Summary
The mechanisms by which bacterial cells generate helical cell shape and its functional role are poorly understood. Helical shape of the human pathogen Helicobacter pylori may facilitate penetration of the thick gastric mucus where it replicates. We identified four genes required for helical shape: three novel LytM peptidoglycan endopeptidase homologues (csd1–3) and a ccmA homologue. Surrounding the cytoplasmic membrane of most bacteria, the peptidoglycan (murein) sacculus is a meshwork of glycan strands joined by peptide cross-links. Intact cells and isolated sacculi from mutants lacking any single csd gene or ccmA formed curved rods and showed increased peptidoglycan cross-linking. Quantitative morphological analyses of multiple-gene deletion mutants revealed each protein uniquely contributes to a shape-generating pathway. This pathway is required for robust colonization of the stomach in spite of normal directional motility. Our findings suggest that the coordinated action of multiple proteins relaxes peptidoglycan cross-linking, enabling helical cell curvature and twist.
Introduction
The abundant morphological diversity present among bacteria has long been appreciated by microbiologists. Yet only recently has much progress been made toward understanding the mechanisms responsible for generating distinctive cell shapes (Young, 2006; Cabeen and Jacobs-Wagner, 2007). To date, most studies have focused on a select group of model organisms representing the most common shapes: rod (Escherichia coli and Bacillus subtilis), coccoid (Staphylococcus aureus and Streptococcus species), and vibrioid (or curved rod; Caulobacter crescentus). The machinery giving rise to cell shape in helical bacteria remains largely unknown. An exception is the Spiroplasma cytoskeletal apparatus, comprised predominantly of Fib, a protein found in only a few prokaryotic species (Williamson et al., 1991), and MreB, a common prokaryotic cytoskeletal protein homologous to eukaryotic actin (Jones et al., 2001; Bove et al., 2003; Daniel and Errington, 2003). Fib, possibly in conjunction with MreB, forms bundles of filaments in a ribbon-like helix, which attaches to the inner surface of the membrane along the shortest (inner) helical line and twists the cell into a helix (Trachtenberg, 2004; Trachtenberg et al., 2008).
Spiroplasma, however, belong to an unusual class of bacteria, the Mollicutes, which lack the rigid cell wall known as the peptidoglycan (PG, or murein) sacculus. The exocellular sacculus surrounds the cytoplasmic membrane of most bacteria and is composed of stiff glycan strands cross-linked by flexible peptide bridges to form a mesh structure (Vollmer and Bertsche, 2008). PG prevents cell lysis due to turgor pressure and is required to maintain bacterial cell shape. Isolated PG sacculi also retain the morphology of the intact cell (Vollmer and Bertsche, 2008). A group of walled bacteria in which some progress has been made toward understanding helical shape determination are the Spirochetes. These bacteria house flagella in the periplasmic space between the inner and outer membranes that are sometimes required for helical cell shape (Goldstein et al., 1994). Periplasmic flagella presumably act as force-generating cytoskeletal elements that bend the cell into a helix (Wolgemuth et al., 2006). However, some Spirochete species retain helical morphology in the absence of periplasmic flagella and the cellular machinery involved in generating their helicity has yet to be identified (Bromley and Charon, 1979; Ruby et al., 1997).
Here we investigate the helical shape of Helicobacter pylori. H. pylori is a member of the Epsilonproteobacteria, a class of bacteria composed almost exclusively of helical and curved organisms. H. pylori’s habitat is the human stomach, which it colonizes in approximately 50% of the world population. H. pylori infection is associated with the development of chronic gastric inflammation that can lead to ulcers and gastric cancer in a subset of those infected (Kusters et al., 2006). H. pylori’s helical cell shape is conserved in human isolates (Goodwin et al., 1985), although the pitch of the helix varies among laboratory strains (L. Sycuro, unpublished observations). This has given rise to the hypothesis that H. pylori’s helical shape serves an important function in pathogenesis (Montecucco and Rappuoli, 2001; Cover and Blaser, 2009). The prevailing theory is that helical shape enhances H. pylori’s flagellar motility through the viscous epithelial mucus layer in which it resides by a cork screw mechanism (Hazell et al., 1986).
Helical cell shape can be thought of as the sum of three morphogenic components: cell elongation, curvature, and twist. Recent studies of the Gram(−) model organisms E. coli and C. crescentus have provided significant insight into the genes and mechanisms these species use to elongate the cell body and generate curvature. While H. pylori encodes some of these genes, others appear to be absent or too highly divergent for sequence-based identification. Specifically, H. pylori encodes all three of the high molecular weight penicillin binding proteins (PBPs) required for PG glycan synthesis (transglycosylation via PBP1) and peptide cross-linking (transpeptidation via PBP1, PBP2, and PBP3) (Tomb et al., 1997; DeLoney and Schiller, 1999). However, low molecular weight PBPs with endopeptidase and/or carboxypeptidase activities that contribute to PG hydrolysis and post-synthetic modification of cross-linked and uncross-linked peptide chains have not been identified in H. pylori. In E. coli and C. crescentus, cell body elongation requires the actin homologue MreB, which, in conjunction with other cytoskeletal filaments and scaffolding proteins (i.e. FtsZ, MreC, MreD, RodA, and RodZ), appears to spatially position PG synthesis in roughly helical bands or patches along the cell body resulting in rod elongation (Cabeen and Jacobs-Wagner, 2007; Alyahya et al., 2009; Bendezu et al., 2009). Although H. pylori encodes all of these proteins, their role in elongating the H. pylori cell body has not been confirmed. CreS, an intermediate filament homologue in C. crescentus, is required for that organism’s cell curvature (Ausmees et al., 2003). Through interactions with MreB, the CreS filament is tethered longitudinally along the inner membrane, creating a growth differential that limits PG synthesis rates along the CreS-lined sidewall and enables faster rates along the opposite sidewall, thus forming a curved cell body (Cabeen et al., 2009). Intermediate filament-like proteins that may influence cell shape maintenance were recently identified in H. pylori (Waidner et al., 2009).
Biophysical modeling has recently suggested an alternative pathway to generating cell curvature and twist through local alteration of PG cross-link number or length (Huang et al., 2008). Here we present biological evidence supporting this model with the identification of four proteins that function to generate H. pylori’s helical shape through alterations in PG cross-linking. We show that these proteins are conserved in other Epsilonproteobacteria as well as curved and helical Gammaproteobacteria, suggesting this approach to generating cell shape may be common among Gram(−) bacteria. We also examine the fitness of non-helical H. pylori mutants in the mouse stomach and find they are deficient despite apparently normal motility in vitro.
Results
A putative metallopeptidase, Csd1, is required for the helical cell shape of H. pylori
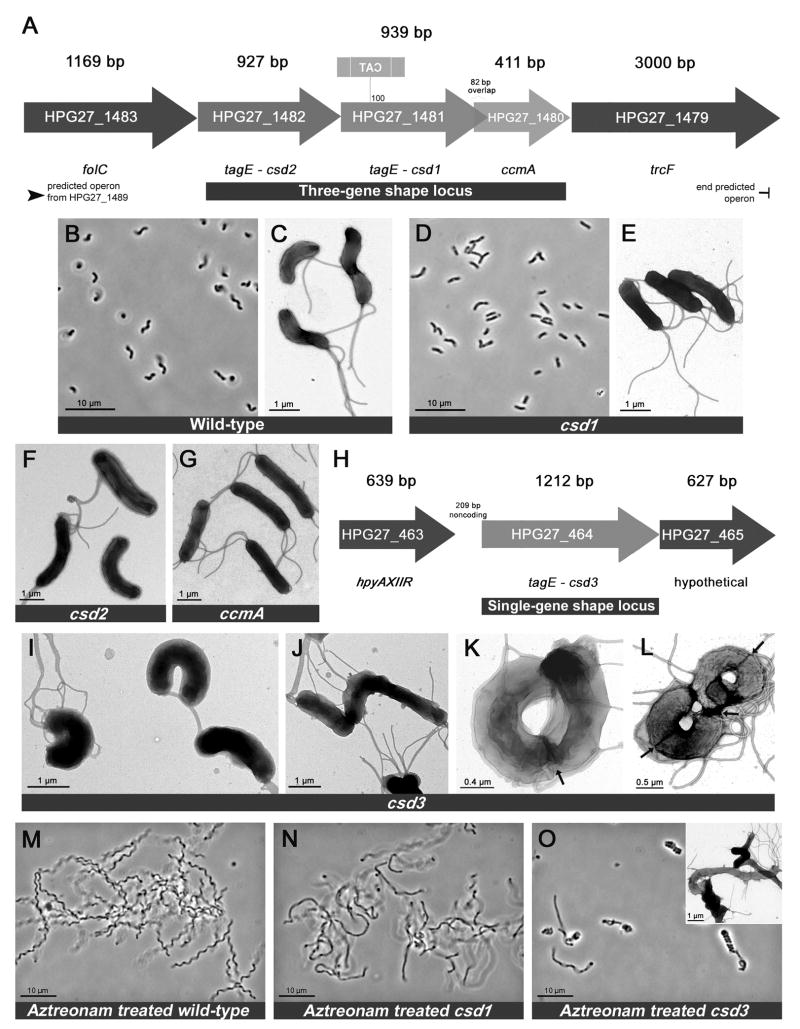
To identify genetic determinants of H. pylori’s conserved helical cell shape, we visually screened a library of random transposon insertion mutants constructed in strain G27 (Salama et al., 2004) for clones with shape defects. Of 2,000 clones screened, nine with altered morphology were selected, including one with curved rod (vibrioid) rather than helical morphology. Amplification of the DNA sequence flanking the transposon revealed this clone’s insertion site was within HPG27_1481 (Figure 1A), a gene annotated as tagE due to its homology to a ToxR-activated gene in Vibrio cholerae (Kovach et al., 1994). V. cholerae contains multiple TagE homologues and although one has been crystallized and shown to contain a metallopeptidase active site similar to that of the Staphylococcus aureus endopeptidase LytM (peptidase family M23; proteins with this domain will be referred to as LytM peptidases for clarity), their function remains unknown (Ragumani et al., 2008). Protein structure queries (Bennett-Lovsey et al., 2008) revealed HPG27_1481 threaded onto the tertiary structures of the crystallized V. cholerae TagE protein (E-value 3.9E−25) and S. aureus LytM (E-value 7.3E−23). Using these models we identified conserved LytM active site residues in the HPG27_1481 protein (Figure 3A). Targeted deletion of HPG27_1481 coding sequence recapitulated the morphological phenotype of the transposon mutant (Figure 1D–E and S1C) and was complemented by reintroduction of HPG27_1481 at a distal chromosomal locus, rdxA, which is often used for complementation in H. pylori (Smeets et al., 2000) (Figure S2B–C). Having identified HPG27_1481 as a putative LytM peptidase involved in producing helical versus curved rod morphology, we named this gene csd1 (cell shape determinant).
Figure 1.
H. pylori cell shape loci and their associated mutant morphologies. A) Three-gene shape locus identified in visual screen. The insertion site of the curved rod shape mutant’s transposon encoding chloramphenicol acetyltransferase (cat) in reverse orientation occurred approximately 100 bp into the HPG27_1481 open reading frame. B–G) Phase contrast and transmission electron microscopy (TEM) images of wild-type and mutant cells. H) Single-gene shape locus containing csd3. I–L) TEM images of single csd3 mutant cells (I–J) and csd3 mutant cells that are elongated and preparing to divide (K–L, septa indicated by arrows). M–O) Phase contrast and TEM images (inset) of wild-type and mutant cells treated with the filamenting drug aztreonam. Strains used: LSH13, KGH8, KGH10, LSH112. See also Figures S1 and S2.
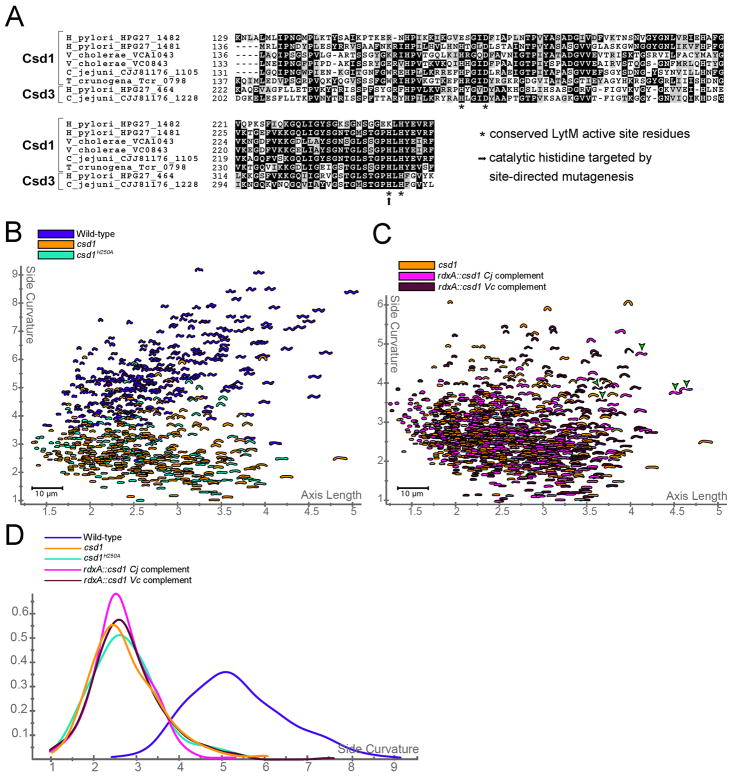
Figure 3.
Conservation of Csd proteins in Epsilonproteobacteria and Gammaproteobacteria. A) Multisequence alignment of LytM domains in H. pylori, C. jejuni, V. cholerae, and T. crunogena Csd1 and Csd3 protein homologues listed by species and locus tag. Stars indicate positions of conserved LytM active site motifs HXXXD and HXH (Ragumani et al., 2008). The arrow shows histitidine residue mutated to alanine (H250A). B) Distribution of wild-type, csd1 null, and csd1H250A point mutant populations. C) Distribution of csd1 null population compared to cross-species complementation of csd1 from C. jejuni (Cj) and V. cholerae (Vc). Semi-helical shapes rarely observed in cross-species complementation strains and never in the parent csd1 null are marked with green arrowheads. D) Smooth histograms showing the side curvature distribution for the above populations. All mutant and complement populations are significantly different from wild-type (bootstrapped K–S p-value < 0.00001), but indistinguishable from csd1 null (p > 0.9). Strains used: NSH57, LSH13, LSH153, NSH142, NSH144..See also Table S1 and Figure S3.
Csd1 is part of a three-gene locus required for helical cell morphology
All sequenced H. pylori strains encode a second LytM peptidase (HPG27_1482) immediately upstream of csd1 that is 53% similar and 29% identical to Csd1 (Figure 1A). We designated this gene csd2 upon discovering its deletion also yielded cells with curved rod morphology (Figure 1F) and complementation restored helicity (Figure S2C). The hypothetical protein encoded downstream of csd1 is a homologue of ccmA (curved cell morphology, HPG27_1480, Figure 1A), a gene of unknown function shown to be important for determining straight rod versus curved rod morphology in Proteus mirabilis (Hay et al., 1999). Deletion of H. pylori ccmA again resulted in curved rod morphology (Figure 1G) and could be complemented (Figure S2B–C).
Microscopic examination of the three curved rod mutants suggested slight variations in the degree of cell curvature, with csd2 the most curved and ccmA the least. No helical cells were seen in any of the mutant cell populations. To distinguish whether the mutants had completely lost the ability to form helices or their helical turns had a longer period such that individual cells only appeared curved, we induced cell filamentation with the drug aztreonam, an inhibitor of septal PG synthesis. In comparison to the regular pitch of helical wild-type cells, there did not appear to be any pattern to the mutants’ slight bends and curves (Figure 1M–N, Figure S1D, and data not shown). In addition to the loss of helicity, all three were slightly wider than wild-type (7–14%), but not significantly different in cell length (Figure S2D–E). In conclusion, deletion of any single gene in this three-gene locus led to impaired helical twist and gene-specific reductions of curvature.
Loss of csd3 results in variable curved rod morphologies
All sequenced H. pylori strains encode a third LytM peptidase at a separate locus (HPG27_464, Figure 1H) that is 39% similar and 21% identical to Csd1. Deletion of this gene, designated csd3, resulted in curved rods with varying degrees of curvature. Although most csd3 mutant cells appeared highly curved or “c”-shaped (Figure 1I), some had little or no curvature (Figure 1J). Elongated cells (those readying to divide or filamented with aztreonam) appeared as concatenated “c”-shapes that sometimes wound into coils (Figure 1K, 1O, S1E) or figure-eights (Figure 1L). Of 100 cells inspected by TEM, we observed 17% straight or bent, 53% “c”-shaped, 25% coiled or figure eight, and 5% coccoid. Csd3 mutant cells were slightly wider (13%) and shorter (10%) than wild-type cells (Figure S2D–E), and normal helical morphology was restored with complementation (Figure S2C). In sum, loss of csd3 resulted in a myriad of highly aberrant cell morphologies stemming from variable curvature and loss of normal helical pitch.
All four shape genes uniquely contribute to a helical shape-generating program
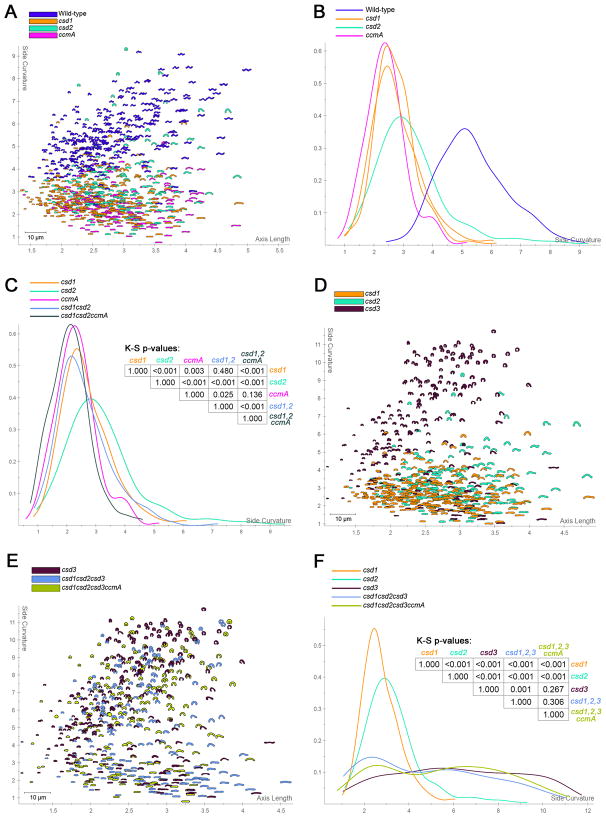
Wild-type cells display a range of morphologies and the differences in shape between our mutant strains were subtle. To further explore functional relationships among these shape genes we constructed a series of multiple-gene deletion mutants and developed software tools for quantitatively analyzing their morphologies. By capturing images of hundreds of cells and systematically measuring shape parameters we can rigorously establish differences among strains. Figure S2A illustrates our shape analysis methodology, which consists of collecting high magnification phase contrast images of each strain and recording each cell’s shape as a polygonal outline. The “central axis” of each cell, running from pole to pole, is then determined algorithmically and used to measure cell length, while the degree of cell body curvature is determined directly from the cell outline (excluding the poles) (Lacayo et al., 2007). These morphological parameters clearly delineate wild-type from mutant populations and the subtle differences in the cell body curvature among csd1, csd2, and ccmA mutant populations (Figure 2A–B).
Figure 2.
Morphological characterization of single and multiple-gene deletion mutants. A, D, E) Scatter plots arraying wild-type and/or mutant populations by cell length (x-axis) and cell curvature (y-axis). A small proportion (~14–18%) of cells in mutant strains containing a csd3 deletion were so highly curved as to erroneously appear coccoid after image processing; these cells were manually removed from the analysis. B, C, F) Smooth histograms displaying population cell curvature (x-axis) as a density function (y-axis) and p-values from bootstrapped Kolmogorov–Smirnov statistical comparisons. Replicate csd1 mutant populations used to define the null distribution for statistical comparisons of curved rod strains are shown in panel B. Strains used: NSH57, LSH13, KGH8, KGH10, LSH112, LSH129, LSH130, LSH136, LSH143.
We constructed a csd1csd2 double mutant and a csd1csd2ccmA triple mutant by deleting the appropriate coding regions. Visually, the csd1csd2 mutant’s morphology appeared most similar to that of the csd1 mutant and the csd1csd2ccmA mutant’s morphology appeared most similar to that of the ccmA mutant. The curvature distributions shown in Figure 2C confirmed these observations. Taken together, these results suggest these three genes act in the same pathway to generate helical cell shape.
We next constructed the triple csd1csd2csd3 mutant and the quadruple csd1csd2csd3ccmA mutant. While the csd3 mutant population is clearly distinguishable from the other single gene csd mutants (Figure 2D), the triple csd mutant and quadruple mutant populations overlap completely with the csd3 mutant, showing the same range of morphological variability and high degree of maximal curvature (Figure 2E–F). These findings suggest csd3 functions in the same or a related shape-generating pathway as csd1, csd2, and ccmA, but csd3 may antagonize the activity of the other three genes. Furthermore, while each of the four genes is required for generating H. pylori's normal helical shape, none are absolutely required for cell curvature.
Csd proteins are conserved in Epsilonproteobacteria, as well as vibrioid and helical members of the Gammaproteobacteria
Having established an important role for the Csd proteins and CcmA in H. pylori’s cell shape we investigated their conservation in other Epsilonproteobacteria and more broadly throughout the Proteobacteria. Proteins containing a conserved C-terminal LytM peptidase domain are widespread in bacteria leading us to focus on Csd1 and Csd3 BLAST hits with good sequence alignment over at least 75% of their length and a minimum of 50% total amino acid similarity. Csd1 showed the broadest conservation with a homologue present in most Epsilonproteobacteria and in 14 species representing five different orders within the Gammaproteobacteria, including all members of the Vibrionaceae and Thiomicrospira crunogena, a curved-to-helical Gammaproteobacterium (Table S1, Figure 3A). csd1 is adjacent to ccmA in most of the curved and helical species. Some Vibrionaceae genomes contained two well-conserved Csd1 homologues, but each was more closely related to Csd1 than Csd2, suggesting the two Vibrionaceae peptidases arose from a separate duplication or transfer event than that which gave rise to Helicobacter Csd1 and Csd2. Indeed, Csd2 homologues were found only in H. pylori and H. hepaticus. Csd3 homologues, which contain an N-terminal extension not present in Csd1 and Csd2, were well conserved in all five strains of Helicobacter and throughout the Epsilonproteobacteria, but were not identified in other classes of Proteobacteria.
All the csd1 and csd3 homologues identified preserve conserved residues predicted to function in catalysis (stars Figure 3A). Mutation of one such residue, H250A (arrow Figure 3A), in the endogenous csd1 gene resulted in a slightly curved rod morphology indistinguishable from the null allele (Figure 3B–C, S3A). We attempted to complement the shape phenotype of the csd1 null mutant with the C. jejuni and V. cholerae homologues. Unlike the csd1 gene from H. pylori (Fig. S2B), neither homologue fully restored normal shape (Figure 3C–D), but occasional cells displayed additional bends or an S-like shape never seen in the null allele strain (Figure 3C–D, S3B–C). Based on the heterogeneity of the shapes of the cross-species complementation strains it is difficult to say whether Csd homologues contribute to cell morphology in other Proteobacteria, but in H. pylori, peptidase activity is required for Csd shape-generating function.
Global peptidoglycan analyses suggest helical cell shape is specified by peptidoglycan peptide cross-linking
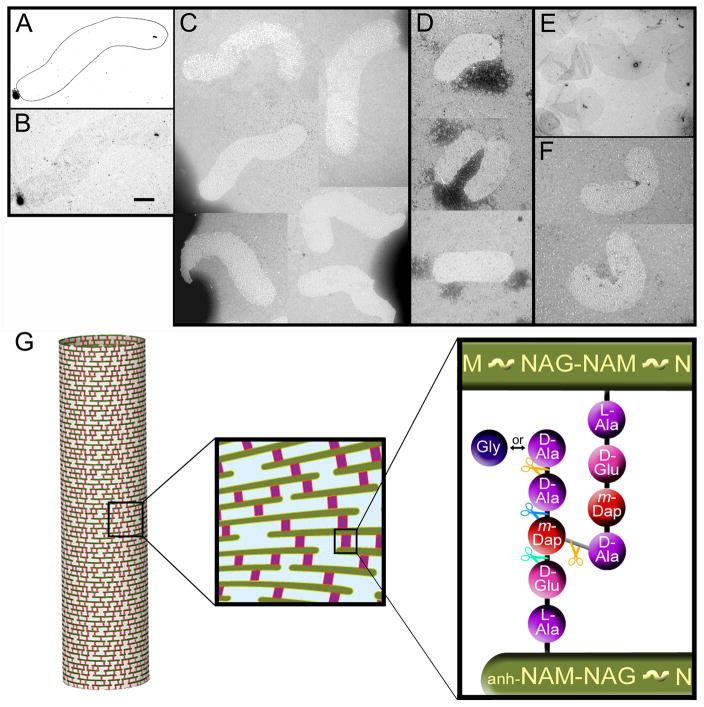
The presence of LytM domains in the Csd proteins suggests they are PG endopeptidases or carboxypeptidases. We first established that the cell shape changes observed in the csd mutants coincided with changes in the shape of isolated mutant PG sacculi (Figure 4A–F). We observed that sacculi from H. pylori behaved differently than those from E. coli or C. crescentus in that they contrasted very poorly with uranyl acetate for visualization by electron microscopy (Figure 4B, E). Hence, the H. pylori sacculi required detection by negative staining (Figure 4C–D, F). The reason for this difference is not known, but H. pylori PG may have wider pores and fewer sites for binding of heavy metal ions than PG from other species. The isolated sacculi nevertheless displayed the shape of intact cells: helical rod for wild-type (Figure 4A–C) and rods with varying curvature for csd1 and csd3 mutants (Figure 4D–F). Thus, as in straight and curved rod bacteria with a PG cell wall, the structural determinant of cell shape in H. pylori is the PG sacculus.
Figure 4.
Transmission electron microscopy (TEM) images and schematic illustration of peptidoglycan (PG) sacculi. A–B) Schematic drawing (A) outlining the poorly visible uranyl acetate-stained wild-type H. pylori sacculus depicted in the image directly below (B). C) Negatively stained wild-type H. pylori sacculi. Morphological variability among the sacculi is a product of their two-dimensional flattening during fixation on the grid, but the apparent wave form of the sacculi approximates the helical morphology of intact cells as observed by TEM (Figure 1C). D–F). csd1 (D) and csd3 (E–F) mutant sacculi illustrating their slightly (csd1) or strongly (csd3) curved rod morphologies, as seen in the intact cells (Figure 1E and 1I). Panel E shows sacculi contrasted with uranyl acetate, others are negatively stained. The scale bar (0.5 μm) applies to all images. Strains used: LSH108, LSH115, LSH119. G) Schematic illustration of PG mesh structure that envelops the cell and is composed of glycan strands (green) interconnected by peptide cross-links (red). Glycan consists of repeating N-acetylglucosamine (NAG or GlcNAc)–N-acetylmuramic acid (NAM or MurNAc) disaccharides with a 1,6–anhydro ring (anh) on NAM sugars at strand termini. Peptide chains extend from each NAM. Shown is the tetra–penta-peptide muropeptide with the peptide cross-link highlighted in grey. m-Dap refers to meso-diaminopimelic acid. Peptide bonds marked with orange scissors may be hydrolyzed by DD-endopeptidases (cleaving DD-peptide bridges) or DD-carboxypeptidases (producing tetra-peptide), those marked with blue scissors may be hydrolyzed by LD-endopeptidases (producing tri-peptide), and those marked by green scissors may be hydrolyzed by DL-endopeptidases or DL-carboxypeptidases (producing di-peptide). Altogether, these modification activites give rise the other muropeptides depicted in Figure S4A.
We then analyzed the global muropeptide composition (described in Figures 4G and S4A) of csd1, csd2, and csd3 null mutants and the csd1H250A point mutant compared to wild-type, as well as the csd1 and csd3 complemented strains. While glycan strand length was unchanged, the content of the peptide side chains in each mutant was significantly altered from wild-type (Table 1, Figure S4B, and Table S2). The most dramatic change was a 26–49% increase in tetra–penta-peptide cross-linked dimers, which coincided with a 8–33% decrease in tetra-peptide monomers in all mutants. In the two complemented strains these muropeptides were unchanged from wild-type levels or the inverse change was observed. The latter effect is likely due to higher expression levels from the rdxA locus relative to the native loci (qRT-PCR data not shown). For these changes to be perfectly balanced we would have expected a decrease in penta-peptide monomers in the mutants, but this was only observed in the csd2 mutant. This discrepancy may result from trimming of penta-peptide monomers by other endopeptidases or carboxypeptidases, yielding tetra-, tri-, and/or di-peptide monomers. Accordingly, we observed decreases of greater than 10% in the tri-peptide monomer content of the csd1H250A, csd2 and csd3 mutants, and the dipeptide monomer content of the csd1 null mutant; these changes were reversed in the csd1 and csd3 complemented strains. We also tested the profile of the ccmA mutant and found that it too contained significant alterations in its muropeptide content relative to wild-type, including the same increase in tetra–penta-peptide cross-linked dimers and decrease in tetra-peptide monomers as the csd mutants. Since CcmA does not contain any recognizable peptidase motif, it may function through interaction with Csd peptidases. Together these data suggest the Csd proteins could be endopeptidases that cleave tetra–penta-peptide PG cross-links.
Table I.
Summary of global peptidoglycan composition in mutant and complemented strains.
| Muropeptide species and structural features | Area – % of of each muropeptide1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild-type | csd1 | csd1 rdxA:: csd1 | csd3 | csd3 rdxA:: csd3 | csd2 | ccmA | Wild-type2 | csd1H250A2 | |
| Monomers (total) | 56.97 | 52.84 | 54.16 | 50.54 | 57.29 | 51.03 | 51.55 | 53.06 | 51.04 |
| monomer di | 1.90 | 1.65 | 2.98 | 2.14 | 3.38 | 2.21 | 3.00 | 2.06 | 2.24 |
| monomer tri | 4.87 | 4.57 | 3.96 | 3.35 | 5.13 | 4.17 | 3.99 | 4.37 | 3.65 |
| monomer tetra | 9.35 | 7.11 | 9.59 | 6.31 | 11.72 | 7.12 | 7.82 | 8.93 | 8.22 |
| monomer penta | 40.85 | 39.50 | 37.63 | 38.75 | 37.06 | 37.52 | 36.74 | 37.69 | 36.94 |
| monomers anhydro | 2.05 | 1.43 | 1.61 | 1.48 | 1.62 | 1.55 | 1.49 | 1.41 | 1.32 |
| Dimers (total) | 38.01 | 43.79 | 40.20 | 41.64 | 37.50 | 43.42 | 42.68 | 39.18 | 44.74 |
| Dimers tetra–tri | 4.72 | 3.83 | 4.84 | 3.18 | 4.78 | 4.54 | 4.44 | 4.33 | 3.80 |
| Dimers tetra–tetra | 13.58 | 13.98 | 15.42 | 9.17 | 15.29 | 14.10 | 15.16 | 15.07 | 16.05 |
| Dimers tetra–penta3 | 19.71 | 25.98 | 19.94 | 29.30 | 17.43 | 24.78 | 23.08 | 19.77 | 24.89 |
| Dimers anhydro | 12.67 | 13.29 | 15.58 | 14.47 | 15.25 | 14.42 | 13.95 | 16.32 | 17.43 |
| Di-peptides (total) | 1.90 | 1.65 | 2.98 | 2.14 | 3.38 | 2.21 | 3.00 | 2.06 | 2.24 |
| Tri-peptides (total) | 7.23 | 6.49 | 6.38 | 4.94 | 7.52 | 6.44 | 6.21 | 6.54 | 5.54 |
| Tetra-peptides (total) | 35.14 | 35.99 | 37.40 | 31.71 | 38.12 | 35.88 | 36.74 | 36.06 | 38.61 |
| Penta-peptides (total) | 50.70 | 52.50 | 47.61 | 53.40 | 45.78 | 49.91 | 48.28 | 47.57 | 49.39 |
| Glycine (total) | 5.51 | 6.52 | 4.83 | 7.30 | 5.03 | 6.35 | 5.95 | 4.30 | 5.88 |
| glycine monomer | 4.48 | 5.06 | 3.97 | 5.65 | 4.40 | 4.99 | 4.70 | 3.51 | 4.57 |
| glycine dimer | 2.05 | 2.93 | 1.72 | 3.30 | 1.26 | 2.72 | 2.51 | 1.57 | 2.63 |
| % Peptides in cross-links | 38.01 | 43.79 | 40.20 | 41.64 | 37.50 | 43.42 | 42.68 | 39.18 | 44.74 |
| Mean glycan chain length | 11.93 | 12.38 | 10.64 | 11.47 | 10.82 | 11.42 | 11.82 | 10.45 | 9.97 |
| % Anhydro termini at cross-links | 75.52 | 82.25 | 82.90 | 83.01 | 82.45 | 82.31 | 82.45 | 85.27 | 86.85 |
Percentages calculated as per (Glauner et al., 1988). Underlined, value differs by more than 10% from that of wild-type; underlined and bold, value differs by more than 20% from that of wild-type. Strains used were LSH108, LSH110, LSH115, LSH119, LSH121, LSH126 (experiment 1), LSH100, LSH154 (experiment 2)
Analysis performed in a separate experiment.
Inclusive of tetra–penta, tetra–penta(Gly5), and tetra–penta anhydro peaks from Table S2.
For csd1, csd2, and ccmA, the increases in tetra–penta-peptide cross-linking correlated with a loss of helical twist and cell curvature. The csd3 mutant also lost normal helical twist, but many cells displayed a higher degree of cell curvature. The csd3 mutant exhibited other significant changes in cross-linked muropeptide content; both tetra–tetra-peptide and tetra–tri-peptide dimers were decreased by over 30% and restored with complementation. It is thus possible that Csd3 also exhibits a carboxypeptidase activity, converting tetra–penta-peptide cross-links into tetra–tetra-peptide cross-links, which are further degraded to tetra–tri-peptide cross-links by another carboxypeptidase. The highly curved morphology observed for csd3 may result from the loss of tetra–tetra-peptide and tetra–tri-peptide cross-links observed uniquely in this mutant’s PG. In sum, each of our mutants exhibited changes in glycan strand connectivity associated with variation in cell curvature and twist.
Changes in the csd mutants’ cell shape and cell wall composition minimally affect motility and do not alter growth or sensitivity to stomach environment stresses
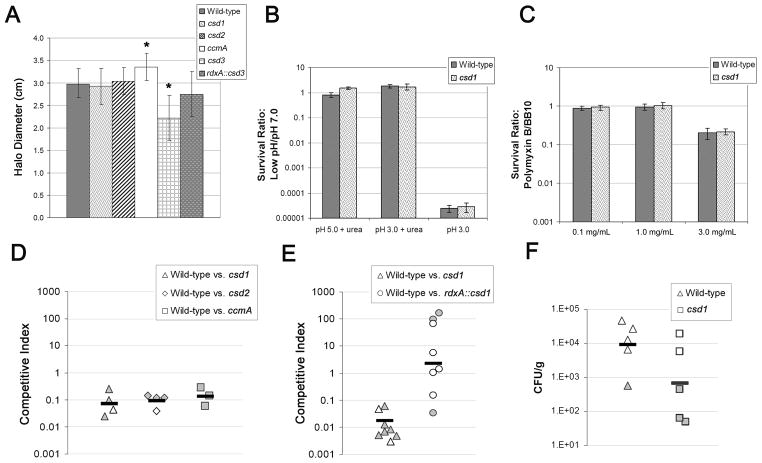
All the mutants described here were normally flagellated and motile in broth (data not shown), but the cork screw model for helical shape posits that they should be impaired for motility in gel-like media. We characterized their ability to swim directionally in a soft agar motility assay and found each performed as well or better than wild-type with the exception of the csd3 mutant, which formed smaller halos (Figure 5A). This motility defect might be explained by the observation that the coiled and nearly-coiled csd3 mutant cells swam in tight circles in broth, while cells with less curvature displayed normal directional motility. To further explore a relationship between motility and loss of normal helical twist we used video microscopy to compare the swimming velocity of wild-type and csd1 mutant cells in media with and without methylcellulose. We observed the wild-type swimming 22.0 μm/sec on average in both media (broth 22.0 ± 13.7 μm/sec, 0.5% methylcellulose 22.3 ± 15.8 μm/sec, mean of 20 cells ± SD) and the csd1 mutant swimming 16 μm/sec on average in both media (regular broth 15.8 ± 8.9 μm/sec, 0.5% methylcellulose 15.9 ± 8.5 μm/sec, mean of 20–21 cells ± SD). Thus the curved rod mutant was not hampered in swimming velocity compared to wild-type in either broth or viscous methylcellulose solution (two-sided Student’s t-tests p > 0.05).
Figure 5.
Phenotypes of non-helical mutants and complemented strains in the mouse stomach and related in vitro conditions. Complementation strains have the native locus disrupted by deletion and insertion of cat and the indicated gene expressed from the rdxA locus. A) Motility phenotype of mutant and complemented strains in soft agar (mean halo diameter inclusive of two or three independent experiments totaling 40–50 stabs per strain ± SD in 0.3% soft agar after four days). Stars indicate significant difference from wild-type (p < 0.05, ANOVA with Bonferroni correction). B–C) Survival in stomach stress conditions from two independent experiments, each with four replicates per strain and condition (mean survival ratio ± SEM). D–E) One week C57BL/6 mouse competition data. Each data point represents a single mouse. Data are plotted as a competitive index: [CFU/mLMUT:CFU/mLWT stomach output]/[CFU/mLMUT:CFU/mLWT inoculum]. Filled points indicate mice from which only one strain was recovered, causing the competitive index to be driven largely by the load of the infecting (wild-type) strain. Mean loads were 560 CFU/stomach, range 100–1050 (D) and 6600, range 2100–19100 (E). Black bars are the geometric mean. F) One week C57BL/6 single strain colonization data. Each data point represents the load (CFU/g) from a single mouse. Filled points indicate mice from which no bacteria were recovered and are plotted at the detection limit. Black bars are the geometric mean. Strains used: A, D, and F) LSH100, LSH113, LSH141, LSH142, LSH119, LSH126; B–C) NSH57, LSH13; E) NSH57, LSH11, LSH102. See also Figure S5.
Each of the mutant strains grew as well as wild-type through log and into stationary phase in single-strain cultures and during 72 hours of log phase growth in co-culture with wild-type (Figure S5A–H). While H. pylori in log phase display helical morphology, the cells undergo a shape transition to a non-culturable coccoid form during prolonged culture in stationary phase (Benaissa et al., 1996; Andersen and Rasmussen, 2009). All four mutants were capable of fully transforming into non-culturable coccoids, but csd3 mutants were slightly delayed in the timing of the process (Figure S5I and data not shown).
Since all the mutants exhibited similar alterations in their peptidoglycan that might alter the integrity of the cell wall, we subjected csd1 cells to stresses H. pylori encounters in the stomach: antimicrobial peptides, acid, and medically administered amoxicillin, an antimicrobial that targets the cell wall. The csd1 mutant survived as well as wild-type upon exposure to the antimicrobial peptide polymyxin and low pH (Figure 5B–C). We also found the csd1 strain was slightly more resistant to amoxicillin than wild-type (minimum inhibitory concentration (MIC): csd1 0.079 ± 0.020 μg/ml, wild-type 0.045 ± 0.014 μg/ml, mean ± SD). Collectively, these data show that the cell wall changes produced in these mutants do not appreciably alter cell wall integrity.
Curved rod H. pylori mutants are outcompeted by helical wild-type cells in the mouse stomach
While we observed no apparent enhancement of motility mediated by helical shape, we wondered whether helical cell shape might have some other selective advantage in the gastric niche. We thus tested the fitness of the csd1, csd2, and ccmA mutants alongside wild-type in a mouse stomach colonization assay. All three mutants were strongly outcompeted by wild-type at one week, with mutant bacteria recovered from only a few mice, and the csd1 phenotype was complemented (Figure 5D–E). In order to determine whether the cell shape mutants were capable of colonizing and surviving in the stomach in the absence of wild-type bacteria we infected mice with the csd1 mutant alone. Although only two of the five csd1-infected mice were colonized (compared to four of five mice infected with wild-type), both were colonized to wild-type levels (Figure 5F). These data indicate that normal helical shape and/or peptidoglycan cross-linking promotes efficient stomach colonization by H. pylori.
Discussion
Here we have identified four genes (csd 1–3 and ccmA) that each uniquely affect bacterial cell morphogenesis but not septation or growth of the helical bacterium H. pylori. All four proteins influence peptide cross-linking within the peptidoglycan sacculus and three encode LytM peptidase homologues suggesting they may be endopeptidases and/or carboxypeptidases that directly hydrolyze PG cross-links or otherwise modify PG. The total extent of peptide cross-linking in the mutants did not change by more than 15%, but the abundance of specific cross-linked muropeptides changed as much as 50%. This combined with the observation that isolated mutant sacculi displayed the same alterations in shape as intact cells suggests that even subtle perturbations of PG cross-linking are sufficient to disrupt helical cell shape in H. pylori.
To our knowledge an association between PG cross-linking and cell curvature or twist has not previously been established in a biological system. Rod-shaped C. crescentus creS mutants did not show alterations in PG cross-linking (Cabeen et al., 2009). Also, studies of the straight rod bacterium E. coli found no obvious change in cell shape upon deletion of any of the low molecular weight PBP endopeptidases and carboxypeptidases, either alone or in combinations that left only DacD and a few penicillin-insensitive peptidases active (Denome et al., 1999). Varma et al. induced spirillum-like morphology in E. coli by inhibiting FtsZ in a mutant lacking PBP5 and PBP7, the latter a DD-endopeptidase that hydrolyzes tetra–penta-peptide cross-links (Varma and Young, 2004). However, the authors did not report whether PG cross-linking was altered in these cells and the mechanism responsible for this observation remains unknown.
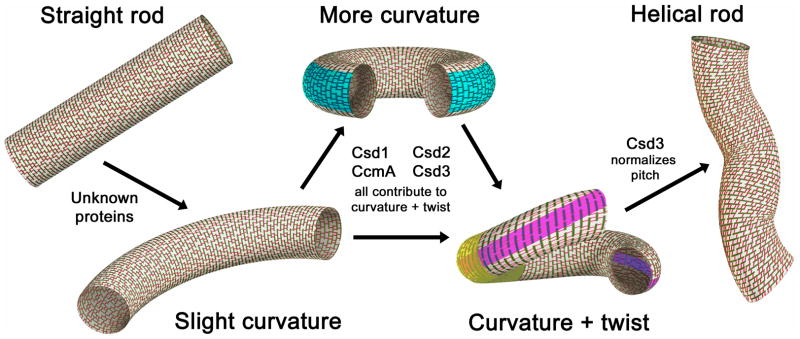
Recent biophysical modeling suggested that hydrolysis of PG cross-links on the outer edge of the cell can induce cell curvature and that a shortening of the length of the cross-links along a helical path might induce helical morphology (Huang et al., 2008). As H. pylori PG lacks the shorter type of cross-link found in some bacteria, (LD)-mDap–mDap (Costa et al., 1999), we propose that this organism could achieve both curvature and twist by hydrolyzing PG cross-links at specific points along the cell. Curving the cell body might be achieved by hydrolyzing cross-links, and thus relaxing the connectivity of the rigid glycan strands, along a straight axis on the outer curvature (Figure 6, blue). Twist may be achieved by shifting this axis diagonally (Figure 6, pink) and/or hydrolyzing cross-links near the inflection point of the curve (Figure 6, yellow), enabling the glycan hoops in that region to slip a little such that the ends of the curve are off-set and a regular helical pitch is defined. This model implies PG hydrolytic activities are precisely localized, perhaps along multiple axes. We have shown that H. pylori indeed expresses multiple proteins that are each required for helical shape maintenance and result in unique morphological perturbations upon deletion. This coupled with the fact that at least three of the proteins seem to be redundant in the major PG biochemical alterations they cause supports the theory that these proteins may be spatially regulated. However, the task of positioning Csd/CcmA activities would likely require the involvement of other proteins, perhaps cytoskeletal proteins such as MreB, FtsZ, or CreS, and these mechanisms remain to be elucidated. Another hypothesis to be tested is whether the morphogenic effect of changes in PG cross-linking is amplified by the induction of gradients in PG synthesis rates as a direct result of cross-link hydrolysis (i.e. stressing of the remaining transpeptide bonds could locally facilitate PG synthesis).
Figure 6.
Model of how H. pylori’s peptidoglycan sacculus forms helical curvature and twist. Glycan strands are green and peptide cross-links red. Blue, pink, and yellow highlights indicate possible zones where relaxation of peptide cross-linking may contribute to the indicated curved and twisting morphologies.
Little is known about the structural and functional diversity of LytM peptidases aside from the fact they are multi-domain proteins with a four-stranded antiparallel β-sheet active site in the C-terminal half of the sequence (Odintsov et al., 2004; Ragumani et al., 2008). Some Gram(−) proteins, such as NlpD and the crystallized V. cholerae protein VCO503, contain PG-binding LysM domains in the N-terminal half of the protein (Ragumani et al., 2008; Uehara et al., 2009), which could be important for substrate recognition or localization. A recent study inactivated all four of the E. coli LytM peptidases, EnvC, NlpD, YgeR, and YebA, finding that the envC mutant had a slight septation defect, which became more severe when nlpD was also deleted (Uehara et al., 2009). The global PG composition of all of these mutants was unchanged from wild-type and no precise biochemical activity has yet been assigned to any Gram(−) LytM peptidase.
Our analyses of the csd null and point mutants’ total muropeptide composition indicated that each protein contributes to the hydrolysis of mDap–D-Ala cross-links and is thus possibly a DD-endopeptidase. Alternatively, the Csd proteins may activate other endopeptidases resulting in the observed changes in muropeptide profile. The csd3 mutant was unique in that it showed ~30% decreases in tetra–tetra-peptide and tetra–tri-peptide cross-linking in addition to the 20–50% increase in tetra–penta-peptide cross-linking evident in all of the mutants. Thus, Csd3 may have multiple activities or may influence the activities of other endopeptidases and carboxypeptidases. The additional relaxation of PG cross-linking in the csd3 mutant may also explain why all of the mutant strains in which csd3 was deleted displayed high degrees of curvature. CcmA bears no sequence or structural homology to any known enzyme, yet apparently it also is important for tetra–penta-peptide hydrolytic activity. CcmA is peripherally associated to the inner membrane in P. mirabilis (Hay et al., 1999) and may exert its function by interacting with the Csd proteins, providing stabilization and/or localization.
Csd1, Csd3, and CcmA are well-conserved throughout the Epsilonproteobacteria, though Csd1 and CcmA homologues were not identified in all species with curved or helical morphologies. Expression of putative csd1 homologues from the Epislonproteobacterium C. jejuni and the Gammaproteobacterium V. cholerae in a csd1 null background appeared to restore some curvature and helical twist in a small subpopulation of cells, supporting a similar function for these proteins. The lack of robust cross-species complementation may result from poor expression, inappropriate localization, or inefficient interactions with the other Csd and CcmA proteins.
The csd and ccmA mutants’ loss of helicity allowed us to begin to explore the functional role of helical shape in H. pylori. Flagellar-based motility is absolutely required for H. pylori infection (Eaton et al., 1996) and helical shape was proposed to allow the bacteria to escape the acidic stomach lumen by boring into the gastric mucus like a screw through a cork (Montecucco and Rappuoli, 2001). Our observations question the model that helical shape is required for H. pylori motility in gel-like media. Another study has also challenged this dogma by demonstrating urease activity dependent reduction in the viscoelasticity of gastric mucus from the gel-like state at low pH to a solution state at more neutral pH (Celli et al., 2009). In spite of their apparently normal motility, three cell shape mutants were each attenuated for stomach colonization in our mouse infection model. We further showed that the increase in tetra–penta-peptide cross-linking observed in csd and ccmA mutant PG did not lead to an obvious weakening of the cell wall to environmental stresses encountered in the stomach. We thus provide evidence that helical shape or some other PG-related property is required for robust stomach colonization by H. pylori, though perhaps not through enhancement of motility.
In this work we have established H. pylori as an excellent model to elucidate molecular determinants of helical cell shape in the Proteobacteria and the selective role of shape during host colonization. Our discovery of a novel family of LytM peptidase homologues required for efficient stomach colonization by H. pylori suggests new targets for antimicrobial therapy that may have efficacy in other pathogens that utilize these proteins, including Vibrio and Campylobacter species.
Experimental Procedures
Bacterial strains and growth
Strains used are described in the Supplemental Experimental Procedures strain table. H. pylori were grown on horse blood (HB) agar plates (Humbert and Salama, 2008) or in Brucella broth (BD Biosciences) containing 10% fetal bovine serum (Hyclone) but no antimicrobials (BB10). Cultures were incubated microaerobically at 37°C in a gas pack jar containing a CampyGen sachet (Oxoid) or in an incubator equilibrated with 14% CO2 and 86% air. For resistance marker selection, HB plates were supplemented with 15 μg/mL chloramphenicol, 25 μg/mL kanamycin, 36 μg/mL metronidazole, or 60 mg/mL sucrose. For culturing bacteria from mouse stomachs, 200 μg/mL bacitracin was added to eliminate normal mouse microbiota growth. Cell filamentation was induced by diluting overnight liquid cultures to 0.2 optical density at 600 nm (OD600) into BB10 containing 2 μg/mL aztreonam and shaking for 4–9 hrs. For plasmid selection and maintenance in E. coli, LB agar or broth was supplemented with 30 μg/mL kanamycin or 100 μg/mL ampicillin.
Phase contrast and TEM microscopy of H. pylori cells
For phase contrast microscopy, cells were grown in shaken liquid culture to mid-log phase (OD600 0.2–1.0), fixed in a 4% paraformaldehyde/PBS solution containing 10% glycerol, and mounted on glass slides. Cells were imaged with a Nikon TE 200 microscope equipped with a 100x oil immersion objective and Nikon CoolSNAP HQ CCD camera controlled by MetaMorph software (MDS Analytical Technologies). TEM was performed as described (Lowenthal et al., 2009). Image cropping and brightness/contrast adjustments were made using Adobe Photoshop Elements 3.0.
Preparation of PG and analysis of muropeptides
H. pylori cells were expanded daily on HB plates lacking antimicrobials to maintain log-phase growth and obtain 100–500 OD600 per strain. Cells were collected with sterile polyester swabs, suspended in 6 mL chilled PBS and lysed by dropwise addition to 6 mL boiling 8% SDS. PG was prepared from cell lysate as described (Glauner et al., 1988; Costa et al., 1999). Muropeptides were released from PG by the muramidase Cellosyl (Hoechst, Frankfurt am Main, Germany), reduced by sodium borohydride, and separated on a 250×4.6 mm 3 μm Prontosil 120-3-C18 AQ reversed phase column (Bischoff, Leonberg, Germany) as described (Glauner et al., 1988). The eluted muropeptides were detected by their absorbance at 205 nm. The muropeptide profile of the wild-type was similar to the published profile of Helicobacter muropeptides (Costa et al., 1999) allowing unambiguous assignment of known muropeptide structures to the peaks detected.
Visualization of sacculi by TEM
Sacculi were immobilized for 15 min on glow-discharged carbon-pioloform coated copper grids (400 mesh). Excess liquid was removed onto filter paper and the grids air-dried (de Pedro et al., 1997). Grids were then incubated with 1–2% uranyl acetate for 1 min, briefly washed with water, and air-dried. Because uranyl acetate treatment gave poor contrast for H. pylori sacculi, samples were also visualized by negative staining. A drop of the sacculi suspension was mixed with a drop of staining solution (3% phosphotungstic acid adjusted to pH 7 with potassium hydroxide prior to use) and added to the same grids. After 30 sec the excess liquid was removed and the grid air-dried. Samples were examined on a Philips CM 100 Compustage (FEI) Transmission Electron Microscope and digital images collected using an AMT CCD camera (Deben).
Motility, growth, and stress testing
Soft agar motility experiments were performed as described (Lowenthal et al., 2009). For velocity measurements, cells at 0.2 OD600 were mixed with an equal volume BB5 with or without 1% methylcellulose and imaged on a hanging drop slide with a 40x objective at 100 msec intervals using phase contrast as described above. Cells were tracked using the ImageJ Manual Tracker (Rasband, 1997–2009) and curvilinear velocity calculations performed with Intercooled Stata 10.0 (StataCorp). Growth and stress testing was accomplished using 100–200 μL BB10 mini-cultures grown in a 96-well plate. For growth experiments, liquid cultures were grown overnight to OD600 < 1 and diluted to 0.005 OD600/mL (0.0025 OD600/mL for each strain in co-culture experiments). At desired intervals cell aliquots were diluted serially and plated to non-selective and (for co-culture experiments) selective HB plates to enumerate total and mutant CFUs. For pH and polymyxin stress testing, 1×108 cells from overnight cultures were added to pH-adjusted BB10 or BB10 containing 0.1–3.0 mg/mL polymyxin, incubated for 1 hr, and plated to HB plates. Amoxicillin sensitivity was determined by plating 200 μL of overnight culture on HB plates lacking antimicrobials and applying E-test strips (AB Biodisk). Plates were incubated for 2–3 days and read according to the manufacturer’s instructions.
Mouse colonization
Female C57BL/6 mice 24–28 days old and certified free of endogenous Helicobacter were obtained from Charles River Laboratories. Mice were housed and infected as described (Amundsen et al., 2008) using 5×107 cells/strain in the inocula for competition experiments and 1×108 cells in the inocula for single strain experiments. The whole stomach was homogenized in 1 mL BB10 and dilutions were plated to non-selective and selective (for competition experiments) HB plates to enumerate total and mutant bacteria. If no mutant bacteria were recovered we set the number of colonies on the lowest dilution plated to 1. All studies were done under practices and procedures of Animal Biosafety Level 2. The facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and all activities were approved by the FHCRC Institutional Animal Care and Use Committee.
Quantitative morphological and bioinformatic analyses
A detailed description of these methods is provided in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI054423 and AI082026 (NRS), a National Science Foundation Graduate Research Fellowship (LKS), and grants from the Biotechnology and Biological Sciences Research Council (grant No. BB/F001231/1) and the European Commission (EUR-INTAFAR project) (WV). The contents are solely the responsibility of the authors and do not necessarily represent the official views of these funding agencies. We thank J. Fero, C. Tung, G. Cromie, S. Talarico, B. Stoddard, and D. Vollmer for technical assistance and consultation, P. Born for peptidoglycan analysis, R. Burmeister for graphic design, E. Gaynor and S. Miller for strains, and B. Schneider and staff (FHCRC Electron Microscopy Shared Resource), as well as V. Thompson and T. Davies (Newcastle University Electron Microscopy Research Service), for assistance with TEM. The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci U S A. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen LP, Rasmussen L. Helicobacter pylori-coccoid forms and biofilm formation. FEMS Immunol Med Microbiol. 2009;56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Benaissa M, Babin P, Quellard N, Pezennec L, Cenatiempo Y, Fauchere JL. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect Immun. 1996;64:2331–2335. doi: 10.1128/iai.64.6.2331-2335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. Embo J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Bove JM, Renaudin J, Saillard C, Foissac X, Garnier M. Spiroplasma citri, a plant pathogenic molligute: relationships with its two hosts, the plant and the leafhopper vector. Annu Rev Phytopathol. 2003;41:483–500. doi: 10.1146/annurev.phyto.41.052102.104034. [DOI] [PubMed] [Google Scholar]
- Bromley DB, Charon NW. Axial filament involvement in the motility of Leptospira interrogans. J Bacteriol. 1979;137:1406–1412. doi: 10.1128/jb.137.3.1406-1412.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. Embo J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J Cell Biol. 2007;179:381–387. doi: 10.1083/jcb.200708001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa K, Bacher G, Allmaier G, Dominguez-Bello MG, Engstrand L, Falk P, de Pedro MA, Garcia-del Portillo F. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol. 1999;181:3710–3715. doi: 10.1128/jb.181.12.3710-3715.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Holtje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoney CR, Schiller NL. Competition of various beta-lactam antibiotics for the major penicillin-binding proteins of Helicobacter pylori: antibacterial activity and effects on bacterial morphology. Antimicrob Agents Chemother. 1999;43:2702–2709. doi: 10.1128/aac.43.11.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Goldstein SF, Charon NW, Kreiling JA. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc Natl Acad Sci U S A. 1994;91:3433–3437. doi: 10.1073/pnas.91.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin CS, McCulloch RK, Armstrong JA, Wee SH. Unusual cellular fatty acids and distinctive ultrastructure in a new spiral bacterium (Campylobacter pyloridis) from the human gastric mucosa. J Med Microbiol. 1985;19:257–267. doi: 10.1099/00222615-19-2-257. [DOI] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazell SL, Lee A, Brady L, Hennessy W. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153:658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci U S A. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert O, Salama NR. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008;36:6893–6906. doi: 10.1093/nar/gkn718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Hughes KJ, Everiss KD, Peterson KM. Identification of a ToxR-activated gene, tagE, that lies within the accessory colonization factor gene cluster of Vibrio cholerae O395. Gene. 1994;148:91–95. doi: 10.1016/0378-1119(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacayo CI, Pincus Z, VanDuijn MM, Wilson CA, Fletcher DA, Gertler FB, Mogilner A, Theriot JA. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5:e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. Functional Analysis of the Helicobacter pylori Flagellar Switch Proteins. J Bacteriol. 2009 doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C, Rappuoli R. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol. 2001;2:457–466. doi: 10.1038/35073084. [DOI] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M. Latent LytM at 1.3A resolution. J Mol Biol. 2004;335:775–785. doi: 10.1016/j.jmb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Ragumani S, Kumaran D, Burley SK, Swaminathan S. Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae. Proteins. 2008;72:1096–1103. doi: 10.1002/prot.22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, Maryland, USA: U.S. National Institutes of Health; 1997–2009. [Google Scholar]
- Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets LC, Bijlsma JJ, Boomkens SY, Vandenbroucke-Grauls CM, Kusters JG. comH, a novel gene essential for natural transformation of Helicobacter pylori. J Bacteriol. 2000;182:3948–3954. doi: 10.1128/jb.182.14.3948-3954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Trachtenberg S. Shaping and moving a spiroplasma. J Mol Microbiol Biotechnol. 2004;7:78–87. doi: 10.1159/000077872. [DOI] [PubMed] [Google Scholar]
- Trachtenberg S, Dorward LM, Speransky VV, Jaffe H, Andrews SB, Leapman RD. Structure of the cytoskeleton of Spiroplasma melliferum BC3 and its interactions with the cell membrane. J Mol Biol. 2008;378:778–789. doi: 10.1016/j.jmb.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J Bacteriol. 2004;186:6768–6774. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, Speth V, Kist M, Graumann PL. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS pathogens. 2009;5:e1000669. doi: 10.1371/journal.ppat.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DL, Renaudin J, Bove JM. Nucleotide sequence of the Spiroplasma citri fibril protein gene. J Bacteriol. 1991;173:4353–4362. doi: 10.1128/jb.173.14.4353-4362.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth CW, Charon NW, Goldstein SF, Goldstein RE. The flagellar cytoskeleton of the spirochetes. J Mol Microbiol Biotechnol. 2006;11:221–227. doi: 10.1159/000094056. [DOI] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.