor. J Cell Biol. 1993;123(5):1149–1160. doi: 10.1083/jcb.123.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luton F, Mostov KE. Transduction of basolateral-to-apical signals across epithelial cells: ligand-stimulated transcytosis of the polymeric immunoglobulin receptor requires two signals. Mol Biol Cell. 1999;10(5):1409–1427. doi: 10.1091/mbc.10.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apodaca G, Katz LA, Mostov KE. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J Cell Biol. 1994;125(1):67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutation of the major site of N-glycosylation of a recombinant human pIgA does not affect its trafficking in polarized MDCK cells.
A) The glycosylation status of the recombinant pIgA heavy chain was analyzed by probing the membrane with the O-glycan specific lectin Jacalin (top panel) or the N-glycan specific lectin Lens culinaris (bottom panel). For each panel, the arrows point to the upper glycosylated and lower unglycosylated pIgA.
B) Wild-type and NQ mutant pIgA preparation were iodinated and tested for their capacity to bind to pIgR-expressing MDCK cells.
Basolateral endocytosis (C), recycling (D) and transcytosis (E) were assayed. All experiments were performed at least three times in triplicate, and one representative experiment is shown.
In F), the pIgA preparations were analyzed for their ability to stimulate the third step of pIgR apical transcytosis. The data represent the average values (± SD) from three independent experiments. For each pIgA preparation, data were compared to the control sample without pIgA by using a paired Student’s t-test. We determined a statistically significant difference with p<0.005 for both WT and NQ pIgA.