Abstract
We have expanded the application of antibody phage display to a new type of antigen: ribonucleoprotein (RNP) complexes. We describe a simple and efficient method for screening antibodies specific for large intact RNPs and individual components. We also describe a fast and easy method to overcome the abundance of amber stop codons in the positive phage clones. The resulting antibodies have been used in ELISA and Western blot analysis.
Keywords: Antibody phage display, scFv, Macromolecular complexes, ribonucleoprotein
1. Introduction
Antibody phage display is an attractive and powerful alternative to hybridoma technology. Beyond generating antibodies against a wide variety of proteins (Dong et al., 2003; Smith et al., 2003; Chang et al., 2006; Marcus et al., 2006), it has also been used to produce antibodies specific for unconventional antigens including small peptides (Rodriguez-Diaz et al., 2004), DNA photoproducts (Zavala et al., 2000), snRNAs (Teunissen et al., 1998) and lipids (Takkinen et al., 1996). However, this technology has not been efficient in producing antibodies that target subunits of macromolecular complexes (Rubinstein et al., 2003). Here we describe an optimized protocol to generate multiple monoclonal scFvs against a large ribonucleoprotein, the E. coli signal recognition particle-receptor complex.
Signal recognition particle (SRP) and its receptor (SR) are conserved in all organisms (Pool, 2005) as part of the molecular machinery that guides integral membrane and secretory proteins to the cellular translocation apparatus during translation (for details refer to Pool, 2005). E. coli SRP has two components: a protein subunit called Ffh (Bernstein et al., 1989; Romisch et al., 1989) and an 114-nucleotide RNA called 4.5S RNA (Poritz et al., 1990). E. coli SR is a single polypeptide molecule called FtsY (Luirink et al., 1994; Miller et al., 1994). Ffh and FtsY each contain a homologous NG domain (Freymann et al., 1997; Montoya et al., 1997) which has GTPase activity and these domains associate to form a heterodimer upon GTP binding. Ffh contains an additional domain, the M domain, which binds to 4.5S RNA, whereas FtsY includes an additional domain - the A domain-which helps anchor FtsY to the membrane (de Leeuw et al., 1997). A three-dimentional model of SRP-SR predicts that the 4.5S RNA lies at the interface of Ffh and FtsY in the complex (Spanggord et al., 2005) (Fig. 1A). In this report we optimized the phage display protocol and generated scFv antibodies specific for the intact SRP-SR RNP complex or its components.
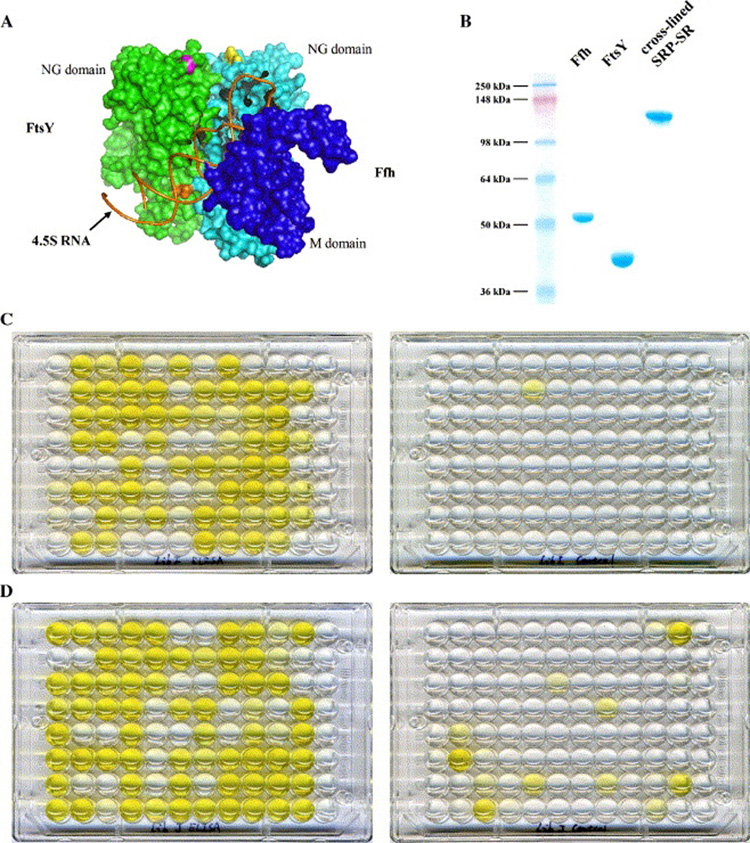
Fig. 1.
Panning for scFvs specific for SRP-SR. (A) 3D model of SRP-SR complex showing the spatial relations of Ffh, FtsY and 4.5S RNA. (B) SDS-PAGE analysis of cross-linked SRP-SR. (C) Monoclonal ELISA plate of Tomlinson I library (left) and its control (right). (D) Monoclonal ELISA plate of Tomlinson J library (left) and its control (right).
2. Materials and methods
Preparation of cross-linked SRP-SR complex
Equal molar amounts of Ffh and FtsY were mixed at a final concentration of 20 µM in binding buffer (20 mM HEPES pH7.5, 100 mM KCl, 2.5 mM MgCl2, 1mM DTT and 2.5 mM GMPPCP) and incubated at room temperature (RT) overnight. The next day, the same molar amount of 4.5S RNA was heated at 80 °C for 8 minutes, cooled on ice for 10 minutes, and added to the Ffh-FtsY binding mixture. After a one-hour incubation at 37°C, glutaraldehyde was added to a final concentration of 0.01%. After incubating at RT for 30 minutes, Tris pH7.5 was added to a final concentration of 0.1 M to quench the reaction. The cross-linked SRP-SR complex was purified on a MonoQ column (GE Healthcare), concentrated to 10 µM and stored at −80 °C.
Improved method to select SRP-SR specific phage clones
Human single-fold scFv libraries Tomlinson I+J were obtained from the Medical Research Council (Cambridge, UK) which came with E. coli strains TG1 (for phage amplification), HB2151 (for scFv production), KM13 helper phage and one copy of a protocol (http://www.geneservice.co.uk/products/proteomic/datasheets/tomlinsonIJ.pdf). All incubations were performed at RT unless specified. Immunotubes (Nalge Nunc International USA) were coated with 100 pmol of cross-linked SRP-SR complex for 2 hrs, then blocked with blocking reagents (see below) at 4 °C overnight. 1013 phage were mixed with the blocking reagent (see below) and added to the tubes. After incubating for two hours, the tubes were washed 20 times with PBST (see below), and bound phage were eluted with 1 mg/ml trypsin. E. coli TG1 was infected with eluted phage and KM13 helper phage to amplify phage for the next round of panning. Four rounds of panning were carried out in total.
To reduce the number of false positive phage clones, we devised the following modifications to the protocol. First, we added a pre-absorption step at each round. The phage solution was incubated with a blocked tube (no antigen) for 30 minutes then transferred to the antigen coated and blocked tube. We also used immunotubes with two kinds of surface materials: maxisorp and polysorp, and prepared two blocking reagents: 3% BSA in PBS and SuperBlock® in TBS (Pierce). Alternative pairs of tubes and blocking reagents were used between consecutive rounds of panning. For example, if maxisorp tubes and BSA were used in the first round, polysorp tubes and SuperBlock® were used in the second round. The third modification we introduced was to increase the concentration of Tween-20 in PBST by 0.05% in each round of panning. For example, we used 0.1% Tween-20 in the first round, 0.15% in the second round, 0.2% in the third round, and so on.
Monoclonal phage ELISA
Monoclonal phage ELISA was performed according to the provided protocol. Briefly, 88 clones from each of the two libraries were inoculated into column 1 through column 11 of 96-well plates, and KM13 helper phage were added to amplify phage particles. Additional plates (ELISA plates) were coated with 10 pmol cross-linked SRP-SR per well and blocked with 3% BSA in PBS. Phage-containing supernatants were transferred from the phage-producing plates to the corresponding wells of the ELISA plates. Column 12 of the ELISA plates showed the background signal because no phage were applied to it. Bound phage were detected by HRP/Anti-M13 monoclonal conjugate (GE Healthcare) (1:2000) and 1-Step™ Ultra TMB-ELISA (Pierce). Parallel ELISA assays using blocked plates (no antigen) were performed to identify any false positive clones.
Sequence analysis
Double strand phagemid DNAs were extracted from TG1 culture expanded from each positive wells of monoclonal phage ELISA plates, and sequenced with LMB3 (5’ CAGGAAACAGCTATGAC 3’) and pHEN seq (5’ CTATGCGGCCCCATTCA 3’) primers.
Expression and purification of scFvs
To solve the difficulties posed by extra amber condons in positive phage clones, scFv coding sequences were cut out of pIT2 phagemid with Nco I and Not I (New England Biolabs) and inserted into pET26b vector (EMD Biosciences) cut with the same enzymes. The pET26b-scFv plasmids were then transformed into RosettaBlue(DE3) (EMD Biosciences), which is an amber suppressor strain. Transformed RosettaBlue(DE3) cells were shaken at 37°C in M9 media with 50 µg/ml kanamycin, 12.5 µg/ml tetracycline and 34 µg/ml chloramphenicol until O.D.600 reached 0.6, then induced with 1 mM IPTG at 30 °C for 16 hours. Harvested cells were subjected to osmotic shock to produce periplasmic extract.
For every liter of induced culture, harvested cells were resuspended in 20 ml ice cold TES (0.2 M Tris pH8.0, 0.5 M EDTA pH 8.0, 0.5 M sucrose), then mixed with 33 ml ice cold 20% TES, and incubated on ice for 30 minutes. The spheroplasts and the periplasmic fraction were separated by centrifugation. Active soluble scFvs enriched in the periplasmic fraction were first purified on Ni-NTA agarose (QIAGEN) columns, then further purified on ImmunoPure® immobilized protein L (Pierce) columns. The purified scFvs were concentrated to 100 µg/ ml and stored at −80 °C.
ELISA with scFvs
The layout of the ELISA is shown in Table I. 10 pmol per well of the following targets were coated on 96-well plates: cross-linked SRP-SR, Ffh, NG domain of Ffh (hNG), M domain of Ffh (hM), FtsY, NG domain of FtsY (yNG) and 4.5S RNA. The plates were then blocked with 3% BSA. Purified scFvs (100 µg/ ml) were diluted 1:100 in 3% BSA before application to ELISA plates. ImmunoPure® protein L-HRP (Pierce) (1:1000) was used to detect bound scFvs. HRP substrate TMB was then added, the reaction was stopped by 1 M H2SO4. The absorbance at 450 nm of each well was measured with Spectra MAX 190 plate reader (Molecular Devices).
Table I.
ELIA of scFvs against SRP-SR and subunits
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IA02 | IB11 | JA01 | JA03 | JA05 | JB05 | JB09 | JC03 | JD02 | JG01 | no scFv | ||
| A | SRP-SR | 10.381 | 10.618 | 7.79 | 9.422 | 10.229 | 11.66 | 10.496 | 11.658 | 11.634 | 10.258 | 0.212 |
| B | Ffh | 0.469 | 10.952 | 12.761 | 0.181 | 0.524 | 7.602 | 0.145 | 0.352 | 0.203 | 0.233 | 0.226 |
| C | hNG | 0.146 | 10.7 | 9.779 | 0.158 | 0.398 | 10.273 | 0.171 | 0.176 | 0.328 | 0.263 | 0.194 |
| D | hM | 0.208 | 0.176 | 0.285 | 0.356 | 0.518 | 0.425 | 0.186 | 0.197 | 0.255 | 0.328 | 0.268 |
| E | FtsY | 0.224 | 0.243 | 10.142 | 9.6 | 10.618 | 8.876 | 1.3 | 14.119 | 12.686 | 14.677 | 0.17 |
| F | yNG | 0.228 | 0.24 | 19.658 | 12.687 | 11.668 | 13.657 | 1.299 | 15.477 | 11.443 | 9.514 | 0.153 |
| G | RNA | 0.238 | 0.176 | 0.469 | 0.294 | 0.208 | 0.365 | 0.351 | 0.279 | 0.344 | 0.237 | 0.301 |
| H | no antigen | 0.246 | 0.166 | 0.531 | 0.396 | 0.444 | 0.363 | 0.381 | 0.352 | 0.348 | 0.407 | 0.202 |
Value in each well is absorbance at 450 nm.
Highlighted wells showed color visible to naked eyes.
2.7. Western blots and dot blots with scFvs
10 pmol of SRP-SR (not cross-linked) complex was either run in each lane of SDS-PAGE and transferred to PROTRAN BA 83 nitrocellulose membrane (Whatman Schleicher & Schuell), or spotted directly on the same type of membrane. The membranes were blocked with SuperBlock® (Pierce). Each lane/dot on the membrane was cut off and incubated with purified scFv diluted 1:100 in SuperBlock®. ImmunoPure® protein L-HRP (Pierce) (1:1000) was used to detect bound scFvs. For the positive control lane/dot, mouse anti-SRP-SR serum (1:2000) was used in place of scFv, and Goat anti-mouse IgG-AP (Santa Cruz Biotechnology) (1:5000) was used to detect bound antibodies.
3. Results and discussion
Preparing cross-linked SRP-SR complex
To ensure that the SRP-SR complex maintained its ternary structure during the panning process, it was cross-linked before bound to the tubes. Purified cross-linked SRP-SR was analyzed in SDS-PAGE side by side with purified Ffh and FtsY (Fig. 1B). The strong single band indicates that the cross-linking and purification procedures worked very well. The presence of 4.5S RNA was also confirmed by running the cross-linked complex in a non-denaturing gel and staining with both ethidium bromide and GelCode® Blue Stain Reagent (Pierce) (data not shown).
Monoclonal phage ELISA
In the early stages of optimizing the panning process, we found that false positive phage clones posed a big challenge. In some cases none of the positive clones from monoclonal phage ELISA bound to SRP-SR specifically (data not shown). After we incorporated the modifications listed above (see 2.2.), the results improved significantly. Fig. 1C and 1D show the ELISA plates of Tomlinson I and Tomlinson J libraries, and the controls. We found that a majority of both the Tomlinson I and Tomlinson J clones tested were positive. Some wells in the control ELISA plates also showed identifiable color, and these samples were immediately deleted from the list of positive clones. We also performed ELISA tests of clones from the original Tomlinson I and Tomlinson J libraries (no panning). While some wells in the Tomlinson J plate gave weak signals, neither the Tomlinson I plate nor either control plate showed any signal (data not shown). All of the monoclonal phage ELISA results confirmed that our improved panning protocol was successful in enriching phage clones that specifically bind to SRP-SR while simultaneously reducing false positive clones.
Sequence analysis
From 90 clones sequenced, a total of 10 different sequences were obtained. Two were from Tomlinson I: IA02, IB11. Eight were from Tomlinson J: JA01, JA03, JA05, JB05, JB09, JC03, JD02 and JG01. Similar to other reports utilizing Tomlinson phage libraries (Yan et al., 2004; Barderas et al., 2006; Marcus et al., 2006), many clones contain premature amber codons. In our case, nine out of 10 clones have amber codons (data not shown). These extra premature amber codons, while still allowing scFv-containing phage production in TG1 cells, prevent the expression of full-length scFvs following the provided protocol. Instead of changing the undesired amber codon (TAG) into a normal codon for glutamine (CAG), we chose a quicker strategy, as described above (see 2.5.).
Expression and purification of scFvs
Soluble scFv was purified from the periplasmic fraction of RosettaBlue(DE3) cells transformed with each of the 10 phage clones. Fig. 2A shows the high purity of three purified scFvs; all other clones were purified to similar purity (data not shown). We found the number of amber stop codons in the coding sequence affected the yield of scFvs. The clone that does not have an amber stop codon, IA02, could produce 0.5–1.0 mg scFv per liter of culture. The eight clones that have one amber stop codon gave similar yields, but the clone that contains two TAG codons, JA05, only produced 0.2–0.5 mg scFv per liter of culture.
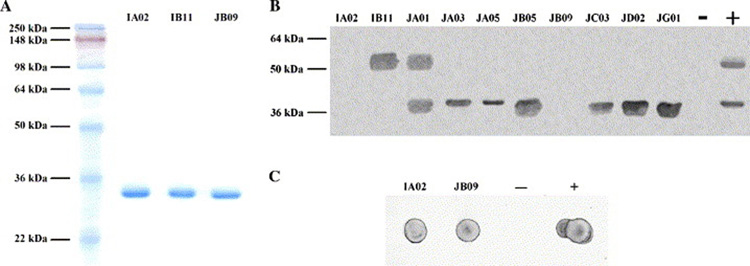
Fig. 2.
Characterization of the scFvs. (A) SDS-PAGE analysis of purified scFvs. (B) Western blot with all scFvs. −: negative control where no primary antibody was used. +: positive control using mouse anti-SRP-SR serum as primary antibody (see 2.7.). (C) Dot blot of IA02 and JB09. Controls are the same as (B).
ELISA with all scFvs
The absorbance reading of the ELISA plate is shown in Table I. All of the scFv clones produced a strong signal with cross-linked SRP-SR, which further proved that they are true positive clones. Based on binding properties revealed by ELISA, the 10 scFvs could be classified into four groups. The first group has one member: IB11, which binds to hNG. The second group includes JA01 and JB05, which bind to both hNG and yNG, suggesting they recognize the highly conserved region of hNG and yNG. The third group includes JA03, JA05, JC03, JD02 and JG01, which bind to yNG. The most interesting group is the last group: IA02 and JB09, which bind only to the intact complex, but not to any of its components. Although JB09 showed some interactions with yNG, the signal was weak compared to the signal observed with SRP-SR. This result suggested that group 4 recognizes structural features that are only present on the whole complex. Considering the diversity of the DNA and protein sequences of the scFvs, we speculate that different members in the same group may bind to different positions on the target molecules.
Western blot with all scFvs
To test whether the scFv clones also bind to denatured Ffh and FtsY, we performed a Western blot. As shown in Fig. 2A, IA02 and JB09 did not react with either Ffh or FtsY, IB11 recognized Ffh only, JA01 bound to both Ffh and FtsY, JA03, JA05, JC03, JD02 and JG01 bound to FtsY only. The results of ELISA (Table I) and Western blot (Fig. 2A) together suggested whereas IA02 and JB09 recognize structural features of SRP-SR, IB11, JA01, JA03, JA05, JC03, JD02 and JG01 recognize protein sequences within the complex. The only scFv that gave inconsistent results when assayed by ELISA and Western blot was JB05, which bound to both Ffh and FtsY in ELISA, but only with FtsY in Western blot.
Dot blot with IA02 and JB09
To further verify that IA02 and JB09 only bind to the intact, non-denatured complex, we also applied them in dot blot analysis (Fig. 2B). SRP-SR complex, when spotted on the membrane directly, should keep its ternary structure. Both IA02 and JB09 bound to the complex on the membrane, which confirmed that these two clones recognize conformational epitopes only present in the SRP-SR complex.
4. Conclusions
Although antibody phage display has been widely used, when it was employed to produce antibodies that target subunits of a macromolecular complex, the success rate was less than 1% (Rubinstein et al., 2003). By keeping the selection target, in our case the SRP-SR complex, the only constant substance in the panning process, and gradually increasing the stringency, we reduced the number of false positives and obtained a good pool of true positive clones. 10 distinct positive clones were identified from 176 clones screened in monoclonal phage ELISA. 80% of the positive clones recognize not only the intact complex, but also its subunits in different assays, while 20% of the positive clones only recognize the intact complex. Therefore, the epitope diversity of the positive clones is excellent.
Extra amber condons within the coding sequences of scFvs is a prevalent problem when using antibody phage display (Yan et al., 2004; Barderas et al., 2006; Marcus et al., 2006), which delays the production and characterization of scFv clones. We devised a new approach to circumvent this problem. Subcloning the scFv coding sequences to a new expression vector by restriction digestion and ligation is much easier and faster than mutagenesis approaches. Not only can our approach be used to the scFv clones one by one, but also be used to a pool of clones in a high throughput environment. We demonstrated that the pET26b-RosettaBlue(DE3) system expressed scFvs very well and all scFvs purified were soluble and fully functional.
Monoclonal antibodies are very useful in the study of the structure and function of large macromolecular complexes. We have devised a fast and efficient protocol to select and produce such antibodies with antibody phage display, which will be helpful to a wide range of research projects.
Acknowledgements
We thank Fai Siu and Rich Spanggord for providing some DNA constructs used in this work, Fang Ding for producing most of the proteins and RNAs involved in the work, Ping Hu and Peter Wang for helpful discussions. This project was supported by the NIH (J. A. D.).
Abbreviations
- scFv
single chain variable fragment
- RNP
ribonucleoprotein
- SRP
signal recognition particle
- SR
signal recognition particle receptor
- ELISA
enzyme linked immunosorbent assay
- DTT
dithiothreitol
- GMPPCP
guanylyl 5′-(beta, gamma-methylenediphosphonate)
- RT
room temperature
- PBST
phosphate buffered saline with Tween-20
- TBS
tris buffered saline
- HRP
horse radish peroxidase
- TMB
3,3’5,5’-tetramethylbenzidine
- AP
alkaline phosphatase
- IPTG
isopropyl-1-thio-β-D-galactopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barderas R, Shochat S, Martinez-Torrecuadrada J, Altschuh D, Meloen R, Ignacio Casal J. A fast mutagenesis procedure to recover soluble and functional scFvs containing amber stop codons from synthetic and semisynthetic antibody libraries. J Immunol Methods. 2006;312:182–189. doi: 10.1016/j.jim.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Chang HW, Nguyen DD, Washington E, Walker ID, Holloway SA. Phage display antibodies to allelic determinants of canine blood cells. Journal of Immunological Methods. 2006;311:1–11. doi: 10.1016/j.jim.2005.11.018. [DOI] [PubMed] [Google Scholar]
- de Leeuw E, Poland D, Mol O, Sinning I, ten Hagen-Jongman CM, Oudega B, Luirink J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- Dong L, Chen S, Bartsch U, Schachner M. Generation of affinity matured scFv antibodies against mouse neural cell adhesion molecule L1 by phage display. Biochem Biophys Res Commun. 2003;301:60–70. doi: 10.1016/s0006-291x(02)02933-9. [DOI] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Luirink J, ten Hagen-Jongman CM, van der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. Embo J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus WD, Wang H, Lohr D, Sierks MR, Lindsay SM. Isolation of an scFv targeting BRG1 using phage display with characterization by AFM. Biochem Biophys Res Commun. 2006;342:1123–1129. doi: 10.1016/j.bbrc.2006.02.073. [DOI] [PubMed] [Google Scholar]
- Miller JD, Bernstein HD, Walter P. Interaction of E. coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- Pool MR. Signal recognition particles in chloroplasts, bacteria, yeast and mammals (review) Mol Membr Biol. 2005;22:3–15. doi: 10.1080/09687860400026348. [DOI] [PubMed] [Google Scholar]
- Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Diaz J, Monedero V, Perez-Martinez G, Buesa J. Single-chain variable fragment (scFv) antibodies against rotavirus NSP4 enterotoxin generated by phage display. J Virol Methods. 2004;121:231–238. doi: 10.1016/j.jviromet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Holt LJ, Walker JE, Tomlinson IM. Use of phage display and high-density screening for the isolation of an antibody against the 51-kDa subunit of complex I. Anal Biochem. 2003;314:294–300. doi: 10.1016/s0003-2697(02)00649-8. [DOI] [PubMed] [Google Scholar]
- Smith KA, Kirkpatrick N, Madden LA, Topping KP, Monson JR, Greenman J. Isolation and characterisation of vascular endothelial growth factor-165 specific scFv fragments by phage display. Int J Oncol. 2003;22:333–338. doi: 10.3892/ijo.22.2.333. [DOI] [PubMed] [Google Scholar]
- Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- Takkinen K, Hakalahti L, Niemi S, Soderlund H, Hemminki A. Isolation of high affinity and specificity testosterone binding antibodies from a phage display library. Immunotechnology. 1996;2:292–293. [Google Scholar]
- Teunissen SW, Stassen MH, Pruijn GJ, Venrooij WJv, Hoet RM. Characterization of an anti-RNA recombinant autoantibody fragment (scFv) isolated from a phage display library and detailed. RNA. 1998;4:1124–1133. doi: 10.1017/s1355838298980633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JP, Ko JH, Qi YP. Generation and characterization of a novel single-chain antibody fragment specific against human fibrin clots from phage display antibody library. Thromb Res. 2004;114:205–211. doi: 10.1016/j.thromres.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Zavala AG, Lancaster T, Groopman JD, Strickland PT, Chandrasegaran S. Phage display of ScFv peptides recognizing the thymidine(6–4)thymidine photoproduct. Nucleic Acids Res. 2000;28:E24. doi: 10.1093/nar/28.7.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]