Abstract
The trimeric Sec61/SecY complex is a protein-conducting channel (PCC) for secretory and membrane proteins. Although Sec complexes can form oligomers, it has been suggested that a single copy may serve as an active PCC. We determined sub-nanometer resolution cryo-electron microscopy structures of eukaryotic ribosome-Sec61 complexes. In combination with biochemical data we found that in both idle and active states, the Sec complex is not oligomeric and interacts mainly via two cytoplasmic loops with the universal ribosomal adaptor site. In the active state the ribosomal tunnel and a central pore of the monomeric PCC were occupied by the nascent chain contacting loop 6 of the Sec complex. This provides a structural basis for the activity of a solitary Sec complex in cotranslational protein translocation.
The protein-conducting channel (PCC) of the canonical secretory pathway is formed in all cells by the Sec61/SecY complex. It engages in the post- and co-translational translocation of secretory proteins across, and the insertion of integral membrane proteins into the membrane of the endoplasmic reticulum (ER) in eukaryotes and the plasma membrane of bacteria (1, 2).
In the co-translational translocation mode the ribosome with an emerging signal sequence is targeted to the membrane by the signal recognition particle (SRP) and its receptor (3). Here, the Sec complex acts as a receptor for the ribosome via its cytosolic loops (4). The alignment of the ribosomal tunnel with a central pore of the PCC allows direct movement of the nascent chain from the ribosomal tunnel exit across or into the membrane (5, 6).
The PCC-forming heterotrimeric Sec complex consists of one large subunit (Sec61α in Mammalia, Sec61p/Ssh1p in yeast, SecY in Bacteria) and two small subunits (Sec61β, γ in eukaryotes and SecE, G in Bacteria). Conflicting models have been presented as to how many of these heterotrimers are necessary to build an active PCC and what the actual path of the polypeptide chain is. The Escherichia coli SecYEG complex forms back-to-back dimers in two-dimensional crystals (7), and low resolution single particle electron microscopic (EM) data revealed a pentagonal ringlike morphology of the PCC interpreted as oligomers (5, 6, 8-12). The monomeric crystal structure of an archaeal SecYEß complex (13), in combination with chemical cross-linking data (14), led to the interpretation that a single copy of the Sec complex is sufficient to serve as an active PCC, even when assembled into a dimer for post-translational translocation (15) or a tetramer for co-translational translocation (8). Recent low resolution cryo-EM data of inactive ribosome-Sec complexes were interpreted to represent single copies of Sec complexes (16, 17) and crystal structures of the bacterial SecY-SecA complex also show a single copy (18). However, because all these structures are either of low resolution or lack translocating peptides, two main questions remain: (i) what conformational and (ii) what oligomeric states can be adopted by the PCC in the different modes of activity, such as signal sequence recognition, vertical and lateral gating.
Cryo-EM and 3D reconstruction
For structure determination by cryo-EM we used digitonin-solubilized purified Ssh1 complex (Sec sixty-one homologue 1 from the yeast Saccharomyces cerevisiae) containing Ssh1p, Sbh2p and Sss1p (19). This complex is active in the co-translational translocation mode only, i.e. when ribosome-bound (20, 21).
We reconstituted the Ssh1 complex with in vitro programmed 80S ribosomes carrying a nascent polypeptide chain (ribosome-nascent chain complexes, RNCs) (6, 22). The peptide includes the first 120 amino acids of the type II membrane protein dipeptidyl aminopeptidase B (DP120) together with its signal anchor sequence, long enough to allow a loop insertion into the PCC (23). As in the case of the Sec61 complex (6), specific and stable binding of the Ssh1 complex to RNCs was observed.
Cryo-EM analysis revealed heterogeneity of the sample and a thorough sorting regimen applied to the dataset (Fig. S1) (24) resulted in a number of structures at sub-nanometer resolution, three of which were analyzed further: the programmed (active) 80S-Ssh1 complex, the non-programmed (idle) 80S-Ssh1 complex without a peptidyl-tRNA and the programmed 80S ribosome with ES27 in exit conformation without Ssh1 complex (Fig. 1, Fig. S2). The 3D reconstruction of all programmed ribosomes resulted in a 6.1 Å map of the yeast 80S ribosome (Fig. S2).
Fig. 1. Cryo-EM reconstructions of 80S ribosome-Ssh1 complexes.

Cryo-EM reconstructions of the idle (A) and the active (B) 80S-Ssh1 complex at 9 Å resolution. (C) Map of the 80S ribosome with ES27 in the exit conformation at 8 Å. Color code: 40S subunit, yellow; 60S subunit, blue; P-site tRNA/nascent polypeptide chain, green; Ssh1 complexes (PCC), red.
As expected, the Ssh1 complex was bound at the exit site of the ribosome similar to the Sec61 complex (6). However, due to apparent flexibility of the ribosome-PCC connection, the PCC density was not as well resolved as the ribosome. Two notable features of the PCC density were observed: (i) the size of the density appeared to be smaller than previously observed and (ii) a central pore was visible in the idle complex.
Visualization of the nascent chain and ribosomal model of the tunnel exit site
When cutting through the densities, the idle complex without a tRNA revealed an empty ribosomal tunnel leading directly to the central pore in the PCC density (Fig. 2A). In contrast, the pore in the active PCC was occupied by additional density. Here, even applying different contour- or filtering parameters did not lead to the appearance of a pore-like feature (Fig. 2B). Notably, the active complexes revealed additional density also in the ribosomal tunnel, representing the nascent polypeptide chain (Fig. 2B-C). For the RNC with ES27 in the exit site conformation the nascent chain density could be traced from the CCA-end of the tRNA almost continuously to the tunnel exit (Fig. 2C and F). For further analysis we generated a molecular model of the tunnel exit region based on the yeast RNC map at about 6 Å resolution (24). It includes models for the proteins rpL4, rpL17, rpL19, rpL25, rpL26, rpL31, rpL35 and rpL39 as well as the rRNA helices H5- H7, H24, H50 and the extension ES24 of H59 (Fig. 2D-E).
Fig. 2. Visualization of the PCC pore, the nascent polypeptide chain and molecular model.

Cut density for the 60S subunit and the idle (A) and active (B) Ssh1 complex is shown as in Fig. 1A. (C) as (A) except ES27 in exit position (dark blue) and P-site tRNA are shown. (D) bottom view of the 6.1 Å RNC map as in Fig. 1. Density for rRNA and ribosomal proteins is highlighted. Asterisk indicates tunnel exit. (E) as (D) showing molecular models. (F) left: isolated density for the P-site tRNA and the nascent DP120 chain (NC) as in (C); right: molecular model for the yeast P-site tRNAAsp and for the nascent DP120 peptide with ribosomal proteins (E).
Oligomeric state and molecular model of the ribosome-bound yeast Ssh1 complex
A double-tag approach (25) was employed for analysis of the oligomeric state of the Ssh1 complex in the cell: a yeast strain was engineered in order to express in similar amounts two differently tagged forms of Ssh1p; T7-Ssh1p and AU1-Ssh1p, both of which are functional (4, 24). Antibodies against one of the tags were used for non-denaturing immunoprecipitation of digitonin-solubilized Ssh1 complex in the presence of RNCs. The second antibody was then used to probe whether the second tag could be co-precipitated indicative of hetero-oligomer formation. Yet, pull-down by the first antibody did not yield any detectable amounts of the second tag, independent of the order of antibody usage (Fig. 3A). Therefore, the stably ribosome-bound Ssh1 complex is likely to exist mainly as a single copy. However, we can not exclude that the Ssh1 complex may assemble into a transient or detergent-sensitive oligomer in the membrane.
Fig. 3. A monomeric ribosome-bound Ssh1 complex.

(A) Native immunoprecipitation (IP) of epitope tagged Ssh1 complexes. Microsomes from yeast cells expressing T7 and AU1 tagged Ssh1p were repopulated with RNCs, solubilized, and subjected to native immunoprecipitation using anti-T7 or anti- AU1 antibodies. Total extract (T), supernatant (S) and immunoprecipitate (P) fractions were analyzed by immuno blot using an anti-T7-goat or an anti-AU1-rabbit antibody to detect T7-Ssh1 and AU1-Ssh1. (B) Top: Close-up side views on idle (left) and active (right) PCC (as in Fig. 2A and B). Bottom: Homology models of a monomeric Ssh1 complex (red) fitted into the densities of idle and active Ssh1 complexes (transparent mesh); The cytosolic loop L8 of Ssh1p (red), Sbh1p (β, dark red) and Sss1p (γ, magenta) are indicated. The nascent chain (NC) is shown in green. (C) as (B) showing top views.
Using this result, we analyzed the cryo-EM densities of the idle and active Ssh1 complexes in order to dock homology models based on the crystal structure of the archaeal SecYEß complex (13). Tetramers, trimers or dimers (12, 26) could not be accommodated in the observed density (Fig. S3). Only a single copy of the Ssh1 complex fit (Fig. 3B), which is in agreement with our pull-down experiment, biochemical data (27-29) and low resolution cryo-EM data of inactive complexes (16, 17). The final models required minor adjustments mainly of cytoplasmic loops L6 and L8 as well as the C-terminal tail (24). While the model accounts for the majority of the observed density, a remaining belt-shaped density that surrounds the fitted molecule (Fig. 3B, C and Fig. S3B) most likely corresponds to the detergent micelle. Taken together, the yeast PCC consists of a single Sec61 (Ssh1) complex when bound to a non-translating or a signal-sequence carrying ribosome. The overall conformation is very similar between the idle and the active PCC suggesting that major structural transitions may not be required for the PCC to switch between these states. The presence of density in the central pore of the PCC bound to the active ribosome suggests that the pore of a single Sec complex is indeed used by the nascent polypeptide chain.
Interaction of the PCC with the ribosome and the nascent chain
In both states we perceived four main connections similar to those observed for the yeast ribosome-Sec61 complex (6) (Table S1). The main connections (C2 and C4) correspond to the L8 and the L6 cytoplasmic loops of the PCC using, similar to inactive ribosome-Sec complexes (16, 17), the universal ribosomal adaptor site (22) comprising mainly the rpL25/rpL35 proteins and rRNA helices H7 and H50 (Fig. 4A and B). We tested the contribution of the two loops of Ssh1p to the ribosome interaction by performing ribosome-binding assays. Mutational analysis by charge inversion of conserved positively charged residues such as R411 in the L8 loop showed that this loop is indeed necessary for ribosome binding (Fig. 4C and D). In contrast to findings for SecYEG (16), a mutation in loop L6 of the conserved R278 did not result in a loss of ribosome binding, indicating that it may not directly participate in establishing the high affinity interaction with the ribosome. Similarly in the yeast Sec61 complex point mutations in L8 lead to severe defects in ribosome binding, while mutations of the basic residues in L6 do not reduce ribosome-binding affinity significantly (4). Two additional connections were C1, established between rRNA helix H59 and probably the N-terminus of Ssh1p, and C3, involving rpL26 and rRNA helix H24 and probably the C-terminus of Ssh1p including TM10.
Fig. 4. Interaction of the PCC with the ribosome and the nascent chain.
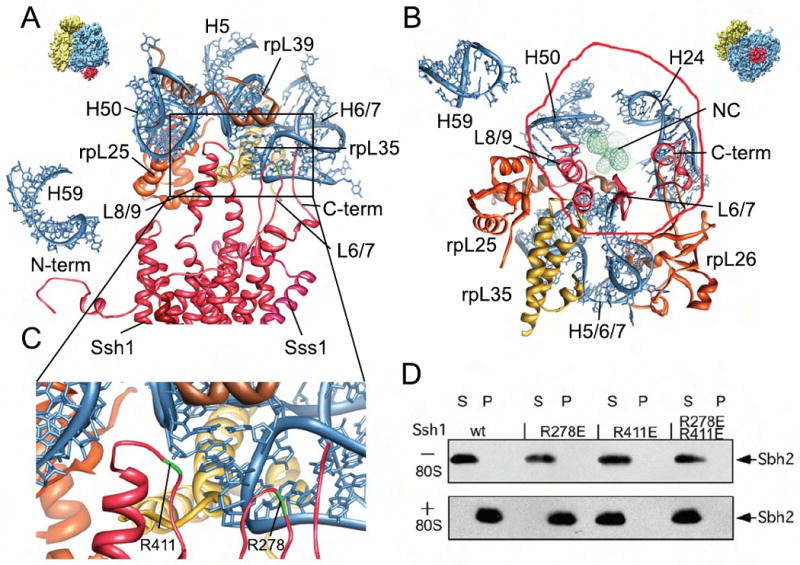
(A-C) Molecular models for rRNA and ribosomal proteins are shown as in Fig. 2 E, for the Ssh1 complex as in Fig 3. Views focus on the cytosolic half of the Ssh1 model (A) and the cytosolic loops L6, L8 and the C-terminus (B). (C) A close-up on interactions of cytosolic loops L6 and L8. The positions of the conserved R278 and R411 are indicated (green). (D) Purified Ssh1 complexes from wild type and L6 and L8 mutants were incubated in the presence or absence of yeast ribosomes before centrifugation yielding supernatant (S) and pellet (P) fractions. After SDS-PAGE, Sbh2p was detected using anti-FLAG antibodies.
The nascent chain was in very close proximity to and probably contacting the L6 loop of the Ssh1 complex (Fig. 4B). Thus loop L6 may function in sensing or guiding the emerging peptide to the pore of the PCC, consistent with its observed role in translocation (4,30).
The mammalian Sec61 complex bound to an active ribosome
We determined the structure of the mammalian Sec61 complex from Canis familiaris bound to an active DP120 signal-anchor containing 80S ribosome (Triticum aestivum) (6, 22) at 6.5 Å resolution (Fig. 5A). It was considerably larger than that observed for the yeast Ssh1 complex and, when filtered to lower resolution, very similar to the previously observed densities of mammalian Sec61 (8, 17) (Fig. 5B).
Fig. 5. The RNC-bound mammalian Sec61 complex is a monomer surrounded by a micelle.

(A) Cryo-EM reconstruction of the 80S RNC-Sec61 complex at 6.5 Å resolution. Top: side view, bottom: bottom view. (B) Isolated density for the Sec61 complex low-pass filtered at 12 Å (top) and at 22 Å (bottom). Connections C1- C4 are indicated. (C) Side view of the ribosome-bound Sec61 complex. The Sec61 model is shown in red, models for ribosomal proteins and rRNA as in Fig. 2 D. The monomeric Sec61 complex fitted into the central portion of the density surrounded by a rim of extra density representing mixed detergent/lipid micelle. (D) Upper left section: side view of the cut densities for the Sec61 complex (red), the surrounding mixed micelle (grey) and the nascent DP120 polypeptide chain and/or the signal anchor sequence (green). Right: Schematic drawing of the mixed micelle of phospholipids (grey) and detergent molecules (blue) surrounding the PCC (red ribbons). Middle section: Isolated densities and schematic drawing as in upper section in a top view (left) or sliced within the plane of the membrane (right). Lower section: Sliced top views, represented as in middle section (left) or as red ribbons for the Sec61 model and transparent mesh for the electron density (right).
Closer inspection, however, revealed distinct structural features such as central rod-like densities surrounded by two belts of weaker and stronger density, respectively. A homology model of the Sec61 complex was calculated and the helices fitted into the central rod-like densities requiring only minor adjustments using Molecular Dynamics Flexible Fitting (MDFF) (31) (Fig. 5C, Fig. S4). We observed the same four major connections to the ribosome (6) (Fig. 5C) with the two central connections representing the cytoplasmic loops L6 and L8 of the Sec61 complex reaching into the ribosomal tunnel exit via the universal adaptor site (Fig. S5, Table S2) similar to the inactive complex (17). Compared to the Ssh1 complex, the loops were somewhat rotated without changing the overall position of the Sec complex (Fig. S5). Thus, the binding mode appears to be well conserved and is basically the same in inactive and active complexes.
The weak proximal and strong distal belt-like density surrounding the central Sec61 complex did not show any rod-like features and apparently represents a mixed detergent/lipid micelle as suggested before at lower resolution (17). As expected in a micelle we observed a characteristic density distribution (32) of regions containing acyl chains or polar head-groups of phospholipids (Fig. 5D; Fig. S4). Significant amounts of the phospholipids phosphatidylcholine and -ethanolamine indeed co-purified with Sec61 in our preparation (Fig. S6) confirming the presence of a mixed micelle. It appears likely that previous reconstructions also represent single copies of the Sec61 or the SecYEG complex in micelles of varying sizes when considering the appearance of the micelle-surrounded single copy Sec61 complex filtered to lower resolution.
The identification of just one copy of the Sec61 complex indicates that also in higher eukaryotes a single complex is stably recruited to the ribosome in the presence of a signal sequence and is probably sufficient to function as the active PCC. This finding is difficult to reconcile with previous interpretations of Sec complex dimers or even larger oligomers bound to the ribosome.
Conformation of the Sec61 complex and interaction with the nascent chain
The conformation of the ribosome-bound Sec61 complex (Fig. 6A) was compared with available crystal structures (13, 18, 33) to address two questions: (i) how does the Sec61 complex, in particular the proposed lateral gate behave in the presence of a signal anchor, and (ii) how is the translocating peptide accommodated in the PCC? The ribosome-bound conformation is most similar to the SecYEß structure of M. jannaschii (13) (Fig. 6B, Fig. S7). The region of the proposed lateral gate around helices 2b and 7 of Sec61α was well resolved and indicated only a small movement of helix 2b of clearly less than 5 Å when compared to the SecYEß structure (Fig. 6B). In contrast, the opening movement observed in the SecYEG-SecA crystal structure (18) and in the Fab-bound SecYE (33) shifted the entire helix more than 5 Å (Fig. S7). We observed density representing the nascent polypeptide in the ribosomal exit tunnel and also in the central aqueous pore of the Sec61 complex (Fig. 6C and D). This density was well defined in the last section of the ribosomal tunnel in which it is contacting the Sec61 L6 loop, but then becomes disordered. In the cytoplasmic vestibule of the Sec61 complex we observed a rod-like density contacting the lateral gate helices 2b and 7, in the lumenal vestibule we found weak and fragmented density. It has been shown previously that also in detergent solution the Sec61 complex can productively engage in polypeptide insertion (6, 34, 35). For the gating event preceding insertion we therefore expect that the signal anchor sequence in our complex is in contact with the PCC. Thus, the rod-like density in the cytoplasmic vestibule (Fig. 6) may resemble the signal anchor sequence, the position of which would be consistent with previous cross-link data (36).
Fig. 6. Conformation and nascent polypeptide chain-interactions of the RNC-bound mammalian Sec61 complex.

(A) Fit of the Sec61 model (red ribbons) into the density (grey transparent mesh). Side views on the lateral gate (top) and on the cytosolic loops L6 and L8 (bottom). (B) Crystal structure of the M. jannaschii SecYEβ complex (grey) (13) superimposed on Sec61 model. The C- and N-terminal halves are shown in red and blue, transmembrane (TM) helix 7 in yellow and TM helix 2 in cyan. β- (dark red) and γ-subunits (SecE, magenta) are indicated. (C) Side view (top, as in A) and top view (bottom, as in B) of the Sec61 model and extra density for the nascent polypeptide chain (green). (D) Side view as in (C) (top) but rotated to focus on the nascent chain (green). Color code as in Fig. 2E. (E) Schematic representation of an actively translating and translocating eukaryotic ribosome-Sec61 complex with a single copy acting as PCC.
The question arises what functional state we observe in our complex. One possibility is that the PCC is in the pre-open state described for the SecYEG-SecA complex (18) in which the lateral gate is partially open but the plug is still occluding the central pore. This appears unlikely because the overall conformation, in particular the lateral gate region, is very different (Fig. S7C). It appears more likely that we have captured a post-insertion state with a closed or nearly closed lateral gate region. Consistent with this, cross-links between helices 2b and 7 revealed a closed lateral gate after insertion of the nascent peptide chain into the SecYEG complex (37).
Based on our data several conclusions concerning co-translational protein translocation can be drawn (Fig. 6 E). (i) Only a single copy of the Sec61 complex is recruited to the non-translating and also the translating ribosome. (ii) In both the yeast Ssh1 complex and the mammalian Sec61 complex, we observed a nascent polypeptide and/or the signal anchor sequence accommodated within this single copy PCC, thus, strongly indicating that its central pore serves as the conduit for the nascent polypeptide chain. (iii) The lateral gate of the PCC can be in a closed or nearly closed conformation after insertion of the translocating peptide. (iv) The mode of PCC binding to ribosomes appears to be conserved between species, and is maintained in the presence or absence of a signal sequence. (v) The main binding site for the PCC is the universal adaptor site at the ribosomal tunnel exit that is contacted mainly by the cytoplasmic loop L8 of the Sec61 complex, whereas loop L6 is also contacting the emerging nascent polypeptide. The observed mode of Sec61 binding fits well with our previous findings that the universal adaptor site also serves to bind SRP (22) – mutually exclusive with the PCC - but is then cleared upon SRP receptor interaction to enable PCC binding (38).
Supplementary Material
Acknowledgments
We thank B. Dobberstein (ZMBH, Heidelberg, Germany) for microsomal membranes, B. Brügger (BZH, Heidelberg, Germany) for lipid analysis, J. Frauenfeld and E. van der Sluis for critical discussions. This research was supported by grants from the Deutsche Forschungsgemeinschaft SFB594 and SFB646 (to R.B. and T.B), SFB 740 (to T.M.), by KA Wallenberg Foundation Stockholm, Sweden (to S.B.) by NIH grants P41-RR05969, R01- GM067887 (to K.S.) and GM35687 (to R.G.), by NSF grant PHY0822613 (to K.S.), by the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum). Computer time for MDFF was provided through an NSF Large Resources Allocation Committee grant MCA93S028. Coordinates of the atomic models and cryo-EM maps have been deposited in the PDB (3XX1 and 3XX2) and in the 3D-EM database (EMD-1651and EMD-1652), respectively.
Footnotes
Supporting Online Material: www.sciencemag.org, Materials and Methods, Supplementary Figs. S1-S7, Supplementary Tables S1 and S2, References.
References and Notes
- 1.Mandon EC, Trueman SF, Gilmore R. Curr Opin Cell Biol. 2009;21:501. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rapoport TA. Nature. 2007;450:663. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 3.Halic M, Beckmann R. Curr Opin Struct Biol. 2005;15:116. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Z, Jiang Y, Mandon EC, Gilmore R. J Cell Biol. 2005;168:67. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann R, et al. Science. 1997;278:2123. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 6.Beckmann R, et al. Cell. 2001;107:361. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 7.Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I. Nature. 2002;418:662. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 8.Menetret JF, et al. J Mol Biol. 2005;348:445. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 9.Hanein D, et al. Cell. 1996;87:721. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 10.Menetret J, et al. Mol Cell. 2000;6:1219. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 11.Morgan DG, Menetret JF, Neuhof A, Rapoport TA, Akey CW. J Mol Biol. 2002;324:871. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- 12.Mitra K, et al. Nature. 2005;438:318. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van den Berg B, et al. Nature. 2004;427:36. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 14.Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. J Cell Biol. 2005;169:219. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborne AR, Rapoport TA. Cell. 2007;129:97. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 16.Menetret JF, et al. Molecular Cell. 2007;28:1083. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 17.Menetret JF, et al. Structure. 2008 Jul;16:1126. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmer J, Nam Y, Rapoport TA. Nature. 2008;455:936. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finke K, et al. EMBO J. 1996;15:1482. [PMC free article] [PubMed] [Google Scholar]
- 20.Prinz A, Hartmann E, Kalies KU. Biol Chem. 2000;381:1025. doi: 10.1515/BC.2000.126. [DOI] [PubMed] [Google Scholar]
- 21.Wittke S, Dünnwald M, Albertsen M, Johnsson N. Mol Biol Cell. 2002;13:2223. doi: 10.1091/mbc.01-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halic M, et al. Nature. 2004;427:808. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 23.Ng DT, Brown JD, Walter P. J Cell Biol. 1996;134:269. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Materials and Methods are available as supporting material on Science online
- 25.Karaoglu D, Kelleher DJ, Gilmore R. Biochemistry. 2001;40:12193. doi: 10.1021/bi0111911. [DOI] [PubMed] [Google Scholar]
- 26.Osborne AR, Rapoport TA, van den Berg B. Annu Rev Cell Dev Biol. 2005;21:529. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 27.Yahr TL, Wickner WT. EMBO J. 2000;19:4393. doi: 10.1093/emboj/19.16.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duong F. EMBO J. 2003;22:4375. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalies KU, Stokes V, Hartmann E. Biochim Biophys Acta. 2008;1783:2375. doi: 10.1016/j.bbamcr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Raden D, Song W, Gilmore R. J Cell Biol. 2000;150:53. doi: 10.1083/jcb.150.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Structure. 2008;16:673. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.le Maire M, Champeil P, Moller JV. Biochim Biophys Acta. 2000;1508:86. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 33.Tsukazaki T, et al. Nature. 2008;455:988. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jungnickel B, Rapoport TA. Cell. 1995;82:261. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 35.Mothes W, Jungnickel B, Brunner J, Rapoport T. J Cell Biol. 1998;142:355. doi: 10.1083/jcb.142.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plath K, Wilkinson BM, Stirling CJ, Rapoport TA. Mol Biol Cell. 2004;15:1. doi: 10.1091/mbc.E03-06-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. J Biol Chem. 2009;284:15805. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halic M, et al. Science. 2006;312:745. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


