Abstract
The expression of Beta, the single-stranded annealing protein (SSAP) of bacteriophage λ in Escherichia coli promotes high levels of oligonucleotide (oligo)-mediated mutagenesis and offers a quick way to create single or multiple base pair insertions, deletions, or substitutions in the bacterial chromosome. High rates of mutagenesis can be obtained by the use of mismatch repair (MMR)-resistant mismatches or MMR-deficient hosts, which allow for the isolation of unselected mutations. It has recently become clear that many bacteria can be mutagenized with oligos in the absence of any SSAP expression, albeit at a much lower frequency. Studies have shown that inactivation or inhibition of single-stranded DNA (ssDNA) exonucleases in vivo increases the rate of SSAP-independent oligo-mediated mutagenesis. These results suggest that λ Beta, in addition to its role in annealing the oligo to ssDNA regions of the replication fork, promotes high rates of oligo-mediated mutagenesis by protecting the oligo from destruction by host ssDNA exonucleases.
Introduction and context
Recombineering
It is now possible to introduce almost any desired type of mutational alteration into the genome, phages, or plasmids of Escherichia coli and other bacteria [1-3]. The procedure, known as recombineering (from recombinogenic engineering) can use either single- or double-stranded DNA (ssDNA or dsDNA) molecules. Recombineering with double-stranded linear DNA molecules is achieved by expressing the Red recombination genes of bacteriophage lambda (λ), designated exo, bet, and gam [4,5]. λ Exo has 5′ to 3′ directionality on dsDNA to produce 3′ single strands to which Beta can bind [6,7]. λ Beta belongs to a class of proteins known as the single-stranded DNA annealing proteins (SSAPs), which are known to promote RecA-dependent or -independent recombination, promote annealing of complementary ssDNA in vitro, and share a unique quaternary structure involving the formation of rings of varying sizes (i.e., oligomers containing 12-18 subunits per ring) [8-11]. In vivo, λ Beta and Exo promote recombination between phage genomes via a RecA-dependent or RecA-independent (ssDNA annealing) mechanism [12]. For efficient dsDNA Red recombination, it is necessary to inactivate the host RecBCD exonuclease either by deleting the recBCD genes from the host chromosome or by expression of the λ Gam protein, which binds to and inhibits RecBCD [13-15]. Polymerase chain reaction (PCR) substrates, containing a drug marker flanked by 40-50 bp of target DNA, are typical dsDNA substrates for recombineering. The use of recombineering with dsDNA PCR substrates has revolutionized the methods employed for gene targeting of mouse cells and metabolic engineering [3,16,17]. Targets for these experiments have typically been bacterial artificial chromosomes (BACs) containing large regions of bacterial or eukaryotic DNA, where conventional cloning procedures using restriction enzymes have proved too difficult or impossible to perform.
λ Red-promoted oligonucleotide-mediated mutagenesis
Oligonucleotides (oligos; typically about 70 nucleotides) can also be used by the λ Red system to produce chromosomal alterations [18]. The mutagenesis protocol is carried out by electroporation of an oligo into cells expressing only the λ Beta protein; since the substrate is already single-stranded, λ Exo is not required. For example, a deletion can be made by electroporating cells expressing λ Beta protein with an oligo having 35 nucleotide homologies on both sides of the target gene or region. Oligos can also be designed to create single or multiple base pair changes, insertions, or deletions. In E. coli expressing λ Beta, the frequency of mutagenesis for 1-bp changes is typically in the order of 0.1% of the viable cells plated (Table 1); a phenotypic selection or screen is typically required to easily find the mutant cell. While this is a respectable frequency as far as gene targeting is concerned, the real value of this method is to generate unmarked mutations (those without an associated antibiotic resistance, for example), which requires higher rates of Beta-promoted oligo-mediated mutagenesis.
Table 1. Frequencies of single-stranded DNA oligonucleotide-mediated mutagenesis from Escherichia coli and Pseudomonas syringae.
| Hosts | SSAP | Oligo concentration | Mismatch | SSO-mediated mutagenesis (recombinants per 108 cells) | Reference |
|---|---|---|---|---|---|
| SSAP-dependent | |||||
| E. coli | Beta | 0.1 μg (70-mer) | T-C or A-G | 0.16-4.7 × 105 | [36] |
| E. coli | Beta | 0.1 μg (70-mer) | C-C | 1.2 × 107 | [36] |
| E. coli (mutS) | Beta | 0.1 μg (70-mer) | T-C or A-G | 0.1-1.6 × 107 | [36] |
| E. coli (mutS) | Beta | 0.1 μg (70-mer) | C-C | 1.3 × 107 | [36] |
| SSAP-independent | |||||
| E. coli | None | 10 μg (70-mer) | C-C | 7.3 × 104 | [38] |
| E. coli | None | 10 μg (70-mer) | T-C | 2.0 × 102 | [38] |
| E. coli (mutS) | None | 10 μg (70-mer) | C-C | 3.5 × 104 | [38] |
| E. coli (mutS) | None | 10 μg (70-mer) | T-C | 3.0 × 104 | [38] |
| P. syringae | None | 1 μg (84-mer) | G-A* | <1.0 × 102 | [38] |
| P. syringae | None | 5 μg (84-mer) | G-A* | 2.4 × 103 | [38] |
*One out of four mismatches created by the oligo (other mismatches include A-G, A-C and A-C). SSAP, single-stranded DNA annealing protein; SSO, single-stranded oligonucleotide.
This mutagenesis frequency is dependent on three conditions: the sequence of the base pair change; which strand of DNA is targeted by the oligo; and the electrocompetence of the cell preparation. By far, the largest effect on the mutation frequency can be observed by the type of change encoded by the oligo. An oligo designed to create a 1-bp change creates a 1-bp mismatch once the oligo is annealed to the chromosome, and thus becomes a substrate for the Dam-directed mismatch repair (MMR) system of E. coli [19,20]. (This is also true for oligos containing 1- to 4-bp insertions or deletions.) Thus, by inhibiting MMR (e.g., by the use of mutS hosts), or by selecting mismatches that are not easily recognized by the MMR system (e.g., C-C), one can drive mutagenesis frequencies to as high as 25% of the survivors of electroporation [21]. A variation of this theme to avoid the MMR system is to flank the intended base pair change with an additional 19 changes in the oligo sequence [22]. Mismatched loops of 5 bp and greater are not easily recognized by the MMR system and transformants containing all the changes are found at a relatively high frequency using pooled transformants. A second electroporation is carried out with an oligo that changes all the bases back to the original sequence, with the exception of the desired base change.
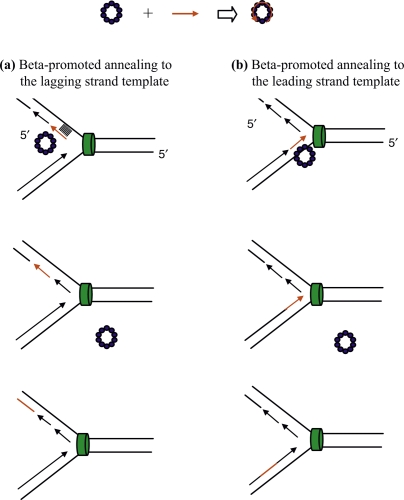
To further increase the mutagenic frequency, one should design the oligo to be complementary to the lagging strand template. It has been observed that oligos targeting the lagging strand template exhibit 3- to 50-fold higher mutagenesis rates relative to oligos that target the leading strand template in E. coli [18]. This bias can reach many thousand-fold in Mycobacterium smegmatis [23] and Pseudomonas aeruginosa (KC Murphy, unpublished observations). These differential rates of targeted gene alteration by complementary oligos have also been observed in mammalian cells, though the results may be influenced in some cases by the presence of transcription through the target site (reviewed by Engstrom et al. [24]). This bias toward the lagging strand led to a proposed mechanism for Beta-promoted oligo-mediated mutagenesis whereby the ‘lagging strand’ oligo is incorporated preferentially at the replication fork, presumably due to the increased presence of ssDNA target sites in this region [18,25] (Figure 1a). In this model, oligos are annealed to regions between Okazaki fragments or perhaps even displace them. Once annealed, the oligos get extended by DNA polymerase I and ligated to the preceding Okazaki fragment. That oligos are physically incorporated into the mutagenized DNA was shown by Huen et al. [26], who showed that biotinylated ssDNA oligos were incorporated into plasmid substrates. The physical incorporation of the oligo was previously seen by Radecke et al. [27] in a mammalian gene targeting study.
Figure 1. Replisome invasion model of single-stranded DNA oligo-mediated mutagenesis.
Top: following electroporation into Escherichia coli host cells expressing bet, the ssDNA oligo (red arrow) is bound by the ring-shaped oligomer of the λ Beta protein (blue). The 3′ end of the primer is designated by the arrowhead. Beta promotes annealing of the oligo to single-stranded regions of the DNA replication fork. The green cylinder represents the replicative helicase DnaB; for clarity, the rest of the replisome is not drawn. The oligo can be complementary to either the lagging strand (a) or leading strand (b) templates. (a) Once annealed to the lagging strand template, the Beta ring is removed, perhaps by some interaction with a component of the replication fork. The oligomer is extended by DNA polymerase I and incorporated into the chromosome by ligation to an adjacent Okazaki fragment. (b) Beta promotes annealing of the oligo just ahead of the polymerase on the leading strand template. After annealing, the Beta ring is removed, perhaps by some interaction with a component of the replication fork. The leading strand polymerase then bumps into the 5′ end of the oligo, leaves its template, and re-initiates DNA synthesis downstream using the 3′ end of the oligo as a primer. Such jumps in leading strand polymerases are thought possible given the gaps in leading strand synthesis that are observed in in vivo studies [33].
Oligos can also target the leading strand template and a mechanism to address this observation is shown in Figure 1b. One imagines that the oligo anneals to the leading strand template ahead of polymerase, though at a lower frequency relative to the lagging strand template because of the limited amount of ssDNA in this region of the fork [28]. In this model, the leading strand polymerase would stop upon encountering the 5′ end of the oligo, disengage the template strand, and reposition itself at the 3′ end of the oligo to reinitiate leading strand synthesis. This process would be analogous to the one encountered by the lagging strand polymerase when it encounters the 5′ end of an Okasaki fragment, where it disengages to reinitiate polymerization at the next priming site. This hopping capability of the lagging strand polymerase has been demonstrated [29] and involves interactions with the clamp loader complex of the replisome (see [30,31] for reviews). Whether the leading strand polymerase possesses the same capability is not known, but there are suggestions it does. For instance, both polymerases are coupled by tethering to the clamp loader [32], suggesting that the leading strand polymerase might possess a recycling mechanism akin to the lagging strand polymerase. Also, both polymerases can leave gaps in their DNA strands following encounters with template lesions (reviewed in [33]), suggesting both polymerases can reinitiate downstream on their templates. Leading strand polymerase reassembly on its template is supported by the ability of DnaG primase to prime synthesis on the leading strand template [34]. Thus, the leading strand polymerase likely has the capacity to disengage its template following an encounter with the 5′ end of an annealed oligo, and reinitiate at the 3′ end, as depicted in Figure 1b. Other models of oligo-mediated recombination, involving the formation of D-loops, annealing of the oligo to the non-transcribed strand of a transcription bubble, and SSAP-directed template switches, have been discussed [24,35] and are not mutually exclusive to the models shown in Figure 1.
Finally, electrocompetent cells should be prepared to maximize DNA uptake. For E. coli, this is easily carried out using standard electrocompetent preparation protocols for recombineering [1,2]. For species of bacteria that are not as electrocompetent as E. coli (e.g., M. smegmatis), van Kessel and Hatfull [23] have reported that co-electroporation of an oligo with a selectable plasmid allows one to screen out cells not competent for DNA uptake. They find that 3-5% of cells that take up the plasmid (which is selectable) also contain the unselected oligo-mediated mutation. The strain can then be easily cured of the sacB-containing plasmid by selection on sucrose.
Major recent advances
Other single-stranded DNA annealing proteins
λ Red-promoted ssDNA oligo mutagenesis is dependent on the expression of only the λ Beta protein. Iyer et al. [11] have previously used computational sequence analyses to classify SSAPs into three evolutionarily distinct superfamilies: λ Beta/RecT, Erf, and Rad52. Each superfamily has its own conserved regions of amino acid sequence and predicted secondary structures, suggesting that each superfamily evolved independently. The large number of potential λ Beta-like recombinases in phage-encoded genomes suggests that other SSAPs might also be found that promote RecA-independent oligo-mediated mutagenesis. Some of these recombinases would be expected to be well adapted for oligo-mediated mutagenesis in their respective hosts. To this end, Datta et al. [36] expressed a number of these Beta-like recombinases from a set of diverse bacteria (including Gram-positives) in E. coli, and examined their ability to promote oligo-mediated mutagenesis. Surprisingly, a number of SSAPs (from Enterococcus faecalis, Legionella pneumophila, and Vibrio cholerae) worked quite well in E. coli, generating recombinants at 25-100% of the level exhibited by λ Beta, while SSAPs from more distantly related non-enteric bacteria (e.g., Bacillus subtilis and M. smegmatis) gave recombinants at a 1000-fold lower rate (though still higher than the rate of spontaneous mutagenesis).
Likewise, van Kessel and Hatfull [23] expressed two mycobacterial SSAPs (Che9c phage gp61 and Halo phage gp43), RecT, and λ Bet in M. smegmatis and measured the relative efficiencies of ssDNA oligo-mediated mutagenesis. Che9c gp61 worked most efficiently, while the Halo gp42 SSAP gave a 10-fold lower rate of mutagenesis. Surprisingly, the E. coli-derived RecT function worked as well as Hal gp42, while λ Beta did not work at all (though this latter result could have been due to poor translation of the E. coli transcript in M. smegmatis). The lower activities of the more distantly related SSAPs in this report and the study by Datta et al. [36] suggest a possible interaction between the phage recombinase and a critical host factor(s). It has been proposed that this putative host factor may be a component of the replicon, as the replication fork has been considered to be the most probable target for the λ Beta protein (though no physical interactions with replication fork proteins have been reported thus far).
Many of these SSAPs are accompanied on their respective genomes by λ-like exonucleases. In the study by Datta et al. [36], four of the SSAPs, when expressed together with their cognate exonucleases, generated recombinants with dsDNA at a rate 1000-fold lower than λ Red, including two SSAPs that showed significant recombination with ssDNA. This result suggests that host proteins may be involved with specific interactions with the phage exonuclease function instead of (or in addition to) the SSAP. Alternatively, it may reflect a higher stringency of the phage recombination systems acting on dsDNA linear substrates relative to ssDNA oligos.
Mechanism of oligo-mediated mutagenesis
It seems reasonable to assume that the ssDNA annealing function of Beta is central to the mechanism of the high level of mutagenesis with ssDNA oligos. This is represented in the models shown in Figure 1 by annealing of the oligo to the ssDNA regions of the replication fork. However, Beta may have another important role. Dutra et al. [37] showed that an E. coli strain deficient for 3′ ssDNA exonucleases (ExoI, RecJ, and EcoVII) was capable of significant oligo-mediated mutagenesis in the absence of any exogenous phage proteins (although at a lower frequency). This result shows that Beta’s role, in addition to its presumed annealing function, likely includes protection of the ssDNA oligo from degradation by host ssDNA exonucleases. As such, this study led to the idea that bacteria that are limited in their ssDNA exonuclease functions may be amenable to some level of ssDNA oligo-mediated mutagenesis in the absence of a phage SSAP.
Recently, a study by Swingle et al. [38] showed that the chromosome of the soil organism Pseudomonas syringae was capable of being modified by ssDNA oligos without expression of an exogenous phage annealing function. Electroporation of P. syringae with 5 μg of an oligo, designed to create a 4-bp change in the rpsL gene that confers streptomycin resistance, generated 2400 StpR colonies per 108 survivors (106-fold greater than the spontaneous rate of StpR). Interestingly, when only 5-fold lower amounts of the oligo were used, the rate of oligo-mediated mutagenesis fell 80-fold, suggesting that excess DNA titrates out an in vivo inhibitor (e.g., ssDNA exonucleases) increasing the lifetime of the oligo. In support of this mechanism, a substrate mixture containing 4 μg of a non-specific carrier oligo and 1 μg of the StpR-targeting oligo gave mutation rates comparable to the use of 5 μg of the targeting oligo alone. Apparently, barriers to oligo-mediated mutagenesis can be overcome by the use of excess oligo via a mechanism that presumably saturates host ssDNA exonucleases.
SSAP-independent and -dependent oligo-mediated mutagenic procedures share similar properties. Mutagenesis rates of both processes show a bias toward oligos targeting the lagging strand of the replication fork, are influenced by the type of mismatch generated (MMR-resistant C-C mismatches show the highest rates), and are independent of RecA, RecB, and RecFOR. These similarities suggests that oligo-mediated mutagenesis is an inherent process that can occur in bacteria at some low level, and that SSAPs like λ Beta can dramatically increase that rate by efficiently protecting the oligo from exonucleolytic attack, and promote annealing of the oligo to ssDNA regions of the replication fork.
One of the main differences between SSAP-dependent and -independent mutagenesis is oligo length requirement. In SSAP-independent analyses, P. syringae showed no differences in the rates of oligo-mediated mutagenesis when oligos between 20 and 120 nucleotides were compared [38]. In a similar test in E. coli, the difference in SSAP-independent mutagenesis between a 20-mer and a 70-mer was only 5- to 10-fold. Conversely, maximal rates of λ Beta-promoted oligo-mediated mutagenesis are observed with a 70-mer and drops at least three orders of magnitude when a 20-mer is used [18]. In the case of the Che9 gp61 SSAP, there was a dramatic 104-fold drop in oligo-mediated mutagenesis when the length of the oligo was reduced from 50 to 36 nucleotides, though interestingly, the authors claim that this did not reflect an inability of gp61 to bind to these oligos [23]. This result is consistent with the reported binding capability of λ Beta to a 36-nucleotide oligo [39]. Clearly, the very high rates of SSAP-dependent oligo mutagenesis require a minimal length of about 50 nucleotides (as opposed to 20 nucleotides for SSAP-independent oligo mutagenesis), which likely reflects some mechanistic requirement for SSAP-promoted protection or annealing of ssDNA in vivo.
The study by Swingle et al. [38] also reported that the chromosomes of E. coli, Shigella flexneri, and Salmonella typhimurium could all be modified by oligos (using rpsL and rpoB target sites) in the absence of exogenous SSAP function, although at much lower frequencies when compared to SSAP-induced mutagenesis (approximately 103- to 104-fold lower). A study by Grogan and Stengel [40] has shown that oligos can also be used to modify the chromosome of the hyperthermophilic archaea Sulfolobus acidocaldarius. These results, and the oligo-mediated gene targeting events that have been observed in eukaryotic systems for many years [41-48], show that exogenous SSAP-independent oligo-mediated mutagenesis is a universal process, and that it likely shares mechanistic similarities across all kingdoms of life.
Future directions
The ability of bacterial chromosomes and BACS to be modified by oligo-mediated mutagenesis allows investigators precise manipulation of genes and regulatory elements down to the single base pair level. Modifications can be induced at a reasonable frequency (0.1%), allowing investigators to find the desired change using pools of transformants and PCR assays. In particular, the mismatch amplification mutation assay (MAMA) PCR has been extremely useful, where an amplicon is generated by the modified target site but not the parental one [49]. Single base pair changes can be inserted in BACs containing bacterial or eukaryotic DNA for downstream applications [22,50]. While the use of MMR-deficient hosts drives the frequency of oligo-mediated mutagenesis higher, future research that allows for increased frequencies of recombineering in MMR-proficient cells would be useful. This notion is based on the concern about the off-target effects of working in an MMR-deficient strain.
Oligo-mediated mutagenesis without the need to express exogenous SSAPs in bacterial species other than E. coli is useful but limited because of the low frequency of mutagenesis (typically a limit of approximately 10-4). However, it is helpful for those species that already have genetic systems in place. For instance, an oligo can be designed to excise a counter-selectable marker (e.g., sacB) from a bacterial chromosome without leaving behind a site-specific scar (i.e., loxP site). Investigators should examine bacterial systems to see if this is possible, especially if DNA sequence analysis suggests a limited number of ssDNA exonucleases are encoded in the host genome. Also, SSAPs from phages other than lambda should be examined for their abilities to promote high levels of SSAP-induced oligo-mediated mutagenesis. In hosts other than E. coli, SSAPs from endogenous phages would likely work well to promote high rates of oligo-mediated mutagenesis, as was recently found for M. smegmatis [23]. A clear application of this technology is to transfer single nucleotide polymorphisms identified in drug-resistant bacterial pathogens to drug-sensitive strains to identify mutations required for bacterial resistance. Such studies could identify the genes involved in the generation of resistance in bacteria and help in the design of new antibiotics.
Finally, a clearer understanding of the molecular mechanism of how the oligo promotes transfer of the mutation into the chromosome is clearly warranted. While the mechanisms in Figure 1 are the most reasonable given the currently available data, other mechanisms may also play a role. Experiments should be designed to test the model further, or perhaps reveal other mechanisms that may be involved, including the formation of D-loops and/or template switch mechanisms [35]. The role of transcription bubbles as targets of the annealing function (one model currently suggested for the mechanism of mammalian targeted gene alteration [24]) is not thought to be a significant contributor to oligo-mutagenesis in bacteria, though it may have a role in some circumstances. One item of special interest would be if a Beta-like analogue exists in eukaryotes, which if transiently induced, would promote the 103- to 106-fold increase in mammalian targeted gene alteration, as is seen when λ Beta is expressed in E. coli cells.
Acknowledgments
Research in the MGM laboratory is supported by grant GM 063790 from the National Institutes of Health.
Abbreviations
- BAC
bacterial artificial chromosome
- dsDNA
double-stranded DNA
- MMR
mismatch repair
- oligo
oligonucleotide
- PCR
polymerase chain reaction
- SSAP
single-stranded DNA annealing protein
- ssDNA
single-stranded DNA
Competing Interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/2/56
References
- 1.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–23. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–99. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 3.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 4.Signer ER, Weil J. Recombination in bacteriophage lambda. I. Mutants deficient in general recombination. J Mol Biol. 1968;34:261–71. doi: 10.1016/0022-2836(68)90251-9. [DOI] [PubMed] [Google Scholar]
- 5.Echols H, Gingery R. Mutants of bacteriophage [lambda] defective in vegetative genetic recombination. J Mol Biol. 1968;34:239–49. doi: 10.1016/0022-2836(68)90249-0. [DOI] [PubMed] [Google Scholar]
- 6.Carter DM, Radding CM. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971;246:2502–12. [PubMed] [Google Scholar]
- 7.Little JW. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J Biol Chem. 1967;242:679–86. [PubMed] [Google Scholar]
- 8.Kmiec E, Holloman WK. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256:12636–9. [PubMed] [Google Scholar]
- 9.Muniyappa K, Radding CM. The homologous recombination system of phage lambda. Pairing activities of beta protein. J Biol Chem. 1986;261:7472–8. [PubMed] [Google Scholar]
- 10.Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci U S A. 1999;96:4279–84. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing vs. invasion in phage lambda recombination. Genetics. 1997;147:961–77. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karu AE, Sakaki Y, Echols H, Linn S. The gamma protein specified by bacteriophage gamma. Structure and inhibitory activity for the recBC enzyme of Escherichia coli. J Biol Chem. 1975;250:7377–87. [PubMed] [Google Scholar]
- 14.Murphy KC. Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–21. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy KC. The lambda Gam protein inhibits RecBCD binding to dsDNA ends. J Mol Biol. 2007;371:19–24. doi: 10.1016/j.jmb.2007.05.085. [DOI] [PubMed] [Google Scholar]
- 16.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–79. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 17.Murphy KC. Chromosomal engineering strategies. In: Smolke C, editor. The Metabolic Engineering Pathway. Boca Raton, FL: CRC Press, Taylor and Francis Group, LLC; 2009. pp. 7.1–7.28. [Google Scholar]
- 18.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:6742–6. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9.0 ExceptionalEvaluated by Martin Marinus 15 Jan 2002
- 19.Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8:154–60. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989;264:6597–600. [PubMed] [Google Scholar]
- 21.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003;100:15748–53. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Ilan Rosenshine 26 Jan 2004
- 22.Yang Y, Sharan SK. A simple two-step, 'hit and fix' method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 2003;31:e80. doi: 10.1093/nar/gng080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 24.Engstrom JU, Suzuki T, Kmiec EB. Regulation of targeted gene repair by intrinsic cellular processes. Bioessays. 2009;31:159–68. doi: 10.1002/bies.200800119. [DOI] [PubMed] [Google Scholar]
- 25.Li XT, Costantino N, Lu LY, Liu DP, Watt RM, Cheah KS, Court DL, Huang JD. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 2003;31:6674–87. doi: 10.1093/nar/gkg844. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Martin Marinus 13 Nov 2003
- 26.Huen MS, Li XT, Lu LY, Watt RM, Liu DP, Huang JD. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34:6183–94. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radecke S, Radecke F, Peter I, Schwarz K. Physical incorporation of a single-stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J Gene Med. 2006;8:217–28. doi: 10.1002/jgm.828. [DOI] [PubMed] [Google Scholar]
- 28.Marians KJ. Replication fork propagation. In: Neidhardt FC, Curtiss RI, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, editors. Escherichi coli and Salmonella Volume 1. Umbarger HE: ASM Press; 1996. pp. 749–63. [Google Scholar]
- 29.Stukenberg PT, Turner J, O’Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–87. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 30.Langston LD, O’Donnell M. DNA replication: keep moving and don't mind the gap. Mol Cell. 2006;23:155–60. doi: 10.1016/j.molcel.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Pomerantz RT, O’Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–64. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Onrust R, Finkelstein J, Turner J, Naktinis V, O’Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. III. Interface between two polymerases and the clamp loader. J Biol Chem. 1995;270:13366–77. doi: 10.1074/jbc.270.22.13358. [DOI] [PubMed] [Google Scholar]
- 33.Wang TC. Discontinuous or semi-discontinuous DNA replication in Escherichia coli? Bioessays. 2005;27:633–6. doi: 10.1002/bies.20233. [DOI] [PubMed] [Google Scholar]
- 34.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–62. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Martin Marinus 24 Feb 2006, Susan T Lovett 01 Mar 2006
- 35.Poteete AR. Involvement of DNA replication in phage lambda Red-mediated homologous recombination. Mol Microbiol. 2008;68:66–74. doi: 10.1111/j.1365-2958.2008.06133.x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Martin Marinus 11 Aug 2008
- 36.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A. 2008;105:1626–31. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutra BE, Sutera VA, Jr, Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci U S A. 2007;104:216–21. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Martin Marinus 19 Feb 2007
- 38.Swingle B, Markel E, Costantino N, Bubunenko MG, Cartinhour S, Court DL. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010;75:138–48. doi: 10.1111/j.1365-2958.2009.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by William Holloman 13 Jan 2010
- 39.Mythili E, Kumar KA, Muniyappa K. Characterization of the DNA-binding domain of beta protein, a component of phage lambda red-pathway, by UV catalyzed cross-linking. Gene. 1996;182:81–7. doi: 10.1016/s0378-1119(96)00518-5. [DOI] [PubMed] [Google Scholar]
- 40.Grogan DW, Stengel KR. Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/lambdaRed and Sulfolobus acidocaldarius recombination systems. Mol Microbiol. 2008;69:1255–65. doi: 10.1111/j.1365-2958.2008.06356.x. [DOI] [PubMed] [Google Scholar]
- 41.Kmiec EB. Targeted gene repair - in the arena. J Clin Invest. 2003;112:632–6. doi: 10.1172/JCI19777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kucherlapati RS. Homologous recombination in mammalian somatic cells. Prog Nucleic Acid Res Mol Biol. 1989;36:301–10. doi: 10.1016/S0079-6603(08)60178-6. [DOI] [PubMed] [Google Scholar]
- 43.Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1988;85:524–8. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen PA, Randol M, Luna L, Brown T, Krauss S. Genomic sequence correction by single-stranded DNA oligonucleotides: role of DNA synthesis and chemical modifications of the oligonucleotide ends. J Gene Med. 2005;7:1534–44. doi: 10.1002/jgm.804. [DOI] [PubMed] [Google Scholar]
- 45.Papaioannou I, Disterer P, Owen JS. Use of internally nuclease-protected single-strand DNA oligonucleotides and silencing of the mismatch repair protein, MSH2, enhances the replication of corrected cells following gene editing. J Gene Med. 2009;11:267–74. doi: 10.1002/jgm.1296. [DOI] [PubMed] [Google Scholar]
- 46.Parekh-Olmedo H, Kmiec EB. Progress and prospects: targeted gene alteration (TGA) Gene Ther. 2007;14:1675–80. doi: 10.1038/sj.gt.3303053. [DOI] [PubMed] [Google Scholar]
- 47.Radecke F, Peter I, Radecke S, Gellhaus K, Schwarz K, Cathomen T. Targeted chromosomal gene modification in human cells by single-stranded oligodeoxynucleotides in the presence of a DNA double-strand break. Mol Ther. 2006;14:798–808. doi: 10.1016/j.ymthe.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Radecke S, Radecke F, Cathomen T, Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol Ther. 2010;18:743–53. doi: 10.1038/mt.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 50.Swaminathan S, Ellis HM, Waters LS, Yu D, Lee EC, Court DL, Sharan SK. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]